Abstract
Background
Porphyromonas gingivalis strains are shown to invade human cells in vitro with different invasion efficiencies, varying by up to three orders of magnitude.
Objective
We tested the hypothesis that invasion-associated interstrain genomic polymorphisms are present in P. gingivalis and that putative invasion-associated genes can contribute to P. gingivalis invasion.
Design
Using an invasive (W83) and the only available non-invasive P. gingivalis strain (AJW4) and whole genome microarrays followed by two separate software tools, we carried out comparative genomic hybridization (CGH) analysis.
Results
We identified 68 annotated and 51 hypothetical open reading frames (ORFs) that are polymorphic between these strains. Among these are surface proteins, lipoproteins, capsular polysaccharide biosynthesis enzymes, regulatory and immunoreactive proteins, integrases, and transposases often with abnormal GC content and clustered on the chromosome. Amplification of selected ORFs was used to validate the approach and the selection. Eleven clinical strains were investigated for the presence of selected ORFs. The putative invasion-associated ORFs were present in 10 of the isolates. The invasion ability of three isogenic mutants, carrying deletions in PG0185, PG0186, and PG0982 was tested. The PG0185 (ragA) and PG0186 (ragB) mutants had 5.1×103-fold and 3.6×103-fold decreased in vitro invasion ability, respectively.
Conclusion
The annotation of divergent ORFs suggests deficiency in multiple genes as a basis for P. gingivalis non-invasive phenotype.
Keywords: periodontitis, oral microbiology, Porphyromonas gingivalis, invasion, genomic polymorphisms, comparative genomic hybridization, RagA
Genomic differences among subspecies account for many important pathogenic traits. With the release of more and more microbial genome databases, it has become clear that limited information related to pathogenic properties can be mined from a genomic database alone. A genomic sequence per se can reveal only a limited number of mechanisms to explain bacterial virulence properties. However, techniques such as gene expression profiling and comparative genomic hybridization (CGH, or also ‘genomotyping’), both utilizing DNA microarrays, can now facilitate genome-wide assessment of relative gene expression levels as well as strain-to-strain comparisons. In an investigation of Campylobacter jejuni, a genome comparison using microbial microarrays revealed loci absence/divergence (1). Genomic analysis using microarrays in Salmonella revealed genes acquired horizontally by the genomic database strain that may be associated with enteric infections in humans (2) and genomic islands that distinguish different serovars (3). These achievements prompted us to utilize CGH of P. gingivalis whole genome microarrays for assessment of genome polymorphisms underlying the different endothelial cell invasion phenotypes. A whole-genome approach such as that used in this study is a practical method to reveal the genetic determinants ‘missing’ from the non-invasive P. gingivalis strains.
Background
Porphyromonas gingivalis has been strongly implicated as an etiologic agent of periodontitis (4) and recent evidence suggests its association with atherosclerosis (5). Although P. gingivalis is also detected in healthy (non-periodontitis) individuals, it is considered an endogenous pathogen (6, 7). Among numerous other virulence properties, P. gingivalis has been shown to invade multiple cell types including animal cell lines, human vascular and oral cell lines (8–11). The intracellular environment shields the organism from host defenses, while allowing it to replicate and modify the host immune response (12).
Invasion of non-phagocytic cells is very likely a key virulence factor for this bacterium as it provides (1) a ‘privileged niche’ with access to host protein (nutritional) and iron substrates, (2) a sequestration from the humoral and cellular immune responses, and (3) a means for persistence that is essential for a chronic pathogen. Oral tissues are likely the primary sites for P. gingivalis infection but this bacterial species can enter the circulation through the microvasculature following tooth brushing and other dental procedures (13, 14). The invasion of human endothelial and epithelial cells by P. gingivalis has been well established (11, 15). There is strong evidence that P. gingivalis disseminates to the large vessels since P. gingivalis DNA can be detected in atheromas by PCR (16). More importantly, it has been reported that only invasive P. gingivalis strains accelerate atherosclerosis in a murine model (17). Accordingly, we have detected viable P. gingivalis in atheromatous vascular tissue (18) (Rafferty et al., in press). We have also shown, using immunofluorescent staining of internalized bacteria, P. gingivalis invasion of and transmission between vascular cell types (19). It is thus our overall hypothesis that P. gingivalis actively interacts with the gingival endothelia in vivo initially, but then a subset of strains or clonotypes disseminate and establish an invasive infection in the systemic vasculature.
In an extensive study of P. gingivalis invasion of human cell types, several strains were demonstrated to be fully competent of invading these cells in vitro (20). However, there was a wide variation in invasion abilities within these strains, varying by as much as three orders of magnitude, as evident with the strain AJW4's (the ‘non-invader’) very low invasion ability (11). The AJW4 had the lowest invasion ability among 27 strains in the tested cell lines thus making it the target of this study. Nevertheless, only a few P. gingivalis determinants of invasion have been identified, with the most studied among them being fimbrillin, the major fimbriae protein (21). Using a genetic approach, several other genes have been investigated for their potential contribution to invasion. A phosphoserine phosphatase mutant of P. gingivalis was shown to be deficient in cell culture invasion (22), as was a mutant of clpB (23), htrA (24), PepO, ATPase, and ABC transporter (25) while a gingipain-null mutant was less potent at invading in vitro mucosal tissue model (26).
Publication of the complete 2,343,476-bp genome sequence of P. gingivalis strain W83 (27) lead to the availability of DNA microarrays for this organism. Seven communications reporting data obtained using these whole-genome micro arrays have been published so far. They have analyzed P. gingivalis genes induced during attachment to the human epithelial cell line HEp-2 (28), identified quorum-sensing genes (29), and genes differentially regulated during accretion of P. gingivalis in heterotypic biofilms with Streptococcus gordonii (30). A comparative genomics study focused on the genomic differences that determine virulence in a mouse model and identified over 150 divergent genes (31). Microarrays were also used to characterize the response of P. gingivalis to H2O2 (32) and identified 62 P. gingivalis W83 genes that were differentially regulated during invasion of primary human endothelial cells (11 up-regulated and 51 down-regulated) (33). No genomotyping of P. gingivalis has been performed related to invasion.
The differential seen in P. gingivalis strains prompted us to initiate the present study, designed to identify genetic determinants that may be responsible for the P. gingivalis invasive phenotype relative to vascular cells. Thus, our hypothesis is that one of the most important P. gingivalis virulence traits, host cell invasion is genetically determined. We examined this by using microbial microarrays, genomic analysis of clinical isolates, bacterial genetics, and in vitro assays.
Materials and methods
Bacterial strains and cell lines
Porphyromonas gingivalis strains W83, 381, A7436, AJW2, and AJW4 as well as the tested clinical isolates were grown anaerobically at 37°C on blood agar plates (BAP) and in Tryptic Soy Broth (Difco/BD, USA) supplemented with 0.5% yeast extract, 0.05% L-cysteine, 0.05 mg/ml hemin and 0.1 mg/ml vitamin K1. DNA from JH16-1, a non-invading P. gingivalis strain (34) was also used, a gift from S. Eick. The P. gingivalis strain 381 was originally isolated by A. Tanner; strain A7436 is a gift from S. Offenbacher; W83 and AJW4 were kind gifts from A. Progulske-Fox; AJW2 and clinical isolates #6, 9, 10, 11, 13, 14, 15, 17, 18, 19, and 25 were kind gifts from V. Haraszthy; and isogenic mutants of PG0185 and PG0186 (35) were kind gifts from Y. Murakami.
HMEC-1, an immortalized non-cancerous human microvascular endothelial cell line (36) was grown in EBM-2 endothelial cell growth medium, supplemented with EGM-2 SingleQuots (Cambrex BioScience, USA). KB (ATCC CCL 17), a HeLa-derived continuous cell line was cultured in Dulbecco MEM (DMEM) medium. The media were supplemented with 10% FBS, 100 units/ml penicillin, and 100 µg/ml streptomycin.
Antibiotic protection assay
Gentamicin/metronidazole killing was performed in biological triplicates essentially as described (8). Briefly, 105 tissue culture cells growing overnight in 24-well plates were infected with the P. gingivalis strain (W83, AJW2, AJW4, W83, or mutants W83ΔPG0185, W83ΔPG0186, and W83Δ0982) at multiplicity of infection (MOI) of 100 for 1.5 hours. After washing with PBS, cells were incubated in DMEM with 300 µg/ml gentamicin and 200 µg/ml metronidazole for 1 hour and then lysed in 1 ml of sterile ddH2O for 20 min at room temperature. The released bacteria were pelleted at 16,000×g for 10 min before plating on blood agar plates (BAP) and incubating anaerobically for 5 days at 37°C. The resulting colonies were enumerated and statistical significance was analyzed using Student's t-test.
P. gingivalis microarray comparative genomic hybridization (CGH)
The P. gingivalis microarray slides based on strain W83 genomic sequence were obtained from the Pathogen Functional Genomics Research Center (PFGRC) at J.C. Venter Institute (JCVI, formerly The Institute for Genomic Research, TIGR). The full genome array consists of 1907 70-mer oligonucleotides that were designed based on predicted ORFs from the annotation of strain W83. Detailed information regarding the slides and the protocols used can be found at JCVI web site http://pfgrc.jcvi.org.
Genomic DNA purification, labeling and hybridization
The P. gingivalis genomic DNA for analysis was extracted from strains W83, 381, A7436, AJW2, and AJW4 using the DNeasy Tissue Kit (Qiagen, USA) according to the manufacturer's instructions. Strain W83 and AJW4 genomic DNA labeling, prehybridization, and hybridization was performed using protocols utilized by JCVI. In short, genomic DNA was digested with BfuCI (New England Biolabs, USA), 1 unit/1 µg of DNA for 5 min on ice to average fragment sizes of 2–5 Kb. For a 39 µl reaction, 4 µg of DNA was combined with 3 µg of random hexamers (Invitrogen, USA), incubated at 100°C for 10 min, and chilled on ice. The volume of reaction was adjusted to 50 µl by adding the components to final concentration as follows: 1× Eco Pol (Klenow) buffer, 0.2 mM dNTP/aa-UTP labeling mix (Invitrogen/Sigma), and 20 units of the Klenow fragment (NEB). After overnight incubation in a 37°C water bath, Klenow fragment was inactivated by adding 5 µl of 0.5 M EDTA. Unincorporated aa-dUTP and free amines were removed using the PCR purification kit (Qiagen). Labeled DNA was eluted with 60 µl of 0.1 M KPO4 buffer, pH 8.5 and dried in a Speed Vac for 1 hour. The dried DNA pellet was dissolved in 4.5 µl of 0.1 M sodium carbonate buffer, pH 9.3 and 4.5 µl of the appropriate Cy dye (Amersham, USA) was added. After incubation in dark for 1 hour at room temperature, dye-coupled DNA samples were purified with a PCR purification kit (Qiagen), eluted with 60 µl of PE buffer, and dried in a Speed Vac for 1 hour. The pellet was resuspended in 60 µl of hybridization buffer containing 50% formamide/5XSSC/1%SDS and 0.5 mg/ml sheared salmon sperm DNA. Before hybridization, the labeled DNA was heated twice at 95°C for 5 min. Independently labeled genomic DNA was used for each slide. Microarray prehybridization, hybridization and stringency washes were performed according to JCVI/TIGR protocol http://pfgrc.jcvi.org/index.php/microarray/protocols.html.
Data generation and analysis
Hybridized and washed slides were scanned for Cy3 and Cy5 fluorescence intensities using Agilent microarray scanner. The TIF format images were analyzed using GenePix 6.0 software (Molecular Devices Corporation, Sunnyvale, USA) producing GPR format image files. Data was assembled from four independent array experiments. Data were log2-transformed and normalized using GINKGO 1.0 software (LOWESS and Histogram Mode Centering from JCVI, at PFGRC, cmr.jcvi.org). Additionally, background correction and the within-array (LOWESS) and between arrays (Aquantile) image data normalization resulting in a table with numerical values were carried out using SAOPMD (Significance Analysis for Oral Pathogen Microarray Data) software tool at www.brop.org/.
An M-value, representing the log2 of control/tester fluorescent intensity signal ratio was assigned to each gene and used to determine the divergence between the control (W83) and the tester (AJW4) strain. GINKGO and SAOPMD were both used for the same purpose and the results from the two analyses were combined for a list of ORFs not found in the non-invasive strain using this approach. The annotation files for these ORFs are available at http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?org=gpg.
PCR verification of W83 genes absent in the non-invasive AJW4 strain
To validate the microarray data, PCR confirmation of selected ORFs not found in the non-invading strain was performed. Five amplifications were performed with gene-specific primers for selected ORFs. For oligonucleotide design, PrimerQuest software (Integrated DNA Technologies, Inc., USA, http://www.idtdna.com/Scitools/Applications/Primerquest/Default.aspx) and Primer-BLAST designing tool (NCBI) were used. The primers are listed in Table 1. Platinum Blue PCR Polymerase Super mix (Invitrogen) was used in a PCR protocol according to the manufacturer's instructions. We amplified across putative deletions identified by the microarray data in both low-invasive P. gingivalis strains AJW4 and AJW2. As positive control, amplification using genomic DNA template from P. gingivalis strains W83, 381, and A7436 was performed. In addition, 11 clinical isolates from periodontal sites were tested for presence of PG0185, PG0186, and PG0982.
Table 1.
Oligonucleotide primers used to confirm the absence of selected ORFs in the genome of non-invading P. gingivalis strains
Primers used in PCR analysis of P. gingivalis genomic DNA
| Locus # | Forward primer (5′-3′) | Reverse primer (5′-3′) | Amplicon |
|---|---|---|---|
| PG0185 | 5′-TTTGCCTGAACACAGAGTCG-3′ | 5′-GACTGCTTTTCCCACGAGAG-3′ | 369 bp |
| PG0186 | 5′-GGAAGCTGCGTTGCAGAATCAA GT-3′ | 5′-AGCA TCTGCTGCACCAATCAAAGG-3′ | 433 bp |
| PG0187 | 5′-AAGATCCTCGTGTTGTAGGTCGCT-3′ | 5′-GCTACGCAAACGCTTGCC ATCTAT-3′ | 306 bp |
| PG0461 | 5′-TGAACCAATCGCACCCACTCTACA-3′ | 5′-TATGCAACTTGGCATCGG TAGGGA-3′ | 429 bp |
| PG0982 | 5′-AGACGGTAAATTCGCCCATGCGTA-3′ | 5′-TGGGCTGAAAGAGGTTGT TCTCCT-3′ | 656 bp |
| PG1261 | 5′-CAAACACTTCACAGGGTGGCAACA-3′ | 5′-TCACTTGGGTGCTGTCCC AACTAT-3′ | 536 bp |
Construction of ΔPG0982::erm mutant in P. gingivalis W83
To determine the potential role in invasion of ORFs identified using this approach, we used PG0186 and PG0186 isogenic mutants (35) and also constructed a mutant of ORF PG0982, a tetratricopeptide domain protein. We followed a gene deletion procedure for ORF PG0982 using the suicide vector pPR-UF1 as described (37). Two fragments (A and B) located at the 5′ and 3′ end of the ORF, respectively, were generated on W83 genomic DNA template through PCR, with restriction sites incorporated into, to allow for cloning into the vector. The 5′ end (Fragment A) primers yielding 400-bp product were: forward, 5′-CCCCCTCGAGAGTCG CCTTACTCCAGGCAAATCA (XhoI site) and reverse, 5′-CCCCGGTACCTCTTGCCGATTAGGTTCAGGCACA (KpnI site) (nt 48-71). The 3′ end (Fragment B) primers yielding 459-bp product were: forward, 5′-CCCCTCTAGAAAACAGCTCTTTGAGTGCAGCGAG (XbaI site) (nt 2167-2190) and reverse, 5′-CCCCGCATGCTTCACGTCCTACTCAGTCCGTTCT (SphI site). Primers for internal PG0982 fragment used for confirmation of knockout (nt 1453-1688) were: forward, 5′-AGCCTGAACGAAACAACTCCCAGT and reverse, 5′-TCTGCCAACTCCTTACGAGCTTCT.
The DNA fragments A and B, flanking PG0982, were digested, cloned into the vector and their presence was verified using restriction digests, following cloning procedures for P. gingivalis (37). After amplification of upstream fragment A, the vector pPR-UF1 and fragment A PCR product were double digested with XhoI/KpnI. After digestion, the fragment was ligated into the vector, E. coli DH5α cells were transformed, and the target plasmid, pPR-UF1A, was isolated from transformants. Following amplification of downstream fragment B, pPR-UF1A and fragment B PCR product were double digested with SphI/XbaI. Fragment B was then ligated into the vector, transformed, and the resulting suicide plasmid pPR-UF1AB was isolated as described above. The plasmid pPR-UF1AB was linearized with PciI and introduced into P. gingivalis W83 via electroporation. The pulsed bacteria were plated on BAP containing 5 µg/ml of erythromycin for selection of transformants carrying the deletion of the ORF. To confirm the deletion, DNA was isolated from two separate transformants and amplified using three primer sets: (1) Fragment A forward primer and fragment B reverse primer (expected product in wild type W83, 2,955 bp, and 3,100 bp in the mutant); (2) Fragment A forward primer and internal fragment reverse primer (expected product in wild type W83, 2,018 bp, and none in mutant); (3) Internal fragment forward primer and fragment B reverse primer (expected product in wild type W83, 1,172 bp, and none in mutant).
Results
Bacterial invasion
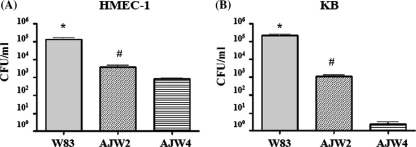
Wide variation in invasion abilities within P. gingivalis strains was observed using the KB cell line, with strain AJW4 with sharply lower invasion ability than the other 26 tested strains (11). To validate the results for endothelial cells, we tested P. gingivalis strains for their invasion ability on HMEC-1, a human endothelial cell line. Differential recovery of P. gingivalis W83, AJW2, and AJW4 is shown in Fig. 1. Among these three strains of P. gingivalis, recovery from host cells was again the lowest for strain AJW4 (Fig. 1A), confirming the difference in strain invasion of this continuous endothelial cell line. The previous observations for KB host cells were also confirmed (Fig. 1B), providing confidence in our choice of strains for this investigation.
Fig. 1.
Antibiotic protection assay. The bars indicate the numbers of intracellular CFU recovered from HMEC-1 cells (A), and control KB cells (B) in three separate experiments (see Materials and Methods). Tissue culture cells were infected in triplicate with P. gingivalis strains W83, AJW2, and AJW4 for 90 min and incubated in medium containing antibiotics for 1 hour prior to lysis. Cell lysates were plated on blood agar plates, incubated anaerobically, and the resulting colonies were enumerated. (A) *p to AJW2 =.0077, *p to AJW4 =.0071, #p to AJW4 =.0061. (B) *p to AJW2 =.0031, *p to AJW4 =.0030, #p to AJW4 =.0147. The error bars indicate standard deviations.
Comparative genomic hybridization
Microarray-based comparative genomic hybridizations were performed with the control, W83, and the tester, low-invasive AJW4 strain. Each microarray slide contains four identical sets of 70-mer oligonucleotides representing all P. gingivalis ORFs. The results represent the common findings of four independent array hybridizations performed with W83 Cy3 and AJW4 Cy5 labeled genomic DNA (for a total of 16 repeats per ORF). In addition, two independent analyses of the images, using SAOPMD and Ginkgo software packages were performed to cross-validate the data. The generated files were imported into Microsoft Excel for final processing. The analysis demonstrated a degree of genetic polymorphism within the 70 bp amplicons on the microarray that the AJW4 hybridization signal was significantly reduced for a total of 120 ORFs, based on M-value (log2 of signal ratio) grading with a cut-off of 0.7. Due to the inherent limitation of the microarray technology used, the results may represent genetic diversity within the oligonucleotide 70-nt sequence leading to reduced hybridization signal rather than absence of the entire ORF, therefore genetic approach was used for ultimate validation of results. Fifty-one of the identified ORFs are annotated as hypothetical proteins, three are involved in carbohydrate metabolism, 4 are putative lipoproteins, 3 are surface proteins, 11 are involved with DNA mobility (insertion sequences, transposons, transposases, or integrases), 4 are conserved domain proteins, and the remaining 43 have other suggested functions. All identified ORFs are listed in the supplement. A file with the experimental data is available at the NCBI GEO site, http://www.ncbi.nlm.nih.gov/geo/, GPL8955.
PCR validation
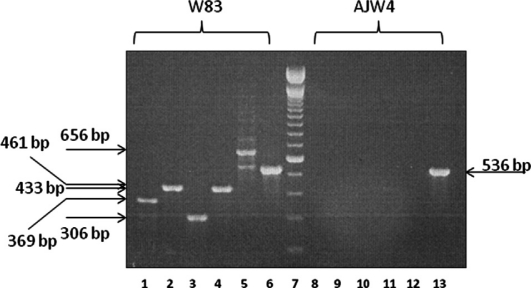
PCR testing of five ORFs not detected in the low-invasive strain AJW4 according to the microarray data was performed using gene-specific primers. The primers were located outside the 70-mer microarray amplicons in order to reduce effects of sequence divergence (http://blast.ncbi.nlm.nih.gov/Blast.cgi). As additional positive controls, DNA from invasive P. gingivalis strains 381 and A7436 were used. We and others have previously demonstrated that these strains are invasive for vascular cell types (9, 38). The PCR verification demonstrated that all five tested ORFs were absent from both non-invasive strains while present in all tested invasive strains (Table 2 and Fig. 2). As an additional control, ORF PG1261, selected with M-value below the cut-off threshold was tested. ORF PG1261 was found in all tested P. gingivalis strains, thus validating the threshold value.
Table 2.
Demonstration of P. gingivalis genetic polymorphism between invasive (W83, 381, A7436) and less invasive (AJW2, AJW4, JH16-1, underlined) laboratory strains. JH16-1 DNA was used (the strain was lost). PCR amplification of selected ORF was used to determine presence (+) or absence (–) of the ORF in genomic DNA templates. PG1261 was used as a control with M-value below the cut-off. M-value (above the cut-off value of 0.7) represents the log2 of the ratio of signal intensity of control to tester strain. The gel image is presented on Fig. 2
Genetic variation between P. gingivalis strains
| W83 ORF (TIGR #) | M-value | W83 | 381 | AJW4 | AJW2 | JH16-1 | A7436 |
|---|---|---|---|---|---|---|---|
| PG0185 ragA | 1.01 | + | + | – | – | – | + |
| PG0186 ragB | 1.43 | + | + | – | – | – | + |
| PG0187 ISpg6 | 1.08 | + | + | – | – | – | + |
| PG0461 ISpg7 | 1.35 | + | + | – | – | – | + |
| PG0982 TPRI | 1.90 | + | + | – | – | – | + |
| PG1261 ISpg4 | 0.60 | + | + | + | + | + | + |
Fig. 2.
PCR verification of selected W83 genes absent in lowinvasive strain AJW4. Lanes 1 and 8, PG0185, 369 bp; Lanes 2 and 9, PG0186, 433 bp; Lanes 3 and 10, PG0187, 306 bp; Lanes 4 and 11, PG0461, 429 bp; Lanes 5 and 12, PG0982, 656 bp; Lanes 6 and 13, PG1261, 536 bp. ORF PG1261 is the positive control, predicted to be present in AJW4. Lane 7 is a 100 bp ladder marker (Invitrogen).
Analysis of clinical isolates
We then analyzed 11 clinical strains isolated from periodontal sites for the presence of suggested invasion-associated ORFs. We used primers for ORFs PG0185, PG0186, and PG0982, as they were already validated in initial tests (Table 2). The PCR analysis of the clinical P. gingivalis isolates demonstrated their presence in the strains, with the exception of strain #6.
Construction of PG0982 isogenic mutant
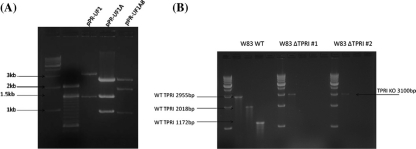
To confirm the presence of the DNA fragments flanking PG0982 coding sequence in the suicide vector, pPR-UF1A (the vector with the upstream fragment A) and pPR-UF1AB (vector with both PG0982 fragments, upstream A and downstream B) were digested with HindIII producing fragments with the expected size (Fig. 3A). After electroporation of competent P. gingivalis W83 with the construct, mutants were selected on erythromycin. Two of the selected transformants were analyzed to confirm the introduced deletion via PCR of genomic DNA as described in Methods. The mutants, W83ΔTPRI#1 and W83ΔTPRI#2, produced the predicted 3,100 bp amplification product only with flanking primers, fragment A forward, and fragment B reverse primer (Fig. 3B). No products were obtained using primers internal for the PG0982 coding sequence.
Fig. 3.
Construction of deletion mutant of PG0982 in strain W83. (A) Suicide vector constructs digested with HindIII. The restriction digests of the suicide vector pPR-UF1 with inserted upstream region A flanking the gene of interest (pPR-UF1A) and then with inserted downstream region B (pPR-UF1AB, final recombinant construct) are presented. Fragment sizes are: pPR-UF1: 3,161 bp and 1491 bp; pPR-UF1A: 2,601 bp, 1,491 bp, and 960 bp; pPR-UF1AB: 2,601 bp, 1,950 bp, and 960 bp. First two lanes, molecular mass markers (size indicated with arrows). (B) Verification of the deletion in the chromosome of two of the selected mutants (ΔTPRI#1 and ΔTPRI#2) by PCR using the primer sets listed in Methods. W83WT, wild type control amplifications yielding products as indicated with arrows, 2,955 bp (upstream fragment A forward primer and downstream fragment B reverse primer), 2,018 bp (A forward primer and internal fragment reverse primer), and 1,172 bp (internal forward primer and downstream fragment B reverse primer). The internal fragment is in the deleted ORF. Both mutants, W83ΔTPRI#1 and W83ΔTPRI#2 only produced 3,100 bp amplification product with fragment A forward and fragment B reverse primer (arrow). Molecular mass marker is 1-kb ladder at left of each analyzed strain.
Effects of gene deletions on bacterial internalization
We used antibiotic protection assay in three independent experiments to assess the ability of the PG0185 (ragA), PG0186 (ragB), and PG0982 (TPRI) mutants to invade. KB cells were used as the wild type strain W83 showed comparable invasion ability in both HMEC-1 and KB cell lines (Fig. 1). The TPRI mutant was not hindered in its ability to invade KB cells. In contrast, the ragA and ragB mutants had decreased invasion ability, 3.9×10−4±5×10−5% invasion (ragA) and 5.6×10−4±1.3×10−4% invasion (ragB) compared to wild-type P. gingivalis strain W83, 2.0±1.0% invasion.
Discussion
P. gingivalis genetic heterogeneity (genome polymorphisms)
Invasion of non-phagocytic cells is a key virulence property of P. gingivalis. Although individual ORFs have been tested for encoded proteins with invasion-related properties and mutants with impaired invasion have been used for studies (17, 39), surprisingly, no systemic study of P. gingivalis invasion at genome level has been communicated so far. Non-invasive P. gingivalis strains may not be common, since in a study surveying the invasive abilities of multiple strains, only one of the tested 27 strains, AJW4, was distinctly less invasive with AJW2 distant second (Fig. 1) (11). A non-invasive strain may be able to survive without invasion determinants in dental plaque, due to the hemorrhage (secured iron) and biofilm (protection) secured by other strains in the biofilm microenvironment, but such a strain wouldn't be able to survive intracellularly and disseminate to other tissues. It is also possible that strains with lower invasive ability than W83 may have a different mechanism of virulence that would be revealed by CGH analysis. In this study, we utilized a non-invading strain to identify the ORFs that are missing in its genome, relative to the invading database strain.
In the study of invasion efficiency of 27 different P. gingivalis strains using antibiotic protection assay, human non-endothelial cells were used (11). To apply the findings to endothelial host cells, we carried out antibiotic protection assays selecting for internalized bacteria using the endothelial cell line HMEC-1. The non-invading strain AJW4 was confirmed as our best choice for genomic analysis since its invasion efficiency was up to four orders of magnitude lower than the invasive control strain W83. As the AJW4 invasion in KB (ATCC CCL-17) cell line was again the lowest, KB cells were utilized for further experiments.
The P. gingivalis genomic heterogeneity has been demonstrated using multiple methods including random amplified polymorphic DNA (RAPD) fingerprinting analysis (40), multilocus enzyme electrophoresis (MLEE) (41), serotyping (42), heteroduplex analysis (43), and RFLP (44). In addition, our data on the interstrain variability of HagA, a major surface adhesin implicated in virulence, demonstrated that this gene falls into three groups depending on the number of adhesin repeats (45). Interestingly, in this study of 23 laboratory and clinical P. gingivalis strains, the only strain found to have the fewest repeats (two) was the non-invasive one, AJW4, used in the present work.
Genomic differences among subspecies account for many important pathogenicity traits. We hypothesize that a subset of ORFs is involved in important pathogenic trait such as invasion of non-phagocytic cells. As we expected, the invasive phenotype seems to be associated with multiple ORFs. This has been the case with well-studied organisms such as Salmonella where the acquisition of the ‘pathogenicity island’ (SPI-1) region may represent the defining genetic event in the separation of the Salmonella and E. coli lineages. In a similar CGH microarray study of virulent M. bovis versus the attenuated Bacille Calmet-Guérin (BCG) vaccine strains, five loci representing 38 ORFs were absent from some or all BCG strains (46). Using CGH of P. gingivalis strains W83 and 33277, multiple divergent features were reported (31). In our study, significant strain-to-strain genetic differences in the P. gingivalis genome were demonstrated between strains with different invasion phenotypes. Using CGH and two software analyses, a total of 120 ORFs were designated as missing in the non-invading strain AJW4. However, the findings may represent genetic diversity within the 70-nt oligonucleotide probes immobilized on the slides, leading to false negatives. Therefore, several of these ORFs were validated using PCR from different templates (invaders and low invaders), and the presence of a control ORF that is below our cut-off value was shown in the low invaders thus validating the methodology (Table 2).
The number of polymorphic ORFs (~5% of genome) identified using CGH of the strains with different invasion phenotype is in line with other studies examining genomic differences between strains. The PFGE analysis of laboratory and clinical isolates has demonstrated that the genome plasticity of P. gingivalis is not due to point mutations and that 55 W83 genes were, on average, missing in the tested isolates that did not differ phenotypically (47). In order to overcome a typical shortcoming of microarray technology, confirmatory analysis of selected genes including control ORF was performed. The results were as predicted in our model and clearly demonstrate the existence of genomic heterogeneity between the isolates. The biological significance of all the interstrain differences can be evaluated by an extended analysis of each divergent gene. The signal ratios in a microarray analysis may be influenced by gene polymorphisms resulting from strain divergence and by gene copy numbers, and this could be clarified in a focused study of an ORF of interest. The strength of CGH is in the identification of particular virulence genotypes followed by investigation of candidate ORFs using functional genomics methodologies to test the hypothesis. Such investigation is discussed below.
Eleven ORFs involved with genetic mobility (insertion sequences, transposons, transposases, and integrases) were identified using CGH and with most clustered in genomic island 7 (PG0827-PG0874) identified in the Oralgen database using base composition analysis and BLAST taxonomy data (www.oralgen.lanl.gov). Another cluster of divergent genes encompass many of the P. gingivalis ORFs 1436–1454, identified as genomic island 12 involved in mobilization functions according to the same database, while divergent ORFs PG1108–PG1113 consist predominantly of ‘hypotheticals’. Flexible self-transmissible mobile genetic elements are shown to disseminate different virulence functions (48). Fittingly, the majority of the divergent ORFs have G+C content different than the chromosome average of 48%, such as PG1439 with G+C content of 32.5%, which is a compositional feature associated with horizontal genetic transfer and pathogenicity islands (49). Genes involved with carbohydrate metabolism were also divergent between P. gingivalis W83 and the low-invasive strain. For example, ORF PG0109 is from a cluster of cell envelope genes (biosynthesis and degradation of surface polysaccharides and lipopolysaccharides) identified as genomic island 2 in the Oralgen database (ORF annotations are from JCVI). Genes involved in carbohydrate metabolism have been associated with invasion in other organisms such as uropathogenic E. coli (50) and Legionella pneumophila (51).
Surface and immunogenic proteins have been suggested to interact with host cell components in a variety of organisms including P. gingivalis. We have previously shown that HagB, a P. gingivalis surface protein (52) is involved in adherence to human primary endothelial cells. Similarly, in the present work, two divergent genes, PG0185 and PG0186, coding for the rag locus surface protein RagA and lipoprotein RagB, were among the ORFs identified as missing in the non-invasive strain using this type of analysis. Further, several of the divergent genes (with hypothetical function) are among those we identified in P. gingivalis using in vivo induced antigen technology (IVIAT) (53, 54). This finding is supportive of our hypothesis because IVIAT proteins are virulence factors only expressed during disease. Therefore, some of the identified divergent genes can be considered as vaccine candidates. Importantly, none of the divergent genes identified here were found to be up-regulated 2.5 hours after endothelial cell invasion (33). This is expected in a model where the expression of genes necessary for host cell invasion is reduced upon completion of the process. Finally, a large part of the identified divergent ORFs were coding for hypothetical proteins that can be objects of interest especially when they share extensive homology with proteins from other pathogens. For example, ORF PG0848 shares extensive homology with genes from Bacteroides, Clostridium, Mycobacterium, Legionella, and other genera and PG1526, featuring an ATP-binding domain, is homologous to genes in Bacteroides and Prevotella.
To extend our findings to the actual periodontal pathology and to test the hypothesis that invasion is a critical virulence property, we examined 11 clinical strains of P. gingivalis and three isogenic mutants. According to our hypothesis, divergent genes are essential for pathogenicity; therefore, they may contribute to the pathology and should be present in disease isolates. To test this, we analyzed P. gingivalis isolates from periodontal sites from patients for the presence of divergent genes PG0185, PG0186 (the ragA and ragB genes), and also for PG0982. It has been suggested that the rag genes code for putative TonB-dependent outer membrane receptor (RagA) and for immunodominant antigen (RagB). Both mutants were significantly less virulent than wild-type strains in a murine model of infection (55) and PG0186 mutant has been reported to have decreased invasion of endothelial cells (56). The PG0982 has GC content of 37%, placing it in the category of lateral genetic flow-acquired genes, features a TPR motif (tetratrico peptide repeat) that functions in a wide variety of cellular processes, and shares homology with Yersinia and Pseudomonas type 3 secretion system low calcium response chaperone LcrH/SycD (TIGRFAM and PFAM databases). The analysis demonstrates the presence of these three divergent genes in 10 of the P. gingivalis clinical isolates.
We also tested isogenic mutants for their efficiency of internalization using bacterial invasion assays. PG0185 and PG0186 had reduced invasion ability, 5.1×103 fold and 3.6×103 fold, respectively, confirming previous observation of impaired invasion of ORF PG0186 (ragB) mutant (56). Interestingly, the available RT-PCR data suggest that gene expression post-invasion may not be indicative of the role of the protein in the invasion process. For example, both PG0185 (ragA) and PG0186 (ragB) mutants had reduced invasion; however, PG0185 (ragA) did not have significant change in expression level 2.5 hours post-invasion, while the expression of PG0186 (ragB) at that time point was down-regulated (33). In contrast, several Shigella dysenteriae type III secretion system effectors of invasion were increased in abundance after infection in vivo (57), as were P. gingivalis transcripts of PepO (PG0159), ATPase (PG1642), and ABC transporter (PG2206); the mutants of these P. gingivalis genes showed impaired invasion (25), although not identified using the microarrays. The non-invasive fimbriae-deficient mutant (in PG2132), a target identified using CGH, is non-invasive and lacks pro-atherogenic response in primary aortic endothelial cells {Roth, 2007 #3204}. Further, P. gingivalis htrA (PG0593) mutant showed increased invasion of epithelial cells (24), while htrA expression was not changed significantly 2.5 hours post-invasion. These differences reflect the variety of roles a protein may play in the sophisticated process of host cell invasion, namely attachment, internalization, intracellular localization, and phagolysosomal escape, and ultimately in intracellular persistence. In addition, the impact of the host cell line is significant (25). Again, in this first global study utilizing unique non-invasive P. gingivalis strain, we aimed at characterizing the interstrain genomic polymorphisms, with the realization that different mechanisms may impact the invasion efficiency (58). Based on the decreased invasion efficiency of the PG0185 mutant, we designated RagA an invasion-related protein.
Conclusion
This work is an important advancement in the genetics of P. gingivalis invasion. We used P. gingivalis microarrays with comparative genomics to specifically address the P. gingivalis invasive genotype using invasive and the only available non-invasive phenotype. The results indicate that more than 100 genes are missing from the genome of non-invading strain. This is the first communication identifying invasion-specific genetic differences of P. gingivalis, a pathogen associated with one of the most prevalent infectious diseases, periodontitis, and possibly with vascular inflammations. Next, the data demonstrates that the true degree of P. gingivalis clonal diversity is significant and is only now beginning to be understood. Interstrain genomic polymorphisms, together with the individual host response hold the key to clarifying the disease initiation and progression in the individual patient. Further, we detected putative invasion-associated genes in 9 of the tested 11 P. gingivalis disease isolates from patients. Finally, based on functional assays with isogenic mutant, we identified PG0185 as an invasion-related gene. Defining the molecular basis of invasive P. gingivalis-host cell interactions is an important step toward a more complete characterization of P. gingivalis virulence mechanisms and the identification of a subset of genes that can be used as targets for the development of the next generation of diagnostic tools and/or intervention strategies.
Supplementary Material
Access the supplementary material to this article: Supplement, table (see Supplementary files under Reading Tools online).
Acknowledgements
We thank W. Ian Lipkin for providing access to Axon scanner, Moritz Kebschull for the HMEC-1 cell line, Sigrun Eick for strain JH16-1 DNA, Yukitaka Murakami for PG0185 and PG0186 mutants, Vladan Miljkovic for microarray facility support, and all contributors of strains to our collection. NIDCR and NIAID sponsored the microarrays.
References
- 1.Dorrell N, Mangan JA, Laing KG, Hinds J, Linton D, Al-Ghusein H, et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001;11:1706–15. doi: 10.1101/gr.185801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porwollik S, Wong RM, McClelland M. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc Natl Acad Sci USA. 2002;99:8956–61. doi: 10.1073/pnas.122153699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porwollik S, Santiviago CA, Cheng P, Florea L, McClelland M. Differences in gene content between Salmonella enterica serovar Enteritidis isolates and comparison to closely related serovars Gallinarum and Dublin. J Bacteriol. 2005;187:6545–55. doi: 10.1128/JB.187.18.6545-6555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998;36:3239–42. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desvarieux M, Demmer RT, Rundek T, Boden-Albala B, Jacobs DR, Jr, Sacco RL, et al. Periodontal microbiota and carotid intima-media thickness: the Oral Infections and Vascular Disease Epidemiology Study (INVEST) Circulation. 2005;111:576–82. doi: 10.1161/01.CIR.0000154582.37101.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang HW, Huang YF, Chou MY. Occurrence of Porphyromonas gingivalis and Tannerella forsythensis in periodontally diseased and healthy subjects. J Periodontol. 2004;75:1077–83. doi: 10.1902/jop.2004.75.8.1077. [DOI] [PubMed] [Google Scholar]
- 7.Loos BG, Van Winkelhoff AJ, Dunford RG, Genco RJ, DeGraaff J, Dickinson DP, et al. A statistical approach to the ecology of Porphyromonas gingivalis. J Dent Res. 1992;71:353–8. doi: 10.1177/00220345920710020201. [DOI] [PubMed] [Google Scholar]
- 8.Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–85. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Progulske-Fox A, Kozarov E, Dorn B, Dunn W, Jr, Burks J, Wu Y. Porphyromonas gingivalis virulence factors and invasion of cells of the cardiovascular system. J Periodontal Res. 1999;34:393–9. doi: 10.1111/j.1600-0765.1999.tb02272.x. [DOI] [PubMed] [Google Scholar]
- 10.Deshpande RG, Khan M, Genco CA. Invasion strategies of the oral pathogen Porphyromonas gingivalis: implications for cardiovascular disease. Invasion Metastasis. 1999;18:57–69. doi: 10.1159/000024499. [DOI] [PubMed] [Google Scholar]
- 11.Dorn BR, Burks JN, Seifert KN, Progulske-Fox A. Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis . FEMS Microbiol Let. 2000;187:139–44. doi: 10.1111/j.1574-6968.2000.tb09150.x. [DOI] [PubMed] [Google Scholar]
- 12.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis . Microbiol Mol Biol Rev. 1998;62:1244–63. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinane DF, Riggio MP, Walker KF, MacKenzie D, Shearer B. Bacteraemia following periodontal procedures. J Clin Periodontol. 2005;32:708–13. doi: 10.1111/j.1600-051X.2005.00741.x. [DOI] [PubMed] [Google Scholar]
- 14.Bahrani-Mougeot FK, Paster BJ, Coleman S, Ashar J, Barbuto S, Lockhart PB. Diverse and novel oral bacterial species in blood following dental procedures. J Clin Microbiol. 2008;46:2129–32. doi: 10.1128/JCM.02004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yilmaz O, Young PA, Lamont RJ, Kenny GE. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology. 2003;149:2417–26. doi: 10.1099/mic.0.26483-0. [DOI] [PubMed] [Google Scholar]
- 16.Kozarov E, Sweier D, Shelburne C, Progulske-Fox A, Lopatin D. Detection of bacterial DNA in atheromatous plaques by quantitative PCR. Microbes Infect. 2006;8:687–93. doi: 10.1016/j.micinf.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Gibson FC, III, Hong C, Chou HH, Yumoto H, Chen J, Lien E, et al. Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation. 2004;109:2801–6. doi: 10.1161/01.CIR.0000129769.17895.F0. [DOI] [PubMed] [Google Scholar]
- 18.Kozarov E, Dorn B, Shelburne C, Dunn W, Progulske-Fox A. Human atherosclerotic plaque contains viable invasive Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans . Arterioscler Thromb Vasc Biol. 2005;25:e17–e18. doi: 10.1161/01.ATV.0000155018.67835.1a. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Michel R, Cohen J, DeCarlo A, Kozarov E. Intracellular survival and vascular cell-to-cell transmission of Porphyromonas gingivalis. BMC Microbiol. 2008;8:26–36. doi: 10.1186/1471-2180-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorn BR, Dunn WA, Jr, Progulske-Fox A. Invasion of human coronary artery cells by periodontal pathogens. Infect Immun. 1999;67:5792–8. doi: 10.1128/iai.67.11.5792-5798.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato T, Kawai S, Nakano K, Inaba H, Kuboniwa M, Nakagawa I, et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol. 2007;9:753–65. doi: 10.1111/j.1462-5822.2006.00825.x. [DOI] [PubMed] [Google Scholar]
- 22.Tribble GD, Mao S, James CE, Lamont RJ. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc Natl Acad Sci USA. 2006;103:11027–32. doi: 10.1073/pnas.0509813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan L, Rodrigues PH, Belanger M, Dunn W, Jr, Progulske-Fox A. The Porphyromonas gingivalis clpB gene is involved in cellular invasion in vitro and virulence in vivo. FEMS Immunol Med Microbiol. 2007;51:388–98. doi: 10.1111/j.1574-695X.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 24.Yuan L, Rodrigues PH, Belanger M, Dunn WA, Jr, Progulske-Fox A. Porphyromonas gingivalis htr A is involved in cellular invasion and in vivo survival. Microbiology. 2008;154:1161–9. doi: 10.1099/mic.0.2007/015131-0. [DOI] [PubMed] [Google Scholar]
- 25.Park Y, Yilmaz O, Jung IY, Lamont RJ. Identification of Porphyromonas gingivalis genes specifically expressed in human gingival epithelial cells by using differential display reverse transcription-PCR. Infect Immun. 2004;72:3752–8. doi: 10.1128/IAI.72.7.3752-3758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrian E, Grenier D, Rouabhia M. In vitro models of tissue penetration and destruction by Porphyromonas gingivalis . Infect Immun. 2004;72:4689–98. doi: 10.1128/IAI.72.8.4689-4698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosogi Y, Duncan MJ. Gene expression in Porphyromonas gingivalis after contact with human epithelial cells. Infect Immun. 2005;73:2327–35. doi: 10.1128/IAI.73.4.2327-2335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan L, Hillman JD, Progulske-Fox A. Microarray analysis of quorum-sensing-regulated genes in Porphyromonas gingivalis . Infect Immun. 2005;73:4146–54. doi: 10.1128/IAI.73.7.4146-4154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simionato MR, Tucker CM, Kuboniwa M, Lamont G, Demuth DR, Tribble GD, et al. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii . Infect Immun. 2006;74:6419–28. doi: 10.1128/IAI.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen T, Hosogi Y, Nishikawa K, Abbey K, Fleischmann RD, Walling J, et al. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J Bacteriol. 2004;186:5473–9. doi: 10.1128/JB.186.16.5473-5479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz PI, Slakeski N, Reynolds EC, Morona R, Rogers AH, Kolenbrander PE. Role of oxyR in the oral anaerobe Porphyromonas gingivalis . J Bacteriol. 2006;188:2454–62. doi: 10.1128/JB.188.7.2454-2462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrigues PH, Progulske-Fox A. Gene expression profile analysis of Porphyromonas gingivalis during invasion of human coronary artery endothelial cells. Infect Immun. 2005;73:6169–73. doi: 10.1128/IAI.73.9.6169-6173.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eick S, Rodel J, Einax JW, Pfister W. Interaction of Porphyromonas gingivalis with KB cells: comparison of different clinical isolates. Oral Microbiol Immunol. 2002;17:201–8. doi: 10.1034/j.1399-302x.2002.170401.x. [DOI] [PubMed] [Google Scholar]
- 35.Nagano K, Murakami Y, Nishikawa K, Sakakibara J, Shimozato K, Yoshimura F. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J Med Microbiol. 2007;56:1536–48. doi: 10.1099/jmm.0.47289-0. [DOI] [PubMed] [Google Scholar]
- 36.Ades EW, Candal FJ, Swerlick RA, George VG, Summers S, Bosse DC, et al. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Invest Dermatol. 1992;99:683–90. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- 37.Bélanger M, Rodrigues P, Progulske-Fox A. Hoboken, NJ: John Wiley; 2006. Current Protocols, Chapter 13, Unit 13C.3. Genetic manipulation of Porphyromonas gingivalis. . [DOI] [PubMed] [Google Scholar]
- 38.Deshpande RG, Khan MB, Genco CA. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis . Infect Immun. 1998;66:5337–43. doi: 10.1128/iai.66.11.5337-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth GA, Aumayr K, Giacona MB, Papapanou PN, Schmidt AM, Lalla E. Porphyromonas gingivalis infection and prothrombotic effects in human aortic smooth muscle cells. Thromb Res. 2009;123:780–4. doi: 10.1016/j.thromres.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63:2522–31. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loos BG, Dyer DW, Whittam TS, Selander RK. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect Immun. 1993;61:204–12. doi: 10.1128/iai.61.1.204-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slots J. Update on Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease. J Int Acad Periodontol. 1999;1:121–6. [PubMed] [Google Scholar]
- 43.Leys EJ, Smith JH, Lyons SR, Griffen AL. Identification of Porphyromonas gingivalis strains by heteroduplex analysis and detection of multiple strains. J Clin Microbiol. 1999;37:3906–11. doi: 10.1128/jcm.37.12.3906-3911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Califano JV, Arimoto T, Kitten T. The genetic relatedness of Porphyromonas gingivalis clinical and laboratory strains assessed by analysis of insertion sequence (IS) element distribution. J Periodontal Res. 2003;38:411–6. doi: 10.1034/j.1600-0765.2003.00665.x. [DOI] [PubMed] [Google Scholar]
- 45.Kozarov E, Whitlock J, Dong H, Carrasco E, Progulske-Fox A. The number of direct repeats in hagA is variable among Porphyromonas gingivalis strains. Infect Immun. 1998;66:4721–5. doi: 10.1128/iai.66.10.4721-4725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, et al. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–3. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 47.Curtis MA, Fawell SC, Hall LM. Microarray analysis of the P. gingivalis genome demonstrates a dynamic chromosomal structure. J Dent Res. 2003;82:B-315. [Google Scholar]
- 48.Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–86. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 49.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–79. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 50.Muller CM, Aberg A, Straseviciene J, Emody L, Uhlin BE, Balsalobre C. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 2009;5:e1000303. doi: 10.1371/journal.ppat.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies M, Gouyette C, et al. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila . Cell Microbiol. 2006;8:1228–40. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- 52.Song H, Belanger M, Whitlock J, Kozarov E, Progulske-Fox A. Hemagglutinin B is involved in the adherence of Porphyromonas gingivalis to human coronary artery endothelial cells. Infect Immun. 2005;73:7267–73. doi: 10.1128/IAI.73.11.7267-7273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song YH, Kozarov EV, Walters SM, Cao SL, Handfield M, Hillman JD, et al. Genes of periodontopathogens expressed during human disease. Ann Periodontol. 2002;7:38–42. doi: 10.1902/annals.2002.7.1.38. [DOI] [PubMed] [Google Scholar]
- 54.Progulske-Fox A, Hillman J, Handfield M. Identification of Porphyromonas gingivalis virulence polynucleotides for diagnosis, treatment, and monitoring of periodontal diseases. United States Patent 7,416,852 Book Identification of Porphyromonas gingivalis virulence polynucleotides for diagnosis, treatment, and monitoring of periodontal diseases. United States Patent 7416852.
- 55.Shi X, Hanley SA, Faray-Kele MC, Fawell SC, Aduse-Opoku J, Whiley RA, et al. The rag locus of Porphyromonas gingivalis contributes to virulence in a murine model of soft tissue destruction. Infect Immun. 2007;75:2071–4. doi: 10.1128/IAI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodrigues P, Arauz A, Bass D, Progulske-Fox A. Baltimore, MD: IADR; 2005. EpsC, ragB and adhesion/invasion of P. gingivalis to HCAE cells. [Google Scholar]
- 57.Pieper R, Zhang Q, Parmar PP, Huang ST, Clark DJ, Alami H, et al. The Shigella dysenteriae serotype 1 proteome, profiled in the host intestinal environment, reveals major metabolic modifications and increased expression of invasive proteins. Proteomics. 2009;9:5029–45. doi: 10.1002/pmic.200900196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roberts AJ, Williams SK, Wiedmann M, Nightingale KK. Some Listeria monocytogenes outbreak strains demonstrate significantly reduced invasion, inlA transcript levels, and swarming motility in vitro. Appl Environ Microbiol. 2009;75:5647–58. doi: 10.1128/AEM.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Access the supplementary material to this article: Supplement, table (see Supplementary files under Reading Tools online).