Abstract
Microfluidic adhesion-based cell separation systems are of interest in clinical and biological applications where small sample volumes must be processed efficiently and rapidly. While the ability to capture rare cells from complex suspensions such as blood using microfluidic systems has been demonstrated, few methods exist for rapid and non-destructive release of the bound cells. Such detachment is critical for applications in tissue engineering and cell-based therapeutics in contrast with diagnostics wherein immunohistochemical, proteomic, and genomic analyses can be carried out by simply lysing captured cells. This paper demonstrates how the incorporation of 4-arm amine terminated poly(ethylene glycol) (PEG) molecules along with antibodies within alginate hydrogels can enhance the ability of the hydrogels to capture endothelial progenitor cells (EPCs) from whole human blood. The hydrogel coatings are applied conformally onto pillar structures within microfluidic channels and their dissolution with a chelator allows for effective recovery of EPCs following capture.
Introduction
The use of microfluidic devices in adhesion-based separation of cells is an active area of research in both clinical medicine and basic science.1–4 This mode of separation is attractive because no labeling with fluorescent or magnetic tags is needed to drive the separation process unlike conventional fluorescence- or magnet-activated cell sorting (FACS and MACS, respectively). The high surface area to volume ratios of microfluidic channels together with the ability to enhance surface area with microfabricated structures 3, 5, 6 has enabled such devices to capture cells of extremely low concentrations for a broad range of applications. A major challenge in this area, however, is the lack of methods to achieve non-destructive release of cells captured within microfluidic channels.7–10 In a diagnostic context, useful information can be obtained by simple adhered cell counts2, 3 or by lysing cells on chip and performing proteomic and/or genomic analysis.3, 4 However, when isolated cells need to be recovered for therapeutic or scientific purposes, cell detachment must be carried out without causing physical damage and changes in phenotypic identity or function in the cells. These constraints limit the chemical and mechanical forces that can be applied to achieve cell release; for example, enzyme-induced cell detachment is known to cause chemical and phenotypic changes within cells.9, 10 Furthermore, when simplicity is desired for devices designed for point-of-care and disposable use, the use of electrical, thermal, or optical means of cell detachment becomes infeasible.11, 12
In previous work we have described how alginate hydrogel coatings can be formed on the inner surfaces of microfluidic channels12 and utilized for cell capture from flowing suspensions followed by release. These coatings contained cell-adhesive molecules covalently bound to the carboxylic acid groups of alginic acid. While these coatings were able to achieve capture and release of primary rat cardiac fibroblasts from homogeneous suspensions, the adhesion of the cells to alginate hydrogels containing no cell-adhesive molecules was fairly high. High baseline adhesion levels are undesirable when cell capture must be carried out from heterogeneous suspensions of cells, particularly when target cell concentrations are low. As a material, however, alginate hydrogels are easy to create via physical crosslinking in the presence of divalent cations and dissolve using relatively low concentrations of chelator molecules such as ethylene diamine tetraacetic acid (EDTA). In the context of microfluidic devices and as shown in our prior work,12 these hydrogels can be created by adsorbing functionalized alginic acid within the microchannels and then forming the gel by flowing a solution of calcium chloride. The concentration of alginic acid in the initial step must be low enough to enable injection into a narrow channel and the flow rate of calcium chloride in the next step must be high enough to ensure that the gel does not fill the entire channel. These parameters can be easily optimized and the non-covalent nature of the hydrogel-microchannel binding allows extension of the coating process to microchannels made of any material.
This article examines how alginate hydrogels can be modified with 4-arm poly(ethylene glycol) (PEG) molecules to enhance functionalization with cell-adhesive antibodies while simultaneously suppressing non-specific binding. The effectiveness of these functionalized hydrogels as capture/release coatings is demonstrated by targeting endothelial progenitor cells (EPCs) from whole human blood using a pillar-array microfluidic device. EPCs are present at relatively low concentrations in blood, with typical concentrations in the range of 10 000 cells/mL in healthy individuals (as measured in our laboratory). The isolation of these cells from blood is a first step in the growth of blood vessels in vitro13 and is typically carried out using a multiple-cycle technique of centrifugation and plating. Our work is motivated by the need for more rapid and low-cost methods of isolation of EPCs and other rare cells utilized in tissue engineering and cell-based regenerative repair.
The incorporation of PEG within alginate hydrogels for 3-dimensional cell culture or drug delivery is well-established,14, 15 however the use of these hydrogels in cell-affinity chromatography is relatively new. This article demonstrates how 4-arm, amine-terminated PEG molecules can not only increase purity of captured EPCs by suppression of non-specific binding but also enhance capture yield by providing more tether points for capture antibodies.
Materials and Methods
Materials
Glass slides, EDC (1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride), sulfo-NHS (N-hydroxysuccinimide), EDTA, 2-(4-morpholino) ethane sulfonic acid (MES) buffer, a micro bicinchoninic acid (BCA) protein assay kit, and heparin vacuum tubes were purchased from Fisher Scientific (Fair Lawn, NJ). For microfluidic device fabrication, SU-8 photoresist and developer were obtained from MicroChem (Newton, MA); silicone elastomer and curing agent were obtained from Dow Corning (Midland, MI). Phosphate-buffered saline (PBS; 1×, without calcium or magnesium) was purchased from Mediatech (Herndon, VA). The capture antibody, monoclonal mouse anti-human CD34, and goat anti-human Flk-1 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-human CD133-PE, mouse anti-human CD45-FITC, and mouse anti-goat IgG-PerCP antibodies were obtained from eBioscience (San Diego, CA). Rabbit IgG was purchased from Vector Labs (Burlingame, CA). Calcium chloride dehydrate, Trypan Blue solution and alginic acid were purchased from Sigma (St. Louis, MO). Amine-terminated 4-arm PEG (PEG-NH2) with molecular weights of 10,000 (10k MW) and 20,000 (20k MW) was purchased from Laysan Bio (Arab, AL).
Microfluidic Cell Capture Device Design
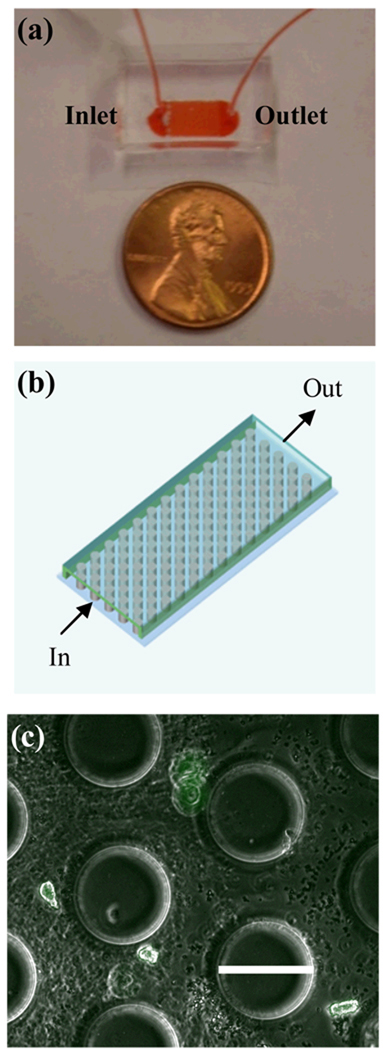
The device uses a post array design similar to that used by Nagrath et al.3 (Fig. 1a-b). To achieve disruption of flow streams and achieve optimal capture, the posts were arranged in a hexagonal layout as described by Gleghorn et al.6 The posts have a diameter of 100 µm and a transverse spacing of 150 µm from center to center (Fig. 1c). Rows have a center to center spacing of 125 µm and each is offset by 50 µm. The post array is 0.7 cm long and 0.5 cm wide. The posts height was approximately 50 µm for the devices fabricated by soft lithography as described below.
Figure 1.
Schematic diagram of the microfluidic post array (a) and image of cell capture device (b). EPCs captured within the device are shown in (c) where green represents staining with fluorescently labeled anti-CD133 (scale bar = 100 µm).
Microfluidic Device Fabrication
A 2-dimensional projection of the cell capture device was drawn using AutoCAD in-house, and the image was imprinted at high resolution onto a chrome mask by FineLine Imaging (Colorado Springs, CO). This photomask was utilized to generate a negative master at the George J. Kostas Nanoscale Technology and Manufacturing Research Center at Northeastern University. In précis, a silicon wafer was coated with SU 8–50 photoresist to a thickness of approximately 50 µm. With the mark overlaid, the wafer was exposed to 365 nm, 11 mW/cm2 ultraviolet light from a Q2001 mask aligner (Quintel Co, San Jose, CA). Unexposed photoresist was then removed using SU 8 developer.
For poly(dimethyl siloxane) (PDMS) device fabrication, the silicone elastomer and curing agent were mixed in a 10:1 (w/w) ratio and poured on top of the negative master wafers, degassed, and allowed to cure overnight at 65 °C. PDMS replicas were then pulled off the wafers prior to punching inlet and outlet holes with a 19-gauge blunt-nose needle. The replicas and glass slides were exposed to oxygen plasma (50 mW with 8% oxygen for 30 s) in a PX-250 plasma chamber (March Instruments, Concord, MA) and then immediately placed in contact with each other. The irreversible bonding between PDMS and glass was completed by baking for 5 min at 65 °C.
PEG/Antibody-Functionalized Hydrogel Synthesis
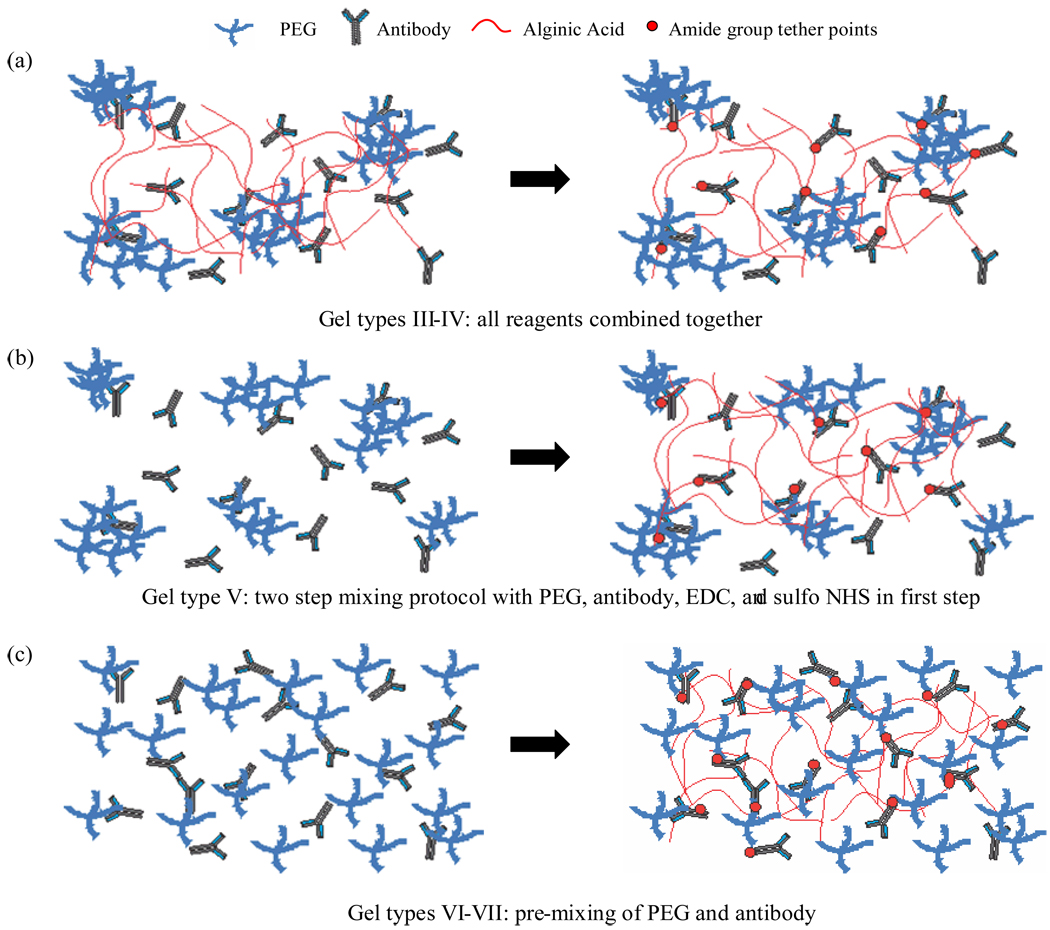
Seven different hydrogel formulations were investigated in this study and these are designated as gel types I-VII. For gel type I, 45 mg of alginic acid, 4.8 mg EDC, 13.2 mg sulfo-NHS, and 20 µL inert IgG (1 g/mL) were added to 2 ml of MES buffer solution and mixed using an IKA Ultra Turrax Tube Disperser (Wilmington, NC) for 29 min and allowed to incubate for 60 min. For gel type II, 45 mg of alginic acid, 4.8 mg EDC, 13.2 mg sulfo-NHS and 100 µL anti-human CD34 (200 µg/mL) were added to 2 mL of MES buffer, mixed as before, and incubated for 60 min. For gel type III, 45 mg alginic acid, 4.8 mg EDC, 13.2 mg sulfo-NHS, 45 mg 20k MW PEG, and 100 µL anti-human CD34 were added to 2 mL of MES buffer, mixed for 29 min, and allowed to incubate for 60 min. Gel type IV consisted of 45 mg alginic acid, 4.8 mg EDC, 13.2 mg sulfo-NHS, 22.5 mg 10k MW PEG, and 100 µL anti-human CD34 added to 2 mL of MES buffer, mixed for 29 min and allowed to incubate for 60 min. Gel type V was created by mixing 4.8 mg EDC, 13.2 mg sulfo-NHS, 22.5 mg 10k MW PEG, and 100 µL anti-human CD34 in 2 ml of MES buffer for 29 min and then adding 45 mg of alginic acid followed by 29 min of mixing and 60 min of incubation. Gels VI and VII were formed by mixing 22.5 mg 10k MW PEG with 100 µL antibody in 2 mL of MES buffer and mixing for 10 min and 29 min, respectively, and incubating for an additional 15 min and 60 min, respectively. 4.8 mg EDC, 13.2 mg sulfo-NHS, and 45 mg alginic acid were then added to the mixture, mixed for 29 min and allowed to incubate for 60 min. All formulations were mixed at room temperature (RT) and stored at 4°C prior to use.
Following the incubation step, each functionalized alginic acid solution for each gel type was injected into a Slide-A-Lyzer Dialysis Cassette 10,000 molecular weight cut-off (Fisher) and dialyzed against MES buffer for 48 hours to remove unreacted sulfo-NHS and EDC. Table 1 summarizes the synthetic steps and components for each gel type. Steps 1 and 2 indicate the sequential nature of the protocol followed for combining the respective reagents.
Table 1.
Summary of Synthesis Protocols for Different Hydrogel Formulations.
| Gel Type |
PEG MW [kDa] |
Components in Each Mixing Sequence† |
Mixing/Incubation Times [min] |
||||
|---|---|---|---|---|---|---|---|
| Alginic Acid |
EDC & Sulfo- NHS |
PEG | Antibody* | Step 1 | Step 2 | ||
| I | none | 1 | 1 | - | 1 | 29/60 | N/A |
| II | none | 1 | 1 | - | 1 | 29/60 | N/A |
| III | 20 | 1 | 1 | 1 | 1 | 29/60 | N/A |
| IV | 10 | 1 | 1 | 1 | 1 | 29/60 | N/A |
| V | 10 | 2 | 1 | 1 | 1 | 29/0 | 29/60 |
| VI | 10 | 2 | 2 | 1 | 1 | 10/15 | 29/60 |
| VII | 10 | 2 | 2 | 1 | 1 | 29/60 | 29/60 |
“1” denotes reagent added in step 1; “2” denotes reagent added in step 2.
Inert IgG was used for gel type I; anti-human CD34 was used in all other gel types.
Infrared Spectroscopy
Functionalized alginic acid samples were spread on poly(tetrafluoroethylene) (PTFE) sample cards (Crystal Labs, Garfield, NJ) using a spatula and allowed to thicken for 4 h. The cards were then inserted into a Perkin Elmer 1000 Fourier-transform Infrared (FTIR) spectrometer (Waltham, MA). The absorbance at 638 cm−1, corresponding to amide groups, was analyzed and compared for each gel type.
In Situ Hydrogel Formation within Microfluidic Devices
A 1 g/mL solution of CaCl2 in deionized water was injected into each device (by hand, using a 1 mL syringe) and allowed to incubate overnight at RT. The CaCl2 solution was then withdrawn by hand using a 1 mL syringe. The PEG- and antibody-functionalized alginate solution prepared for each gel type was then injected into the devices by hand and allowed to adsorb for 1 h. Next, the devices were rinsed with MES buffer at 10 µl/min for 10 min using a Harvard Apparatus PHD 2000 syringe pump (Holliston, MA), followed by a 100 mM CaCl2 solution in MES buffer at 10 µl/min for 10 min to form a thin layer of hydrogel on the walls of the microchannels. Finally, the devices were rinsed with MES buffer at 10 µl/min for 10 min to remove unreacted CaCl2. All experiments were conducted at RT.
BCA Protein Assay
A BCA protein assay solution was prepared according to manufacturer instructions. The solution was then injected into each device at 5 µl/min for 40 min at RT. The output was collected in a microplate and absorption at 562 nm was measured using a Bio-Tek Powerwave XS spectrometer (Winooski, VT).
Blood Draw
Whole human blood was drawn from healthy volunteers in heparin collection tubes under a protocol approved by the Northeastern University Institutional Review Board.
EPC Capture Protocol
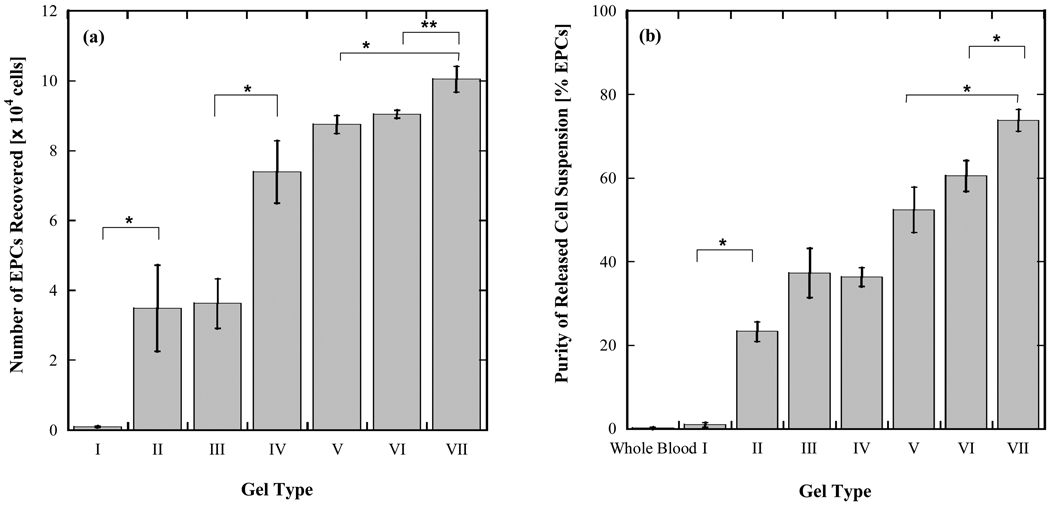
Whole blood was injected into microfluidic capture devices at 5 µl/min for 60 min. Each device was then rinsed with MES buffer at 10 µl/min for 10 min. For release of captured cells, a 50 mM solution of EDTA in PBS was injected at 10 µl/min for 10 minutes and the output was collected in a 1.5 mL microcentrifuge tube. Devices were kept at RT for all cell release steps. Each individual experiment included 10 microfluidic devices. 300 µL of blood was passed through each device, at the rate specified above. The cells released from each device were pooled into a single suspension to allow enumeration by a Beckman Coulter Quanta SC (Brea, CA) flow cytometer. Hence the data reported in Fig. 4a-b represent yield and purity for EPCs recovered from a total blood volume of 3 mL.
Figure 4.
Yield (a) and purity (b) of EPCs captured from whole blood within microfluidic devices coated with PEG- and antibody-functionalized hydrogels. 300 µL of whole blood collected in heparin tubes was directly injected into individual microfluidic devices and 10 devices were run in parallel. Cells released from each device were pooled into a single suspension to allow enumeration by flow cytometry. Data reported in (a) and (b) represent yield and purity for EPCs recovered from a total blood volume of 3 mL. Error bars denote standard deviations based on 3 independent measurements of EPC and total cell counts made with the same sample.
Flow Cytometry
For EPC enumeration, cells released from each device were mixed with 10 µl each of anti-human CD133-PE, anti-human CD45-FITC, goat anti-human Flk-1, and anti-goat IgG-PerCP. The mixture was stored in the dark at RT for 30 min followed by centrifugation at 130 × g for 10 min. The supernatant was decanted and cells were suspended in 200 µL of PBS for enumeration using flow cytometry. While there is some debate as to the precise surface marker profile of EPCs, a general definition based on a number of literature sources16–18 was applied in the present study and released cells that were CD133+, CD45−, and Flk-1+ were counted as EPCs.19–23
Results
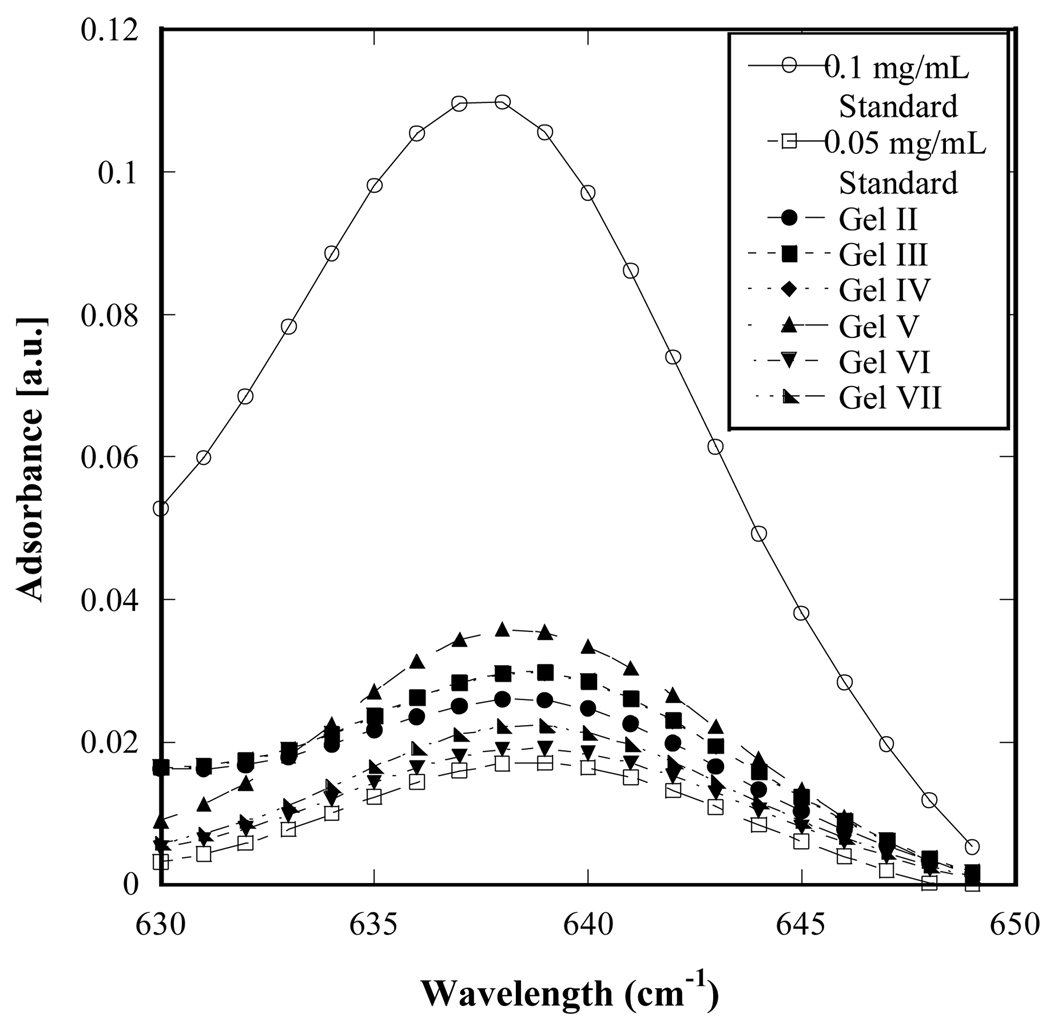
Fig. 2 shows infrared (IR) spectroscopy data for the functionalized alginic acid solutions emerging from the one- or two-step synthesis protocol. The spectra for gel types II-VII show equivalent levels of amide group content. In all of these gels, amide groups can result not only from binding between antibody and PEG molecules but also from antibody-alginic acid and PEG-alginic acid binding. The IR spectroscopy analysis therefore reflects that antibody and amide bond content, taken together, is very similar across all gel types, indicating successful incorporation of antibody and PEG; however no functional distinctions between the different gel types can be predicted from these data.
Figure 2.
Infrared spectra of PEG- and antibody-functionalized hydrogels (gel types II-VII; not normalized to avoid total overlap) compared to standard solutions of anti-CD34 with concentrations of 0.1 mg/mL and 0.05 mg/mL. The relative intensities of the hydrogel sample peaks are comparable, indicating similar levels of amide group content. Note that this measurement is a bulk measurement that does not distinguish between covalently-bound antibody from antibody molecules that are physically trapped within the hydrogels.
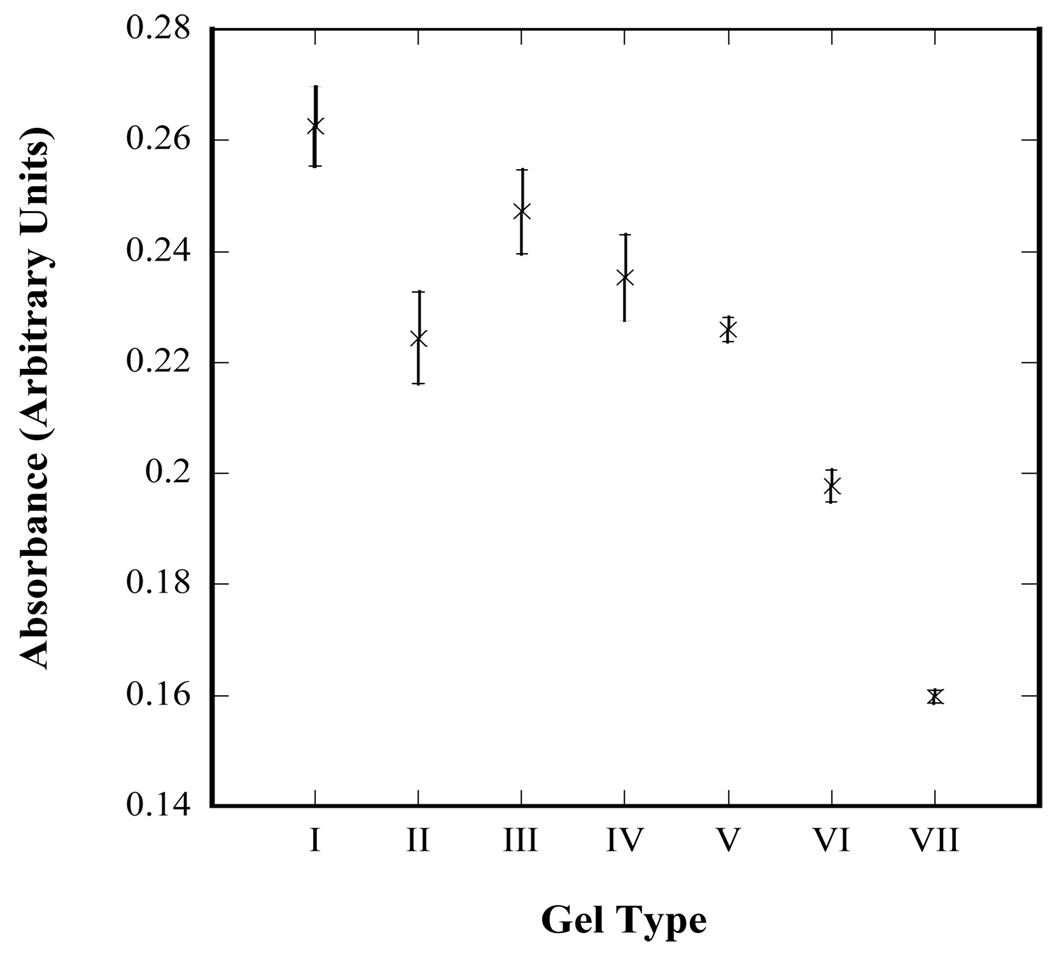
Fig. 3 shows relative total protein measurements made using a BCA assay kit. The BCA solution becomes changes from green to blue as it comes in contact with proteins such as antibodies. Hence, by flowing this solution through hydrogel-coated microfluidic devices, the amount of accessible antibody on each gel type can be compared. The color change of the solutions exiting the devices is shown as a function of gel type in Fig. 3 and is expressed in arbitrary units of absorbance at 562 nm rather than as a calibrated mass or concentration. Such a calibration cannot be performed as the interaction between the BCA solution and protein is a flow-surface interaction whereas standard calibrations would be carried out by mixing and incubating together the BCA solution and a solution of known protein concentration. The relative measurement nevertheless allows comparison of the accessible anti-CD34 capture antibody between each gel type. Fig. 3 shows an increase in accessible antibody from gel types I-VII while the total antibody and amide content of the mixture remains constant (Fig. 2), indicating an increase in the efficiency of conjugation between the gelled surface and the antibody.
Figure 3.
Qualitative measurement of accessible antibody within hydrogel-coated microfluidic devices. A bicinchoninic acid (BCA) assay kit was utilized to measure the relative amount of antibody accessible to a solution flowing through each device. A lower absorbance is associated with a greater amount of accessible antibody. Error bars denote standard errors based on 8 independent measurements for each gel type.
Fig. 4 shows yield and purity data for the capture of EPCs from whole blood using the hydrogel-coated microfluidic devices. In Fig. 4a, gel type I, which has an inert antibody conjugated to it, shows negligible EPC adhesion as expected. Gel type II, which contains the anti-CD34 antibody, shows significantly higher EPC adhesion relative to gel type I (p < 0.005), albeit with a high degree of scatter. The purity of capture achieved with gel type II is, however, relatively low (~23%; Fig. 4b). The effect of adding the 4-arm PEG to the hydrogel structure is shown clearly by comparing gel types II and IV, whose synthesis protocol is otherwise identical. The branched amine termini of the 4-arm 10k MW PEG molecules provide an opportunity for a greater level of antibody conjugation, as reflected in the higher overall EPC adhesion (Fig. 4a). The suppression of non-specific binding results in an increase in purity (Fig. 4b; gel type IV). Interestingly, the use of 20k MW PEG (gel type III) resulted in significantly lower EPC capture yield relative to 10k MW PEG (gel type IV; p < 0.005) under the same synthesis conditions and purity levels were comparable.
In gel types V-VII, a two step protocol for combining reagents was followed. In gel type V, the conjugation of the antibody molecules to the 4-arm PEG is carried out first before introducing alginic acid. This formulation improved yield and purity of EPC capture relative to gel type IV. However, this protocol introduces the possibility of all four arms of the PEG molecules being occupied by antibodies, leaving no amine groups to bind to alginic acid. To address this risk, the two-step protocol was modified such that EDC and sulfo-NHS were added in the second step with alginic acid and the first step was restricted to the mixing together of PEG and antibody. When short times were provided for mixing and incubation for the first step (10 min and 15 min, respectively, for gel type VI), the yield did not improve relative to gel type V, however purity was marginally higher. Higher mixing and incubation times were examined next (29 and 60 min, respectively, for gel type VII) in an effort to achieve greater mixing and entanglement of the PEG molecules with the antibody molecules. This formulation provided significantly higher yields and purity relative to gel types VI and VII (p < 0.005 and p < 0.01, respectively). For all gel types, the viability of captured and released cells, as measured by Trypan Blue exclusion, was approximately 90%.
Discussion
The ability to selectively capture and then release cells within microfluidic channels requires a balance of physical forces and chemical interactions11 while simultaneously maintaining cell viability and function, particularly in the context of tissue engineering and regenerative medicine where cell isolation is a common initial step.24–26 Alginate hydrogels are particularly interesting for this application because of the ability to easily create them by physical crosslinking of alginic acid in the presence of divalent cations and their well-known ability to dissolve in the presence of chelators.27–29 Furthermore, these hydrogels are biocompatible30–32 and can be functionalized with cell-adhesive molecules.12, 33–36 Alginate hydrogels are, however, highly prone to non-specific cell adhesion;31 for example, in a recent study by our group that examined the capture of fibroblasts by alginate hydrogels, there was appreciable cell adhesion to the non-functionalized hydrogel.12 When alginate hydrogels were functionalized with anti-CD34, a capture molecule for EPCs,37 and utilized for EPC capture from whole blood, similar results were obtained in the form of high non-target cell adhesion (results not shown). The present study was motivated by these observations and the hypothesis that the inclusion of PEG within the hydrogel structure would be a possible means to overcome this problem. Another shortcoming associated with functionalization of alginate hydrogels is the limited number of carboxylic acid groups available for carbodiimide-based conjugation. In the context of microfluidic capture devices, this limitation is exacerbated by the fact that flowing cells only “see” the adhesive capture molecules on the surface whereas conjugation protocols are easiest to carry out in the bulk prior to hydrogel formation within the microchannels.12
PEG is well known for its biological non-adhesiveness38–41 and the present study demonstrates how these properties can be harnessed while simultaneously increasing the capture molecule content of the hydrogel. These objectives are achieved by the use of 4-arm PEG molecules with primary amine terminations at the end of each arm. In an ideal case, one arm of each 4-arm PEG molecule would bind to a carboxylic acid group to the alginate hydrogel backbone, leaving up to three primary amine groups for antibody functionalization, provided the 4-arm PEG is sufficiently large to prevent steric hindrance between adjacent antibody molecules. This arrangement would, in principle, triple the antibody content of the hydrogel and provide protection against non-specific cell binding relative to non PEG-y-lated alginate hydrogels. However, such an idealized architecture is difficult to achieve synthetically if the same carbodiimide chemistry is to be utilized for both hydrogel-PEG and PEG-antibody binding. The present study examined how variations of a relatively simple synthetic protocol, made using probabilistic as well as chemical considerations, can overcome this constraint and provide effective capture and release of EPCs from whole blood. Protecting groups, such as fluorenylmethyloxycarbonyl chloride (fmoc),42 can be effectively employed to achieve some level of control over protein or antibody conjugation to primary amine groups, however, here again it is difficult to ensure that only a certain number of amine groups are protected on each PEG molecule.
The need for simplicity in the hydrogel synthesis protocol arises from the intended application of these hydrogels in low-cost microfluidic cell separation systems for tissue engineering and clinical diagnostics. The present work, for example, provides the design basis for a microfluidic separator if EPCs from blood for subsequent use in vascular tissue engineering43, 44 or cell-based regenerative repair of vascular tissue in vivo.43, 45, 46 Hence, there is significant motivation for keeping the number of synthesis steps and reagents to a minimum.
Fig. 4 illustrates the evolution in synthetic protocols from gel types I through VII. The adhesive effect of the anti-CD34 antibody is evident by comparing gel types I and II (Fig. 4a). Increased yield and purity are observed with the incorporation of 10k MW PEG (gel types II vs. IV), while the level of scatter remains relatively large. The large error bars in both instances reflect the random nature of bond formation between the alginate hydrogel and the amine group-containing antibody and PEG molecules. As shown pictorially in Fig. 5a, it is likely that the 4-arm PEG molecules remain entangled in clusters, which would result in a patch like accumulation of these clusters within the hydrogel, with antibody molecules being bonded within the clusters as well as directly to alginic acid. Such irregular distribution of antibody molecules would give rise to relatively large variations in cell adhesion between hydrogel samples that are otherwise identical with respect to reactants and method of combination. Indeed, the alginic acid solutions obtained after the two synthetic steps for gel types II and IV contained small particles of PEG visible to the naked eye (see light micrographs in Supporting Information), corroborating the hypothesis of inadequate mixing.
Figure 5.
Illustration of structural differences in gel types. The left side of the figure is representative of mixing step 1 prior to the formation of amide bonds via EDC chemistry and the right side represents the product resulting from mixing step 2 (see also Table 1). The progressive improvement in EPC capture yield and purity from gel types III-VII is attributed to the two step mixing protocol where PEG and antibody are pre-mixed before the introduction of alginic acid and the coupling reagents EDC and sulfo-NHS. Pre-mixing allows optimal dispersion of antibody molecules among the PEG chains.
Each gel type is injected into a microfluidic device and subjected to identical flow and shear conditions. Slight variations in gel viscosity may impact the gel thickness within the device, however due to the geometry of the post array even a three-fold difference in gel thickness will impact the overall surface area by less than 6% (see detailed calculation in Supporting Information). The differences in capture efficiency between gel types is far greater than 6% and it is therefore reasonable to argue that gel composition plays a much stronger role than gel thickness in the present context.
It is interesting to note that 20k MW PEG did not provide better capture and yield properties. Intuitively, one might expect the larger chains to provide a greater degree of steric freedom to antibody molecules bound to the amine termini and a greater resistance to non-specific binding (i.e. greater purity). However, the results in Fig. 4 indicate lower yield and comparable purity relative to hydrogels containing 10k MW PEG (gel types III vs. IV). Both of these hydrogels have similar levels of antibody accessible to the BCA reagents (Fig. 3), indicating chemical structure similarity. Hence, the more likely explanation for this difference is a physical structure difference wherein antibody molecules are farther apart from each other in the 20k PEG- than in the 10k PEG-containing hydrogel. The antibody molecules are far enough apart that their probability of encountering a flowing EPC and their ability to capture it are both lower with 20k PEG. On this basis, 10k PEG was utilized in all subsequent formulations.
With gel type V, the first step in the synthesis was the combination of PEG and antibody with the coupling agents EDC and sulfo-NHS prior to the addition of alginic acid in the second step. Relative to gel type IV, gel type V provides slightly greater EPC capture but with a lower degree of scatter, indicating better mixing of the antibody molecules with the PEG. The accessible antibody content of gel type V is similar to that of gel type IV (Fig. 3) providing credence to the postulate of better PEG-antibody mixing being the distinguishing factor. Better mixing also allows for more effective interspersing of PEG and antibody molecules on the hydrogel surface, which is consistent with the higher EPC purity obtained with gel type V relative to gel type IV. Fewer PEG particles were observed in the PEG- and antibody-functionalized alginic acid solution, consistent with this postulate.
The two-step synthesis protocol for gel types VI and VII built on the concept of better pre-mixing by providing time for antibody and PEG molecules to mix ‘undisturbed’ without the constraining presence of EDC and sulfo-NHS. Here, the longer mixing time provided alginic acid solutions that were visually clear, indicating good mixing, and the resulting coatings provided better EPC capture performance in terms of yield and purity relative to gel type V. The longer mixing and incubation times provided for gel type VII relative to gel type VI provided the best yield (~104 EPCs recovered) and purity (74%).
Fig. 4a does not show a comparison between EPC capture yields obtained with the hydrogels and the EPC content of whole blood. Flow cytometry measurements in our laboratory indicate a typical EPC count of 33 × 104 EPCs per 3 mL of whole blood for the samples used in this study. Hence the best performing gel type in this work only captures about a third of the total available EPC content. However, it important to note that this level of capture was achieved with only a single pass of the blood sample through the hydrogel-coated microfluidic devices. Series operation of multiple capture devices or sample recycling would offer facile means to increase total yield. Stage-wise separation could be employed to increase the purity of captured EPCs beyond the 74% level achieved with gel type VII in a single pass. These considerations are outside the scope of the present work, where the focus was limited to the design and synthesis of the capture coating material.
Conclusions
Alginate hydrogels functionalized with 4-arm PEG molecules and antibodies can be utilized for effective capture and release of EPCs from whole blood in microfluidic devices. Providing adequate opportunity for the 4-arm, amine-terminated PEG to mix with the anti-CD34 antibody prior to covalent attachment of both species to alginic acid via carbodiimide chemistry is essential to maximize yield and purity of EPC capture. The significance of this work lies in the ability to achieve such capture and release from a complex, heterogeneous cell suspension using a relatively simple synthesis protocol. This capture/release approach can potentially be extended to the isolation of other rare cells from complex solutions, such as stem or progenitor cells from digested tissue.
Supplementary Material
Acknowledgment
The authors gratefully acknowledge support from the National Institutes of Health and the National Science Foundation under grants R01-EB009327 and CBET 0932195, respectively, to S.K.M. The authors also thank Prof. Rebecca Carrier for access to the ultraviolet spectrometer, Prof. Elizabeth Klings for assistance with blood draws, and Prof. Jennifer West for insightful discussions on promoting hydrogel adhesion to PDMS.
Footnotes
Supporting Information Available: Light micrographs of particles in some functionalized alginic acid solutions and sensitivity analysis calculation to examine impact of hydrogel coating thickness on available surface area for cell capture. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Cheng XH, Gupta A, Chen CC, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2009;9(10):1357–1364. doi: 10.1039/b818813k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng XH, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2007;7(2):170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450(7173):1235-U10. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotz KT, Xiao W, Miller-Graziano C, Qian WJ, Russom A, Warner EA, Moldawer LL, De A, Bankey PE, Petritis BO, Camp DG, Rosenbach AE, Goverman J, Fagan SP, Brownstein BH, Irimia D, Xu WH, Wilhelmy J, Mindrinos MN, Smith RD, Davis RW, Tompkins RG, Toner M. Inflammation Host Response, I. Nat. Med. 2010;16(9):1042-U142. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyle PS, Bibette J, Bancaud A, Viovy JL. Science. 2002;295(5563):2237–2237. doi: 10.1126/science.1068420. [DOI] [PubMed] [Google Scholar]
- 6.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Lab Chip. 2010;10(1):27–29. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wankhede SP, Du Z, Berg JM, Vaughn MW, Dallas T, Cheng KH, Gollahon L. Biotechnol. Prog. 2006;22(5):1426–1433. doi: 10.1021/bp060127d. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Jones P, Haswell SJ. Chem. Eng. J. 2008;135 Supplement 1:S82–S88. [Google Scholar]
- 9.Fujioka N, Morimoto Y, Takeuchi K, Yoshioka M, Kikuchi M. Appl. Spectrosc. 2003;57(2):241–243. doi: 10.1366/000370203321535187. [DOI] [PubMed] [Google Scholar]
- 10.Jung K, Hampel G, Scholz M, Henke W. Cell. Physiol. and Biochem. 1995;5(5):353–360. [Google Scholar]
- 11.Murthy SK, Radisic M. Cell Adhesion and Detachment. In: Li D, editor. Encyclopedia of Microfluidics and Nanofluidics. Springer; 2008.. [Google Scholar]
- 12.Plouffe BD, Brown MA, Iyer RK, Radisic M, Murthy SK. Lab Chip. 2009;9(11):1507–1510. doi: 10.1039/b823523f. [DOI] [PubMed] [Google Scholar]
- 13.Kaushal S, Amiel GE, Guleserian KJ, Shapira OM, Perry T, Sutherland FW, Rabkin E, Moran AM, Schoen FJ, Atala A, Soker S, Bischoff J, Mayer JE. Nat. Med. 2001;7(9):1035–1040. doi: 10.1038/nm0901-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gatest GJ, Phillips JA. Biotechnol. Tech. 1992;6(6):517–522. [Google Scholar]
- 15.Sawhney AS, Pathak CP, Hubbell JA. Biomaterials. 1993;14(13):1008–1016. doi: 10.1016/0142-9612(93)90194-7. [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Zeng L, Ding S, Xu K. Transplant. Proc. 2010;42(9):3745–3749. doi: 10.1016/j.transproceed.2010.07.094. [DOI] [PubMed] [Google Scholar]
- 17.Eggermann J, Kliche S, Jarmy G, Hoffmann K, Mayr-Beyrle U, Debatin KM, Waltenberger J, Beltinger C. Cardiovasc. Res. 2003;58(2):478–486. doi: 10.1016/s0008-6363(03)00252-9. [DOI] [PubMed] [Google Scholar]
- 18.George J, Shmilovich H, Deutsch V, Miller H, Keren G, Roth A. Tissue Eng. 2006;12(2):331–335. doi: 10.1089/ten.2006.12.331. [DOI] [PubMed] [Google Scholar]
- 19.Hristov M, Erl W, Weber PC. Arterioscler. Thromb. Vasc. Biol. 2003;23(7):1185–1189. doi: 10.1161/01.ATV.0000073832.49290.B5. [DOI] [PubMed] [Google Scholar]
- 20.Asahara T, Murohara T, Sullivan A, Silver M, vanderZee R, Li T, Witzenbichler B, Schatteman G, Isner JM. Science. 1997;275(5302):964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 21.Urbich C, Dimmeler S. Circ. Res. 2004;95(4):343–353. doi: 10.1161/01.RES.0000137877.89448.78. [DOI] [PubMed] [Google Scholar]
- 22.Hristov M, Erl W, Weber PC. Trends Cardiovasc. Med. 2003;13(5):201–206. doi: 10.1016/s1050-1738(03)00077-x. [DOI] [PubMed] [Google Scholar]
- 23.Leor J, Marber M. J. Am. Coll Cardiol. 2006;48(8):1588–1590. doi: 10.1016/j.jacc.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 24.Sorkin M, Wong VW, Glotzbach JP, Major M, Levi B, Longaker MT, Gurtner GC. J. Am. Coll. Surg. 2010;211(3):S65–S65. [Google Scholar]
- 25.Peterbauer-Scherb A, van Griensven M, Meinl A, Gabriel C, Redl H, Wolbank S. J. Tissue Eng. Regen. Med. 2010;4(6):485–490. doi: 10.1002/term.262. [DOI] [PubMed] [Google Scholar]
- 26.Bosio A, Huppert V, Donath S, Hennemann P, Malchow M, Heinlein UAO. Engineering of Stem Cells. Vol. 114. Berlin: Springer-Verlag Berlin; 2009. pp. 23–72. [DOI] [PubMed] [Google Scholar]
- 27.Couperwh I, McCallum MF. Arch. Microbiol. 1974;97(1):73–80. doi: 10.1007/BF00403047. [DOI] [PubMed] [Google Scholar]
- 28.Kong HJ, Mooney DJ. In: Mechanical Properties of Bioinspired and Biological Materials. Viney C, Katti K, Ulm FJ, Hellmich C, editors. Vol. 844. 2005. pp. 313–318. [Google Scholar]
- 29.Rowley JA, Madlambayan G, Mooney DJ. Biomaterials. 1999;20(1):45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 30.Jeon O, Powell C, Ahmed SM, Alsberg E. Tissue Eng. Part A. 2010;16(9):2915–2925. doi: 10.1089/ten.TEA.2010.0096. [DOI] [PubMed] [Google Scholar]
- 31.Suarez-Gonzalez D, Barnhart K, Saito E, Vanderby R, Hollister SJ, Murphy WL. J. Biomed. Mater. Res. Part A. 2010;95A(1):222–234. doi: 10.1002/jbm.a.32833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu JS, Du KT, Fang QZ, Gu YP, Mihardja SS, Sievers RE, Wu JC, Lee RJ. Biomaterials. 2010;31(27):7012–7020. doi: 10.1016/j.biomaterials.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 33.Alsberg E, Anderson KW, Albeiruti A, Rowley JA, Mooney DJ. Proc. Nat. Acad. Sci. U.S.A. 2002;99(19):12025–12030. doi: 10.1073/pnas.192291499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheetham PSJ, Blunt KW, Bucke C. Biotechnol. Bioeng. 1979;21(12):2155–2168. [Google Scholar]
- 35.Drury JL, Boontheekul T, Mooney DJ. J Biomech Eng. Trans. ASME. 2005;127(5):891–891. doi: 10.1115/1.1865194. [DOI] [PubMed] [Google Scholar]
- 36.Hermanson . Bioconjugate Techniques. Academic Press; 1996. [Google Scholar]
- 37.Plouffe BD, Kniazeva T, Mayer JE, Murthy SK, Sales VL. FASEB J. 2009;23(10):3309–3314. doi: 10.1096/fj.09-130260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, Yuan L, Song W, Wu ZK, Li D. Prog. .Polym. Sci. 2008;33(11):1059–1087. [Google Scholar]
- 39.Heuberger M, Drobek T, Voros J. Langmuir. 2004;20(22):9445–9448. doi: 10.1021/la048384k. [DOI] [PubMed] [Google Scholar]
- 40.Unsworth LD, Tun Z, Sheardown H, Brash JL. J. .Colloid and Interface Sci. 2006;296(2):520–526. doi: 10.1016/j.jcis.2005.09.056. [DOI] [PubMed] [Google Scholar]
- 41.Norde W, Gage D. Langmuir. 2004;20(10):4162–4167. doi: 10.1021/la030417t. [DOI] [PubMed] [Google Scholar]
- 42.Frutos AG, Brockman JM, Corn RM. Langmuir. 2000;16(5):2192–2197. [Google Scholar]
- 43.Rafii S, Lyden D. Nat. Med. 2003;9(6):702–712. doi: 10.1038/nm0603-702. [DOI] [PubMed] [Google Scholar]
- 44.Wu X, Rabkin-Aikawa E, Guleserian KJ, Perry TE, Masuda Y, Sutherland FWH, Schoen FJ, Mayer JE, Bischoff J. Am. J. Physiol. Heart Circ. Physiol. 2004;287(2):H480–H487. doi: 10.1152/ajpheart.01232.2003. [DOI] [PubMed] [Google Scholar]
- 45.Li TS, Hamano K, Nishida M, Hayashi M, Ito H, Mikamo A, Matsuzaki M. Am. J. Physiol. Heart Circ. Physiol. 2003;285(3):H931–H937. doi: 10.1152/ajpheart.01146.2002. [DOI] [PubMed] [Google Scholar]
- 46.Yang C, Zhang ZH, Li ZJ, Yang RC, Qian GQ, Han ZC. Throm. Haemostasis. 2004;91(6):1202–1212. doi: 10.1160/TH03-06-0378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.