Abstract
We have characterized mutations in the early arrest gene, harpy (hrp), and show that they introduce premature stops in the coding region of early mitotic inhibitor1 (Rca1/emi1). In harpy mutants, cells stop dividing during early gastrulation. Lineage analysis confirms that there is little change in cell number after approximately cycle-14. Gross patterning occurs relatively normally, and many organ primordia are produced on time but with smaller numbers of cells. Despite the lack of cell division, some organ systems continue to increase in cell number, suggesting recruitment from surrounding areas. Analysis of BrdU incorporation shows that endoreduplication continues in many cells well past the first day of development, but cells cease endoreduplication once they begin to differentiate and express cell-type markers. Despite relatively normal gross patterning, harpy mutants show a number of defects in morphogenesis, cell migration and differentiation resulting directly or indirectly from the arrest of cell division.
Keywords: zebrafish, endoreduplication, primary neurons, muscle pioneers, neural crest, otic placode, pronephros, DNA replication
INTRODUCTION
Development often progresses through an early phase of proliferation of multipotent progenitors followed by specification and differentiation of specific cell types. During this time, the rhythm of the cell cycle undergoes a dramatic slowing, shifting from rapid cleavage divisions to the long regulated cell cycle typical of differentiated tissues. In most cases, the shift from proliferation to differentiation is gradual and involves prolongation of one or both “gap” phases as cells react to various environmental signals (Kane et al., 1992; Kane and Kimmel, 1993; Zamir et al., 1997; Reviewed in Baker, 2007; Nogare et al., 2007; Ulloa and Briscoe, 2007; Orford and Scadden, 2008). Proliferation is necessary to generate a sufficient number of cells to support larvae and then adult functions, but it also plays a number of direct and indirect roles in tissue patterning. For example, proliferation can promote “community effects” whereby the size of a population modifies responses of individual cells to inductive cues (Gurdon et al., 1993; Schilling et al., 2001). In addition, many organ systems develop from morphogenetic fields, groups of cells coordinated by diffusion gradients of growth factors that, depending on concentration, specify diverse cell fates while spatially regulating the pattern of growth (Mariani et al., 2008). On a more local scale, asymmetric cell division is a common mechanism for generating cell type diversity whereby daughter cells adopt different fates due to partitioning of cytoplasmic determinants during mitosis (Kuang et al., 2007; Knoblich, 2008; Orford and Scadden, 2008). Thus, proliferation and patterning are finely balanced and coordinated during much of early development.
The requirement for early proliferation has been previously studied in Xenopus embryos by using hydroxyurea and aphidicolin to block entry into S-phase at different stages of development (Harris and Hartenstein, 1991; Rollins and Andrews, 1991). Treatment during blastula stage severely perturbs embryonic patterning because there are too few cells to execute normal morphogenetic movements of gastrulation. In contrast, treatment during gastrulation results in development of a relatively normal body plan, with each region comprising fewer cells. Within the neural tube, an intricate array of primary neural cell types still forms, though later aspects of neural development are necessarily compromised without further cell division. Such findings underscore the surprising degree of precision afforded by cell-cell interactions.
Recently, Zhang et al. (2008) described a mutation in zebrafish early mitotic inhibitor 1 (emi1) that blocks mitosis just after the onset of gastrulation. The homologous gene in Drosophila has been identified by the mutation Regulator of Cyclin A (Rca1; Dong et al., 1997). Rca1/emi1 is an inhibitor of Anaphase Promoting Complex/Cyclosome (APC/C), an E3 ubiquitin ligase that targets mitotic regulators for destruction (Reimann et al., 2001a; Reimann et al., 2001b; Miller et al., 2006; Di Fiore and Pines, 2007). In the absence of Rca1/emi1, APC/C remains constitutively active and blocks entry into mitosis. In addition, Rca1/emi1 mutants continue to undergo endoreduplication, repeated cycles of DNA synthesis without mitosis, through at least early segmentation. This likely reflects APC/C-mediated destruction of Geminin, an inhibitor that normally blocks re-replication (Di Fiore and Pines, 2007; Machida and Dutta, 2007). Consistent with data from mitotically-blocked Xenopus embryos, gross patterning is relatively normal in Rca1/emi1 mutants. Somites initially form normally and, indeed, the somitogenic clock operates with greater than normal precision, confirming the hypothesis that mitosis can interfere with the ability of individual cells to synchronize with their local cohort (Horikawa et al., 2006). However, patterning within somites is perturbed, as cells that should form the central mesenchymal domain instead join the outer epithelial layer (Zhang et al., 2008). Likely there are many other patterning defects arising directly or indirectly from the absence of cell division, as might be expected from the known effects of growth factors that mediate the salient cell-cell interactions.
Here we describe two new mutations in Rca1/emi1 and show that they are allelic to the “early-arrest” mutant harpy (hrp; Kane et al., 1996). Using cell counting and lineage analysis, we show that the cell cycle arrest occurs at approximately cycle 14. Using in situ hybridization to analyze markers of specification and differentiation, we show that although global pattern formation is normal, there are many diminutive developmental defects arising from cessation of cell division. Using transplantation between mutant and wild-type embryos, we find that the cycling defects are autonomous although some of the observed phenotypic perturbations are not. And from an analysis of BrdU uptake in the DNA of the mutant embryo, we find that endoreduplication continues well past 24 hours in the cycle arrested cells, with a major exception being cells that normally stop cycling early, e.g., cells that differentiate as specific neural or mesodermal cell types.
RESULTS
Identification of new alleles of harpy
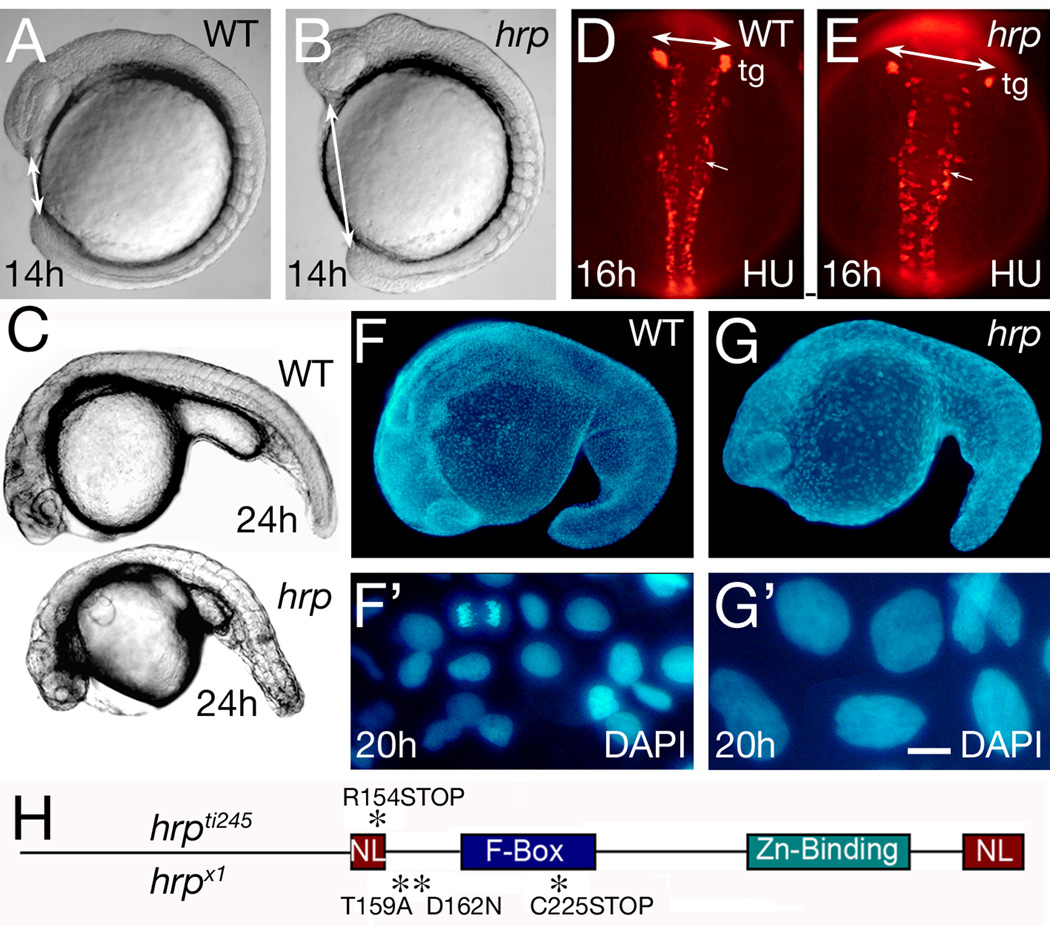
The mutant harpy was initially identified as an early arrest phenotype in the Tübingen zebrafish screen for early morphogenetic mutants, represented by a single recessive allele ti245 (Kane et al., 1996). The earliest visible phenotype seen using a dissecting scope was an abnormally bumpy head (Fig. 1 A,B) accompanied by a shortened body axis, easily seen by comparing the distance between the nose and tailbud around the ventral aspect of the embryo. As development proceeds, mutant embryos were slightly smaller than normal at 24 hours and show little or no growth thereafter (Fig. 1C). In contrast to the other early arrest mutants, cell death and necrosis was not apparent before 24 hours (Kane et al., 1996), although afterwards low numbers of cells appeared to be lysing throughout the embryo, especially in the CNS. (Evidence of low level cellular lysis based on pyknotic nuclei can be seen in Fig. 2H and throughout Fig. 7.)
Fig. 1. General phenotype of harpy mutants.
(A, B) Live wild-type and mutant embryos at the 10 somite stage. Double sided arrows indicate the distinctive nose-tail bud separation in the mutants.
(C) Side views of live wild-type and mutant embryos at 24 hours.
(D, E) Anti-HuC (HU) stained neurons at 16 hours in wild-type and mutant embryos. The trigeminal ganglia (tg) are indicated, and double sided arrows indicate the width of the neural keel. Small arrows indicate single cells; note the larger size of the mutant cells.
(F, G) DAPI-staining at 20 hours in wild-type and mutant embryos. F’ and G’ are whole embryo dissociations. Note the cell in mitosis in the WT field; no cells are found in mitosis in the mutants.
(H) Rca1/emi1 coding region showing individual protein domains and the locations of point mutations (asterisks) found in hrpti245 and hrpx1 mutants.
Scale bar = 50 µm (D, E), 5 µm (F’, G’).
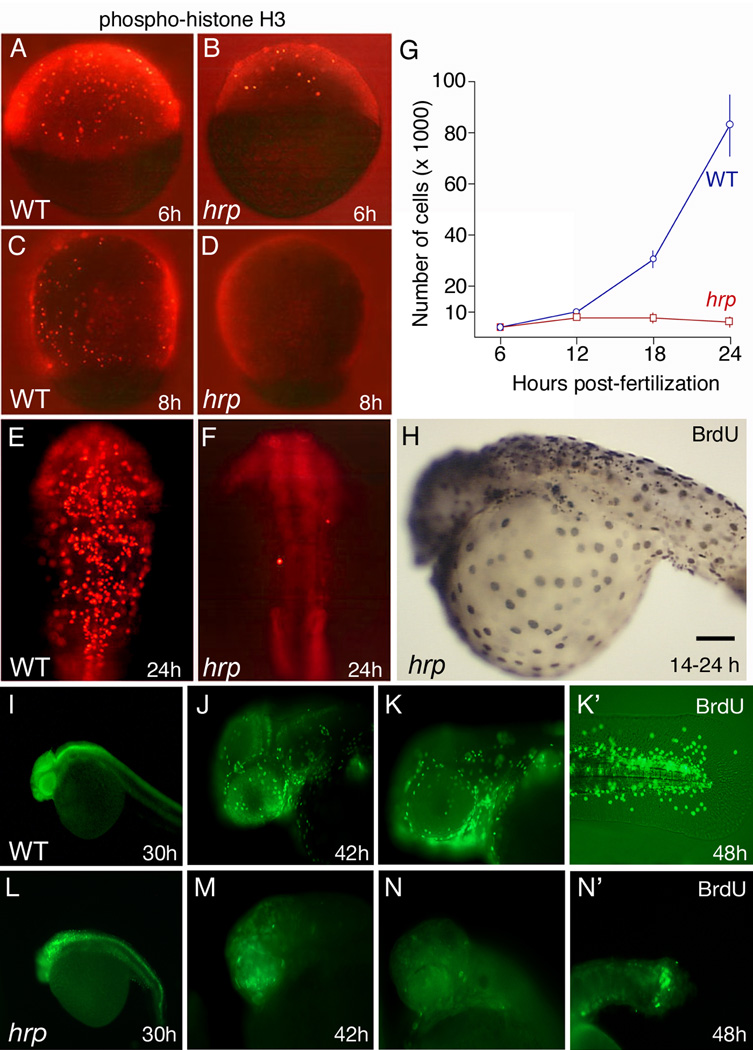
Fig. 2. Cycle characterization of cells in harpy mutants.
(A–F) Anti-phospho-histone H3 (pH3) staining in wild-type and harpy mutant embryos from 6 through 24 hours showed that typically by 8 hours no cells are in mitosis. Only occasional cells were pH3 positive at 24 hours (F).
(G) Mean number of cells per embryo, based on cell counts. Cells were counted after dissociating individual embryos with collagenase. Each time point shows the mean and standard deviation from 10 embryos.
(H–N) BrdU incorporation in wild-type and harpy mutant embryos. H shows an harpy mutant injected at 14 hours and fixed 10 hours later. I–N shows embryos injected and fixed 45 min later. Note that at 30 and 42 hours many cells were positive in mutants; however, at 48 hours, mutants were mostly negative except for a small focus of BrdU incorporation in the tail.
Scale bar = 100 µm (A–F), 85 µm (H, J, K, M, N), 200µm (I,L)
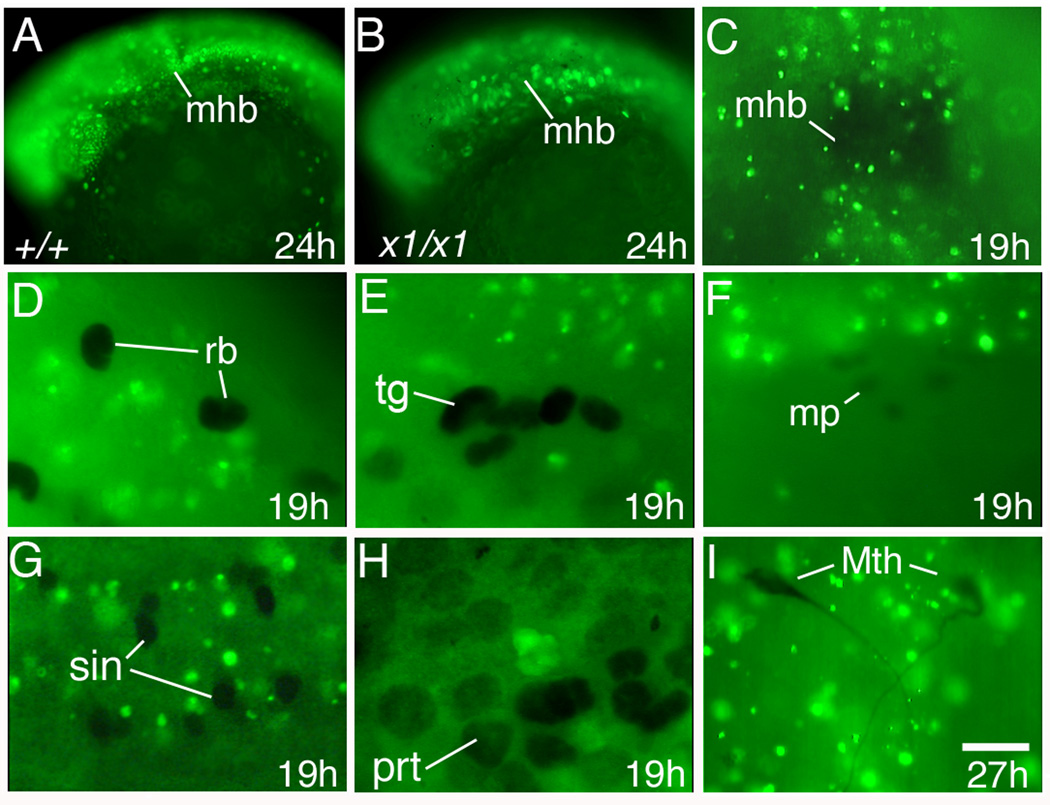
Fig. 7. Patterns of BrdU incorporation.
Images show lateral views with anterior to the left (A, B, E, F, H) or dorsal views with anterior to the top (C, I) or to the left (D, G).
(A, B) Head region of a wild-type embryo and harpy mutant incubated with BrdU from 18 to 24 hours. Note that the majority of cells incorporate BrdU in both mutant and wild-type embryos.
- (C) Anti-Engrailed stained midbrain-hindbrain border.
- (D, E) anti-Isl1/2 staining showing Rohon-Beard sensory neurons and trigeminal ganglia.
- (F) Anti-Engrailed stained muscle pioneers.
- (G) Anti-Pax2 stained spinal interneurons.
- (H) Anti-Pax2 stained pronephric cells.
- (I) 3A10 antibody-stained Mauthner neurons; these were fixed at 27 hours.
Abbr:
mhb, midbrain-hindbrain border;
mp, muscle pioneers;
Mth, Mauthner neurons;
prt, pronephros.
sin, spinal interneurons;
rb, Rohon-Beard sensory neurons;
tg, trigeminal ganglia;
Scale bar = 100 µm (A, B), 75 µm (C), 30 µm (D–G, I), 15 µm (H).
In a screen for neural patterning mutants, one of us (B.B.R.) recovered a recessive lethal mutation, termed x1, which causes a phenotype in which neurons were reduced in number but increased in size (Fig. 1 D,E). Further investigation of DAPI-stained embryos showed similar changes in cell number and size throughout the embryo (Fig. 1 F,G), and DAPI-staining of dissociated cells revealed that cell nuclei were greatly enlarged in cells from mutant embryos (Fig. 1 F’,G’). The early morphological defects of x1 mutants resembled those of hrpti245 and complementation testing confirmed the mutations were allelic. The new allele is henceforth designated as hrpx1.
harpy is a mutation in Rca1/emi1
The altered number and size of cells in harpy mutants suggested a defect in the cell division cycle. The nuclei shown in Fig. 1 G’ are more than twice the diameter of wild-type cells, suggesting that they might be at 8N or greater ploidy. In support, staining hrpx1 mutants for phospho-histone H3 revealed a gradual cessation of mitosis just after the onset of gastrulation. Few if any mitotic cells were detected at any stage after 8 hours (Fig. 2 A–F). Identical results were seen for the hrpt1245 allele (not shown).
The mutation tiy121 of Zhang et al. (2008) that disrupts the mitotic regulator Rca1/emi1 produces a phenotype that closely resembles that of hrp. We confirmed that harpy maps within 1 cM of the map position of Rca1/emi1 (data not shown). On sequencing the alleles (Fig. 1H), both contained nonsense mutations that truncate the Rca1/emi1 protein midway through the transcript: hrpti245 has a stop codon within a putative nuclear localization signal and hrpx1 has a stop codon within the F-box domain, as well as two other changes in the coding region that introduce amino acid substitutions in the N-terminal half of the protein. (The tiy121 allele contains a premature stop codon that truncates the protein prior to the F-box domain. Hereafter, we refer to the tiy121 allele as hrptiy121.) Truncation of the C-terminus, which includes the Zn-binding region and other domains that are indispensable for Rca1/emi1 activity (Reimann et al., 2001a; Miller et al., 2006), argues that all three harpy alleles are likely to be amorphic for Rca1/emi1 function. There is a fourth allele of hrphi2648 that has been identified in the insertional screen in Nancy Hopkin’s Laboratory at MIT (Amsterdam et al., 2004; Wiellette et al., 2004); this allele is caused by an insertion in the first intron and behaves as a hypomorph (Zhang et al., 2008; Rhodes et al., 2009).
The cell cycle in harpy mutants is blocked during gastrulation at approximately division 14
We performed several experiments to better characterize the timing and division characteristics of the cell cycle defect in harpy mutants. To confirm whether cell division is blocked, embryos were dissociated with collagenase at various stages of development and the number of cells was counted directly using a hemocytometer (Fig. 2G). Wild-type embryos showed a steady increase from 4,600 ± 1,100 cells at 6 hours to 82,600 ± 11,800 cells at 24 hours. In contrast, hrpx1 mutants showed little change in cell number during this period, still comprising only 6,400 ± 2,000 cells at 24 hours. These data are consistent with the phospho-histone H3 staining and confirm that cell division ceases soon after 6 hours in harpy mutants.
It was previously reported that hrptiy121 mutants continue to synthesize DNA in endoreduplication cycles through at least 14 hours (Zhang et al., 2008). We examined BrdU incorporation in hrpx1 mutants at later stages to determine the persistence of these cycles. Despite the lack of cell division, many cells continued to synthesize DNA throughout the segmentation period, shown by BrdU incorporation over a 10 hour period (Fig. 2H). To explore the duration of endoreduplication, we used shorter pulses (45 min) at times up until 48 hours. We found that the intensity of label in mutants was slightly lower than wild-type embryos at 30 h (Fig. 2 I,L). Afterwards, at 42 hours, the apparent number of cells labeled decreased (Fig. 2 J,M), and at 48 hours, the only region of substantial labeling was in the tail (Fig. 2 K,N). Thus, endoreduplication continued well into day two of development, but eventually tended to stop in all but a few cells of the embryo. It is interesting to note that for all embryos, both wild-type and mutant, individual cells are either labeled intensely or not at all, suggesting that the cell cycle dynamics in the mutant might be similar to that in wild-type embryos, i.e., cells in the harpy mutants may contain a distinctive S-phase. These results are consistent with the results by Rhodes et al. (2009) using flow cytometry to document the appearance of 4 N and greater nuclei during the segmentation stages.
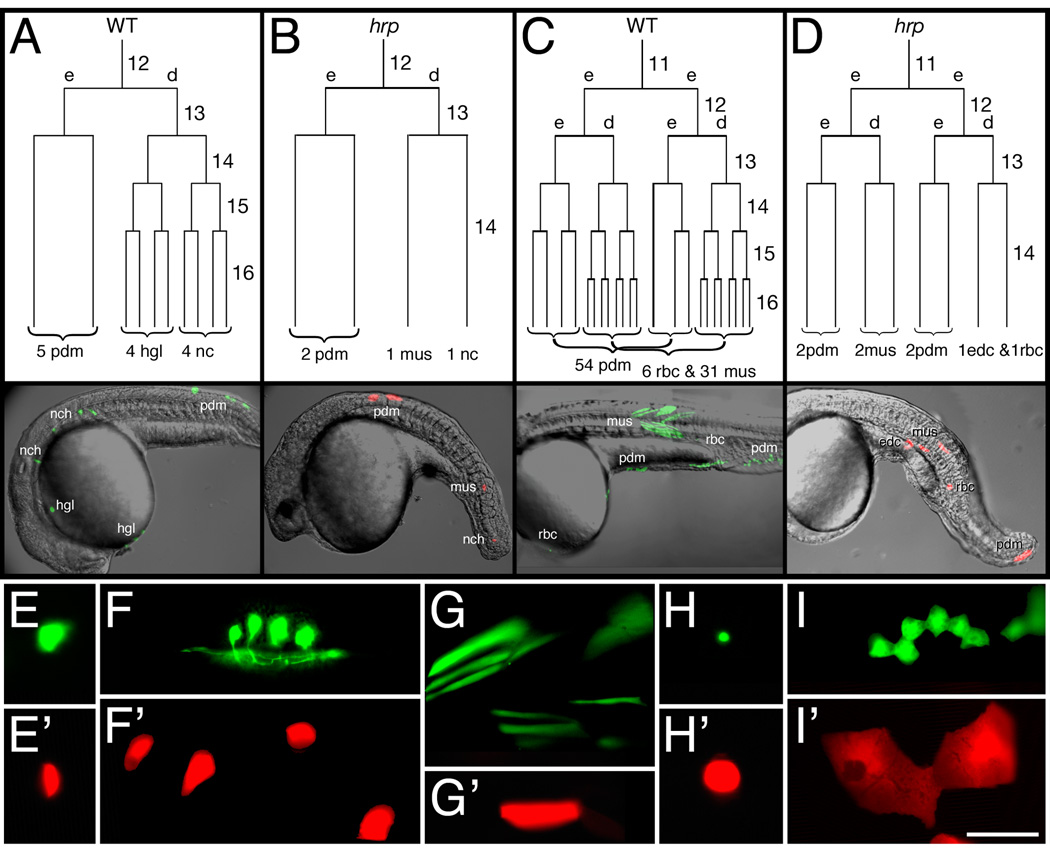
To precisely assess the cycle that is blocked in harpy mutants, we labeled individual cells in mutant and wild-type embryos in divisions immediately after the Midblastula Transition, noting the cell cycle when the cell was labeled. We later counted the number of labeled cells at the beginning of gastrulation (6 hours) and the next day (at approximately 30 hours). In these experiments, we labeled a single exterior blastomere with lineage tracer and followed the progeny over the next day of development. Cells at the surface of the blastula typically alternate between anticlinal and periclinal divisions; hence, after two divisions there are typically 2 enveloping layer cells and 2 deep cells.
Because of the fate differences in enveloping layer cells and deep cells, we counted each cell type separately, and from those counts, using the starting clone cell cycle and the log to the base 2, we derived the average cycle number that was attained by each enveloping layer or deep cell clone. These experiments are summarized in Fig. 3 and Table 1. In normal embryos, deep cells advance to cycle 14 by the beginning of gastrulation and division 14 (into cycle 15) tends to occur during early gastrulation (Kane et al., 1992; Kane and Kimmel, 1993; Kimmel et al., 1994). On the other hand, enveloping layer cells typically arrest or slow their divisions at the beginning of epiboly: this temporary cycle arrest occurs in cycle 13 or, somewhat less frequently, in cycle 14. Within experimental error, these results were recapitulated in our studies. In both mutant and wild-type embryos, deep cells reached mid cycle 14 and enveloping layer cells only just reached cycle 14 at the beginning of gastrulation (Table1, 6 hour time point).
Fig. 3. harpy mutant cells stop dividing at cell cycle 14.
(A, B) Dorsally-derived fates, and (C, D) ventrally-derived fates. In each embryo, an individual blastomere was labeled at cycle 11 or 12 and followed by timelapse video microscopy through early segmentation. Each is restricted to a deep cell lineage (d) or an EVL (e) lineage at the cycle after labeling. The reconstructed cell lineage is displayed by cell cycle. 24 hour histotypes and cell counts for the clones are shown at the bottom of the lineage tree, and left side views of the 24 hour embryos are shown below.
(E–I) Comparison of different cell types at 24 hours. Whereas notochord cells (E) were normally sized in the mutants, neurons (F) and muscle cells (G) were larger, and blood cells (H) and periderm cells (I) were supersized. In our lineage studies we never saw axons sprouted from presumptive neurons in mutants.
Wild-type cells, green; harpy mutant cells, red.
Abbr:
edc, endothelial cell;
hgl, hatching gland;
mus, muscle;
nc, notochord;
pdm, periderm;
rbc, red blood cell.
Scale bar = 100 µm (E–I).
Table 1.
Average number of cycles completed in EVL and deep clones.
| 6 hours | 30 hours | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EVL | deep | EVL | deep | |||||||
| cycle inj. |
n | no. of cells |
calc. cycles |
No. of cells |
calc. cycles |
No. of cells |
calc. cycles |
No. of cells |
calc. cycles |
|
| harpy | 10 | 4 | 6.3 | 13.6 | 9.3 | 14.0 | 6.3 | 13.6 | 9.3 | 14.0 |
| 11 | 4 | 4.0 | 13.9 | 7.5 | 14.9 | 4.0 | 13.9 | 11.0 | 15.3 | |
| 12 | 5 | 2.2 | 14.1 | 4.0 | 15.0 | 2.2 | 14.1 | 9.8 | 16.2 | |
| all | 13.9 | 14.6 | 13.9 | 15.1 | ||||||
| WT | 10 | 4 | 8.5 | 13.9 | 8.8 | 14.0 | 55.5 | 16.8 | 62.0 | 16.9 |
| 11 | 7 | 4.0 | 14.2 | 8.9 | 15.0 | 16.4 | 16.1 | 71.3 | 17.4 | |
| 12 | 7 | 2.7 | 14.1 | 2.1 | 14.5 | 16.4 | 16.7 | 16.3 | 17.6 | |
| all | 14.1 | 14.6 | 16.4 | 17.3 | ||||||
Cell counts the next day revealed nearly a 10× difference between the numbers of cells in clones in mutant and those in wild-type embryos. Clones in wild-type embryos underwent 2 to 3 rounds of division between the beginning of gastrulation and 30 hours, reaching mid cycle 16 for EVL clones and mid cycle 17 for deep clones. Our results at 30 hours are consistent with a normal value of 50 to 100 thousand cells in the one day embryo. In the same time period, EVL clones in mutant embryos did not divide at all and deep cell clones either did not divide or underwent 1 additional round of division. These results predict that there are about 10 thousand cells in the mutant embryo at the time of arrest.
For many of the above experiments, we also timelapse recorded a number of embryos (N=12) from the blastula stage until after the end of gastrulation tailbud stage and reconstructed partial lineages (Fig. 3). Two pairs of wild-type and mutant lineages are shown, matched as ventral and dorsal examples. The wild-type examples were typical of this type of analysis: in the deep lineages there are many early divisions with major tissue restrictions occurring in the gastrula stage; in the enveloping layer lineages, the division rates tended to slow or arrest during epiboly. After gastrulation—and after recording—both lineages went through a number of divisions; we show the 24–30 hour fates and their numbers present on the next day. In the mutants, cell divisions occurred normally before gastrulation and then arrested just after the onset of gastrulation; when we checked the embryos the next day, no more divisions were apparent.
Global patterning was normal in harpy mutants but local patterning was not
In terms of the cell types produced by the above lineages, the lineage relationships in harpy mutant embryos are simple versions of development in wild-type embryos. Notochord lineages are normally related to muscle lineages and blood lineages are normally related to endothelial lineages, and, respectively, such examples are shown in Fig. 3 A,B and Fig. 3 C,D.
At 30 hours, the labeled cells in the clones gave an agnostic glimpse into the morphology of individual cells in the mutant embryos. Cells that normally have their terminal mitosis in the gastrula tended to appear normally sized in the mutants. For example, notochord cells (Fig. 3E) seemed to be completely normal (although the sheath cells surrounding them were larger than normal, data not shown). Neurons (Fig. 3F) and muscles (Fig. 3G) were slightly larger than normal, however, neurons tended not to sprout axons, and muscle cells were often dumpy and did not always span an entire somite. On the other hand, cells that normally divide many times in the embryo, such as blood (Fig. 3H) and periderm (Fig. 3I), became supersized versions of their wild-type counterparts. In the case of the blood cells, these cells were actually bigger than the blastomeres that produced them, suggesting that some process—we suggest endoreduplication—might be occurring in these particular cells that is causing abnormal cell sizing.
Many aspects of early development are relatively normal in harpy mutants: defects do not appear during cleavage, early epiboly, nor the beginning of gastrulation. harpy mutants can first be identified morphologically at 14 hours when they exhibit shortened AP axes (Fig. 1 A,B). This appears to reflect slowing of convergent extension (Fig. D,E) as cells begin to increase in size. Neurogenin staining revealed that many neural progenitors are already enlarged at 10 hours and the neural plate is roughly 30% wider than normal (data not shown); Anti-HuC staining showed that it still appeared wider than normal at 16 hours (Fig. 1 D,E). The neural tube approaches normal width by 24 hours, but in the head region the neural tube becomes thinner than the wild-type especially in the anterior brain regions.
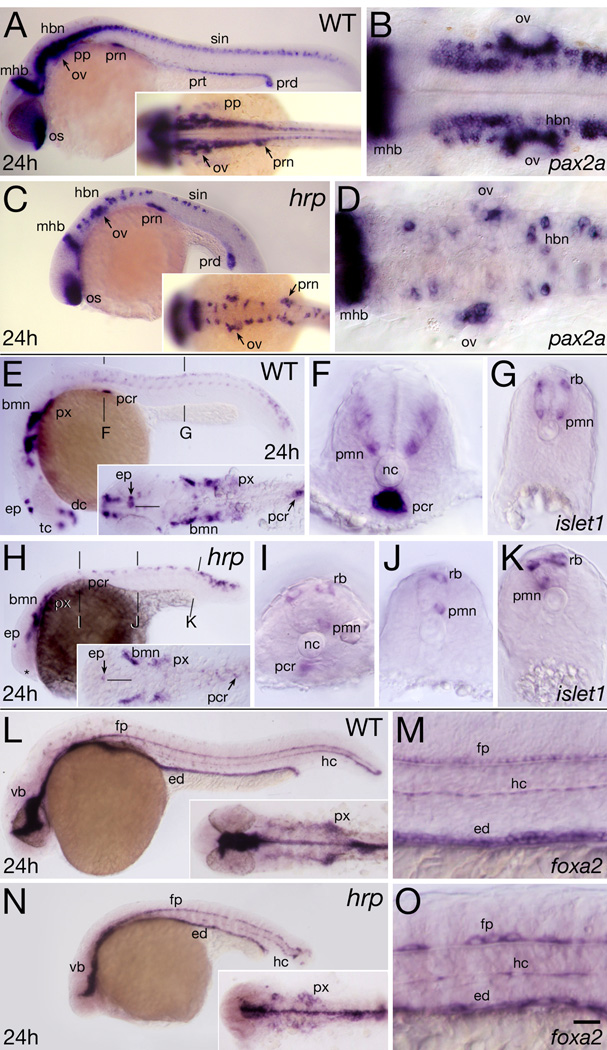
To provide a global view of specification in the mutant, we examined the mRNA expression patterns of pax2a, islet1 and foxa2, genes which are expressed in many different tissues of the embryo (Fig. 4).
Fig. 4. Neural patterning defects in harpy mutants. Panels show wild-type reference embryos and harpy mutants.
- (A, C) Whole mount side views; insets show dorsal views of head-trunk region.
- (B, D) High resolution dorsal views of hindbrain.
- (E, H) Whole mount side views; insets show dorsal views of head region. The midline of the CNS (black line) is indicated near the epiphysis.
- (F,G, I–K) Transverse sections at levels indicated in whole mounts. Note lacking primary motor neurons, normally a bilaterally paired structure.
- (L, N) Show a whole mount side view; insets show dorsal views of head region.
- (M, O) Show high resolution side views of trunk. Note the gaps between the large cells of the mutant floor plate and hypochord.
Abbr:
bmn, branchio-motor nuclei;
dc, diencephalon;
ep, epiphysis;
ed, endoderm;
fp, floor plate;
hbn, hindbrain neurons;
hc, hypochord;
mhb, midbrain-hindbrain border;
nc, notochord;
os, optic stalk;
ov, otic vesicle;
pcr, pancreas;
pp, pharyngeal pouch;
pmn, primary motor neurons;
prn pronephric neck;
prt pronephric tubule;
prd, pronephric duct;
px, pharynx;
rb, Rohon-Beard sensory neuron;
scn, spinal cord neurons.
tc, telencephalon;
vb, ventral brain;
Scale bar = 100 µm (A,C,E,H,L,N), 40 µm (B,D,F,G,I,J,K,M,O).
pax2a mRNA is expressed in cells in the midbrain-hindbrain border, reticulospinal and spinal interneurons, otic placode, and pronephros (Krauss et al., 1991a). In general, all of these cell types were produced at the correct times and locations in harpy mutants (Fig. 4 A–D). As expected, based on the division arrest phenotype, there were fewer cells present and they were larger than normal; however, there was a distinct impression that there was less tissue expressing pax2a. This was best seen in the interneurons of the spinal cord, where many regions in the mutant were devoid of pax2a expressing cells, and also in the hind brain, which normally has almost a continuous layer of expression in the reticulospinal layers (Fig. 4 B,D). There were numerous other defects, notably, the otic vesicle was smaller (discussed below) and expression in the proximal portion of the pronephric tubule was absent.
islet1 is expressed in cells of the neurectoderm, in the epiphysis, in nuclei of the telencephalon and the diencephalon, and in branchiomotor neurons, primary motor neurons and Rohan-Beard sensory cells (Korzh et al., 1993; Inoue et al., 1994). In harpy mutants (Fig. 4 E–K) the general pattern of expression was normal, and, with the caveat mentioned above due to the lack of cell division, tissue expression was not as diminished as in the case of pax2a expression. A nice example of normal expression was that of the branchio-motor nuclei overlying the pharynx, which appeared almost normal in the mutant (Fig. 4 H inset). The major exception to this general observation was the complete absence of islet1 positive nuclei in the telencephalon and diencephalon of the forebrain, and the almost complete loss of the epiphysis (Fig. 4 E,H). Patterning at the cellular level was also abnormal in many regions. Typically, Rohon-Beard cells and primary motor neurons are reiterated paired structures of the spinal cord (Fig. 4 F,G); in the mutant there were many missing cells (Fig. 4 I–K) often represented by one very large cell on only one side of the spinal cord in tissue sections.
islet1 is also expressed in the endodermally derived portions of the pharynx and the pancreas. These tissues are dramatically reduced. The endodermal portion of the pharynx is the light expression underneath and caudalwards of the branchio-motor nuclei, seen in the inset in Fig. 4 E; this was reduced in the mutant. islet1 is expressed in the endocrine portion of the pancreas; this expression was also reduced in the mutant (Fig. 4 F,I). Similar to expression in the pancreas, both of these tissues are among the earliest tissues to proiferate in the developing embryo; in the absence of growth, only founder cells would be present and one would expect the region of expression to appear smaller but not necessarily weaker, and that is what we observed.
foxa2 (also called axial) is expressed in the ventral CNS of the forebrain, the floor plate neurons of the spinal cord, in the mesodermally-derived hypochord, and in the endoderm (Strähle et al., 1993; Odenthal and Nüsslein-Volhard, 1998). Normally the floor plate and hypochord are arranged as rows of single cells, as seen in Fig. 4 M. In the mutant, the two tissues were patterned normally but cellular organization was constrained by the reduction in cell number: both tissues formed a single row of oversized cells which exhibited large gaps (Fig. 4 O). On the other hand, the ventral forebrain (Fig 4 L,N) and the mutant pharynx (Fig 4 N inset) showed defects in line with the lack of growth in both of these tissues.
Neural specification
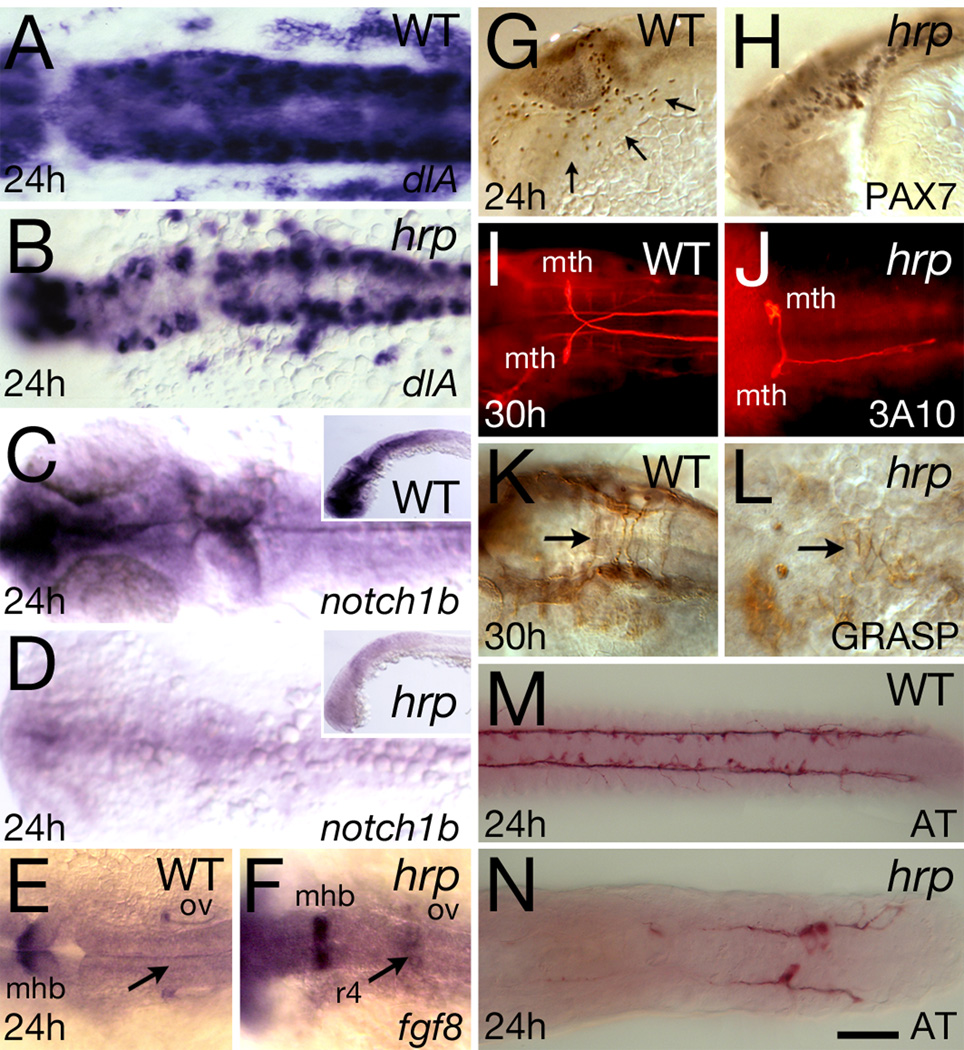
We focused on perturbations in the nervous system using the mRNA expression patterns of deltaA, notch1B, and fgf8, and protein expression patterns of Pax7, 3A10, Grasp, and acetylated tubulin (Fig. 5).
Fig. 5. Neural patterning defects in harpy mutants. Panels show wild-type control embryos and harpy mutants.
(A, B) Expression of deltaA (dlA) at 24 hours. Dorsal view of hindbrain. Note the normal intensity of expression in mutant cells.
(C, D) Expression of notch1b at 24 hours. Dorsal view of head; insets show lateral view. Note expression in mutants is almost absent.
(E, F) Expression of fgf8 at 24 hours. Dorsal view of hindbrain. Note ectopic staining in the 4th rhombomere of the mutant.
(G, H) anti-Pax7 staining at 24 hours. Arrows indicate neural crest cells migrating over the eye and pharyngeal arches; while these cells are present in the mutant, they are not seen on the migration pathway.
(I, J) dorsal view of 3A10-stained Mauthner neurons at 30 hours, displaying abnormal pathfinding and absence of axons in the mutants
(K, L) anti-Grasp staining at 30 hours; dorsal view of the hindbrain. Arrows indicate commissural neurons that in the mutant are chaotic.
(M, N) anti-acetylated tubulin (AT) staining at 24 hours; dorsal view of the trunk. Although axons are present in mutants, they do not express acetylated tubulin except in the tail region.
Abbr:
mhb, midbrain hindbrain border;
mth, Mauthner neurons.
ov, otic vesicle;
r4, 4th rhombomere;
Scale bar = 50 µm (A–D), 20 µm (F, G), 100 µm (E–N).
Notch and Delta control neuronal cell fate and proliferation in the vertebrate nervous system much like in Drosophila. Notch expression is generally associated with proliferative zones of undifferentiated cells; these cells are thought to consist primarily of neural stem cells (Bierkamp and Campos-Ortega, 1993). Delta expression is found in scattered cells within proliferative zones; these cells have become post mitotic and are differentiating into neurons (Appel and Eisen, 1998; Haddon et al., 1998).
We found that the expression pattern of deltaA was mostly normal in the CNS of harpy mutants (Fig. 5 A,B), although the number of positive cells was reduced. The differences that were observed seemed caused by the large cell size and the reduced number of cells in the mutant CNS. However, notch1b expression was severely reduced throughout the CNS of the mutants (Fig. 5 C,D), suggesting that neural stem cell populations diminished in the mutant.
fgf8 is expressed normally in the neural plate during gastrulation (not shown) and continues to be expressed in the midbrain-hindbrain border through 24 hours (Reifers et al., 1998), as seen in Fig. 5 E. However, harpy mutants also showed an ectopic stripe of fgf8 expression in rhombomere 4 of the hindbrain (Fig. 5 F). This represents an expression domain that is normally lost in wild-type embryos by 14 hours (Kwak et al., 2002), but which was aberrantly retained in harpy mutants.
anti-Pax7 marks a population of neural crest cells that migrate extensively and are often seen migrating over the surface of the eye and into lateral regions where cranial placodes and pharyngeal arches form. These cells were produced in harpy mutants, as shown by the presence of migratory Pax7-expressing cells in the head and anterior trunk (Fig. 5 G,H), however, they were never observed beyond the lateral edges of the brain, suggesting that migration is impaired.
Pathfinding
Axon pathfinding relies on stereotyped molecular cues provided by other cells. We asked if axonal pathfinding was perturbed in harpy mutants, since cues are likely to be missing due to early cessation of cell division.
3A10 antibody marks Mauthner cells (Metcalfe et al., 1990); these large neurons normally extend axons across the midline to join the contralateral medial longitudinal fascicle. Mauthner cells always formed in harpy mutants. However, in about a third of the mutants, Mauthner cells failed to extend axons or showed highly aberrant axon-trajectories (Fig. 5 I,J), in line with the almost complete lack of axons in lesser neurons.
Grasp-positive commissural neurons in the hindbrain normally decussate near rhombomere boundaries to form a regular array of commissures (Trevarrow et al., 1990), but in harpy mutants commissural axons formed highly chaotic patterns (Fig. 5 K,L).
Acetylated tubulin antibody labels mature axons, and is strongly expressed in axons of the 24 hour embryo (Fig. 5 M). In harpy mutants, although neurons with axons are present, they only expressed acetylated tubulin in the tail (Fig 5 N) or not at all.
Placodal specification
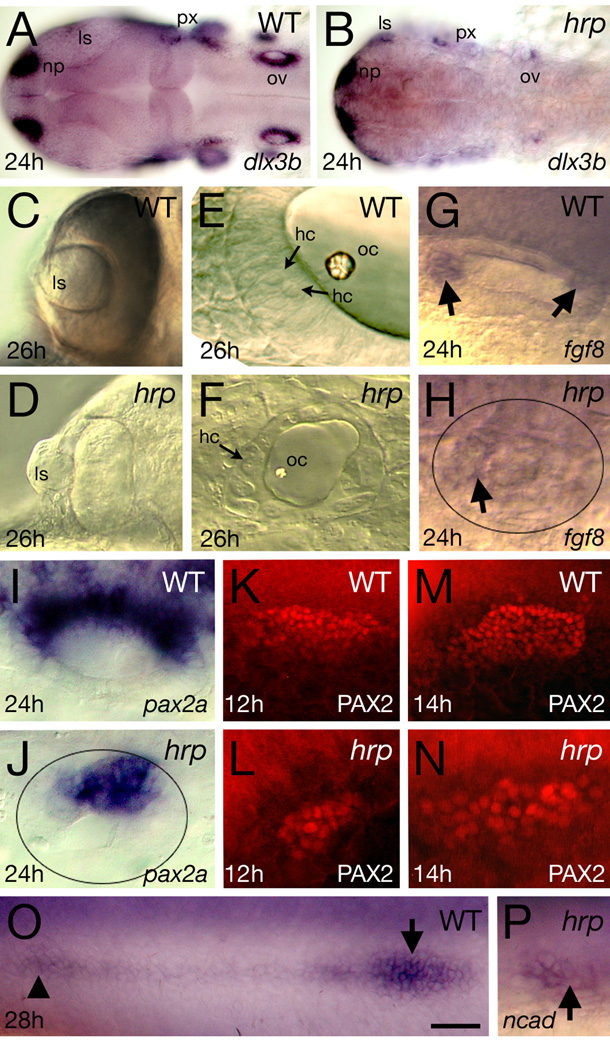
In Fig. 6 we focused on placodal structures and other structures that interact with the epidermis, using the mRNA expression patterns of dlx3b, pax2a, fgf8 and ncad. Most of these structures are induced and their morphogenesis requires many cellular interactions, which could be comprised by the abnormally large size of the mutant cells.
Fig. 6. Placodal patterning defects in harpy mutants. Panels show wild-type reference embryos and harpy mutants.
(A, B) Expression of dlx3b at 24 hours, showing reduced numbers of positive expressing cells in the mutant.
(C, D) Optic vesicles in live embryos at 26 hours, showing extruded lens in the mutant.
(E, F) Otic vesicle in live embryos at 26 hours. Arrows indicate hair cells. Note the absence of layers and miniaturized structures in the mutant.
(G, H) Expression (arrows) of fgf8 at 24 hours in the otic vesicle. Note that expression is almost absent in the mutant vesicle (demarcated by oval).
(I–N) Expression of pax2a in the otic tissue.
(I, J) pax2a mRNA at 24 hours in the otic vesicle. Note that expression reduced in the mutant vesicle (demarcated by oval).
(K–N) staining with anti-PAX2 in the otic placodes, showing expansion of the PAX2 territory between 12 and 14 hours in the mutant.
(O, P) Expression of N-cadherin (ncad) at 24 hours, showing a rosette of the lateral line (arrow head) and the lateral line primordia (arrow). In many mutants, the primordia could not be found.
Abbr:
ls, lens;
hc, hair cells;
np, nose placode;
oc, otocyst;
op, otic placode;
ov, otic vesicle;
px, pharynx;
Scale bar = 100 µm (A,B), 50 µm (C,D), 10 µm (E,F), 20 µm (G–P).
dlx3b mRNA is normally expressed in the nasal, lens and otic placodes (Akimenko et al., 1994), shown in Fig. 6 A. In the harpy mutant, dlx3b was expressed in far fewer cells (Fig. 6 B), indicating that at 24 hours some placodal structures do not contain the correct cell number or that some cells are not yet completely specified. The lens forms in harpy mutants and gives rise to a small but relatively normal lens, but failure of the eye-cup to expand properly leads to extrusion of the lens by 24 hours (Fig. 6 C,D). The otic vesicle remains small and usually produces only one or two hair cells with a single otolith (Fig. 6 E,F). Reliable support cell markers are not available in zebrafish, but the macula remains essentially a single layer rather than stratifying into apical hair cell- and basal support cell-layers. Additionally, the macula shows little or no expression of fgf8 (Fig. 6 G,H), which normally marks both hair cells and support cells of the developing maculae (Leger and Brand, 2002; Millimaki et al., 2007).
pax2a mRNA expression appeared in fewer cells in the mutant otic vesicle at 24 hours (Fig. 6 I,J), consistent with its morphology and dlx3b expression at that time. Nevertheless, early development of the otic placode was surprisingly normal in harpy mutants, as shown by Pax2-staining at 12 and 14 hours (Fig. 6 K–N). Moreover, despite the absence of cell division, the number of Pax2-positive cells nearly doubled between 12 and 14 hours in mutants (19.2 ± 3.8 cells at 12 hours, n=6; 37.2 ± 4.4 cells at 14 hours, n=8), which is similar to the fold-increase seen in wild-type embryos (82.9 ± 7.9 at 12 hours, n = 8; 130.2 ± 7.7 at 14 hours, n=5) This suggests that early expansion of the otic territory occurs by ongoing otic induction or recruitment, not cell division.
Lastly, ncad mRNA is expressed in many regions of the embryo; here we use it to visualize the lateral line primordia. This group of cells moves down the lateral sides of the embryo and leaves small rosettes of cells that develop in the sensory structures of the lateral line (Fig. 6 O). In the mutant, this structure was small and often difficult to find; when we did find it (Fig. 6 P), it was hidden in various locations, somewhat randomly, on the sides of the embryo or the yolk sac.
Relationship between endoreduplication and differentiation
During normal development, cell divisions become asynchronous and slower as cells begin to differentiate (Kane et al., 1992; Kane and Kimmel, 1993; Kimmel et al., 1994; Zamir et al., 1997). We hypothesized that the slowing of endoreduplication might be related to the initiation of cytodifferentiation that occurs beginning in the segmentation stages of development. This must occur in only a fraction of the total number of cells because most cells at these stages continue to show S-phase in both mutant and wild-type embryos (Fig. 7 A,B). To test this, we examined patterns of BrdU incorporation in embryos stained with markers of various specified and differentiated cell types. We used three antibodies known to label early neuronal and mesodermal cell types: Anti-Islet1/2 antibody stains Rohon-Beard sensory neurons, primary motoneurons and trigeminal ganglion neurons by 10 hours (Inoue et al., 1994). Anti-Engrailed antibody labels the midbrain-hindbrain border and muscle pioneer cells (Hatta et al., 1991). Anti-Pax2 antibody, which primarily binds Pax2a protein (Riley et al., 1999), labels cells in the midbrain-hindbrain border, otic placode, pronephros, reticulospinal neurons in the hindbrain, and CoSA spinal interneurons (Krauss et al., 1991b; Mikkola et al., 1992). 3A10 antibody labels Mauthner neurons (Metcalfe et al., 1990).
To assess whether DNA synthesis occurred in these cells during the segmentation stages, BrdU was injected at 12 hours and embryos were fixed and co-stained at 19 hours or later for BrdU and the individual cell type markers. Many of the specific cell types expressing the above markers did not show BrdU incorporation (Table 2, Fig. 7 D–I), though BrdU was detected broadly in surrounding cells. In particular, BrdU incorporation was not seen in muscle pioneers, Rohon-Beard sensory neurons, primary motoneurons, trigeminal cranial ganglia, Mauthner, reticulospinal, and spinal interneurons, all cell types that are known to have early terminal mitoses (Mendelson, 1986; Myers et al., 1986; Mikkola et al., 1992; Hirsinger et al., 2004; Won et al., 2006; Caron et al., 2008; Rossi et al., 2009).
Table 2.
BrdU incorporation in cells of harpy mutants expressing markers of specification and differentiation.
| Marker | Cell Type | Number cells BrdU positive |
Number cells counted |
Number embryos |
|---|---|---|---|---|
| Engrailed | Muscle Pioneers | 0 | 124 | 7 |
| Islet1/2 | Rohon-Beard sensory neurons | 0 | 412 | 29 |
| Primary motoneurons | 0 | 408 | 29 | |
| Trigeminal ganglion | 0 | 243 | 29 | |
| Pax2 | Reticulospinal primary neurons | 0 | 94 | 26 |
| Spinal interneurons | 0 | 272 | 22 | |
| Otic Vesicle | 26 | 451 | 11 | |
| Pronephros | 14 | 293 | 11 | |
| Midbrain-Hindbrain Border | 24 | 290 | 9 | |
In contrast, BrdU was detected in a small fraction of Pax2-staining cells in the developing pronephric duct, within the otic vesicle and in the midbrain-hindbrain border (Table 2, Fig. 7 C and data not shown). Each of these domains generate multiple diverse cell types many of which do not differentiate until later. Together, these findings are consistent with the hypothesis that the process of endoreduplication does not continue after cells have begun terminal differentiation, but may still occur during earlier stages of organogenesis.
harpy acts both cell autonomously and cell nonautonomously
To ask if the division characteristics and the cell morphology of the harpy mutant were cell autonomous, we transplanted mixtures of wild-type and mutant cells into wild-type or mutant host embryos, as described previously (Ho and Kane, 1990). In brief, cells from two donors, one labeled with rhodamine-dextran and one labeled with fluorescein-dextran, were transplanted into one host; donors were genotyped based on their 24 hour phenotypes. We recorded the phenotypes of the individual donor cells as well as the number of cells that were produced by cell division. In general, we found that most of the division and phenotypic characteristics that we identified in our lineage work above were autonomous to the genotype of the transplanted cells. Fig. 8 shows examples of transplanted wild-type and mutant cells placed in either wild-type or mutant hosts. Notably, harpy mutant cells that became neurons were larger than their wild-type counterparts, regardless of the genotype of the host. Muscle cells, as noted above were almost normal in size. However we did note some non-autonomous effects: Normally, harpy mutant neurons tend not to sprout axons. We found that wild-type cells placed in the CNS of harpy mutants also tended not to sprout axons (Fig. 8 B,D), whereas harpy cells in wild-type CNS sometimes did (Fig. 8 C). We interpret this to mean that some developmental environments in the mutant are not normal. This is an interesting result because Rca1 (in Drosophila) might have some role in axogenesis (Dong et al., 1997). At least in vertebrates, this may be a minor effect compared to nonautonomous effects in the embryonic CNS.
Fig. 8. harpy acts both cell autonomously and non-autonomously.
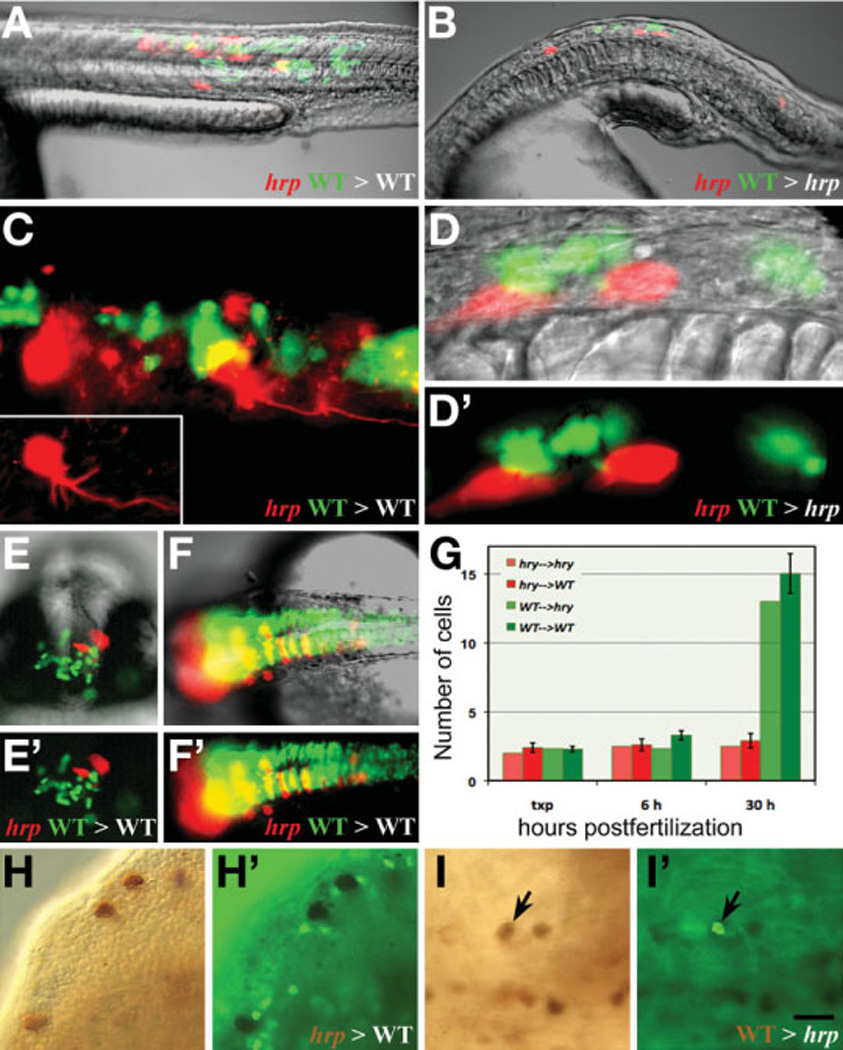
- (A,B) Composite images of a neural and muscle chimera.
- (C) UV image showing a magnified view of neural and muscle chimera in A. Inset shows single harpy interneuron which projected an axon.
- (D) magnified views of neural chimera in B. Note that the wild-type cells did not project axons.
- (E) Facial view of telencephalic region and
- (F) dorsal view of brain neural clones. Note that wild-type cells frequently cross the midline; mutant cells tend to not.
(G) Comparison of number of cells in clones. For each chimera, 2 to 3 cells from two separate individuals were placed into a host embryo at 5 hours, and recorded immediately afterwards (txp), then at 6 hours and again at 30 hours.
- (H, H’) Labeled mutant cells transferred to an wild-type host; mutant cells are not dividing.
- (I,I’) Labeled wild-type cells transferred to an unlabeled mutant host. A dividing wild-type cell is indicated (arrow).
Scale bar =200 µm (A,B,E,F), 50 µm (C,D), 100 µm (F,I).
Normally, cells that enter the neural keel have a tendency to divide at the midline with the daughter sibs often moving to opposite sides of the forming neural tube. We noted that when mutant cells were transplanted into the CNS of wild-type or mutant embryos, the mutant cells tended to stay on one side of the neural keel (Fig. 8 E,F) even when their wild-type co-transplanted cells moved to both sides.
We also counted the number of donor cells at 30 hours in our host embryos. For each chimera, 2 to 3 cells from two separate individuals were placed into a host embryo in the late blastula. The cell cycle number at this time would be about cycle 12 or 13. Immediately following the transplant, each chimeric embryo was recorded. Then at 6 hours, just before gastrulation, the number of cells in the chimera was re-inspected and recorded; similarly at 30 hours. Regardless of host genotype, harpy mutant cells do not tend to proliferate after 6 hours whereas wild-type cells do (Fig. 8 G).
In order to test whether the mitotic block in harpy mutants is cell autonomous, we produced mosaic embryos by transplanting cells from biotin-dextran labeled donor embryos to unlabeled host embryos during blastula stage, fixed the embryos at 24 hours, and double stained them with a horseradish peroxidase reaction product to detect donor cells and anti-phospho histone H3 to detect mitotic cells. When we transplanted cells from a mutant into a wild-type embryo, we found that mutant cells never expressed phospho-histone H3, indicating that they do not enter mitosis (Fig. 8H). Labeled wild-type controls divided normally, even when transplanted into a mutant host (Fig. 8I). These data confirm that disruption of harpy function blocks mitosis cell autonomously.
DISCUSSION
We have shown that harpy and Rca1/emi1 mutations are allelic and have further characterized the cell cycle defect in the mutant as well as its developmental consequences. Mutants show a global block to mitosis beginning early in gastrulation, after which some cells continue to grow in size as they undergo endoreduplication. Despite this cell cycle defect, most aspects of global patterning occur relatively normally, though there are also numerous local defects that are likely attributable to loss of proliferation or abnormal cell size.
The relatively normal global patterning seen in harpy mutants is reminiscent of previous studies in Xenopus showing that early patterning and cell type differentiation are surprisingly normal in embryos blocked in DNA synthesis by exposure to hydroxyurea and aphidocolin (Harris and Hartenstein, 1991; Rollins and Andrews, 1991). In earlier studies on the analysis of early blood specification, we have documented that in harpy mutants differentiation occurs on time for three major early blood histotypes although the numbers of cells are reduced (Warga et al., 2009). These findings underscore the remarkable regulative capacity of vertebrate embryos and further indicate that cell fate is influenced primarily by cell-cell interactions rather than lineage-restricted cell fate determinants.
The basis for local developmental defects
Development with relatively few cells of large size would likely impose significant spatial and geometrical constraints on the morphogenesis of some structures. For example, formation of thin elongate structures such as the floor plate and hypochord requires finer precision than can be accommodated in harpy mutants, possibly accounting for the spatial gaps noted in these structures (Fig. 4O). Additionally, complex organ systems requiring assembly of large numbers of cells is often compromised in harpy mutants, as seen in the severely dysmorphic otic vesicle (Fig. 6F) and extrusion of the lens from the eye (Fig. 6D). Reduced numbers of cells would also tend to limit cell type diversity, thereby disrupting essential local signaling interactions. This could account for abnormal migration of neuronal growth cones and neural crest cells (Fig. 5H,J,L). Indeed, mutant neurons developing within a wild-type host often showed improved axonogenesis, reflecting some degree of rescue from non-autonomous functions within the wild-type environment (Fig. 8C).
Large size alone probably perturbs certain cellular process required for proper development. For example, increased cell size could adversely affect cell signaling by altering rates of diffusion of secreted factors or by hampering the efficiency of signal transduction due to reduced surface:volume ratio. In some cases, defects in migration could also arise from mechanical changes in membrane/cytoskeletal dynamics of enlarged cells. Thus, the delay in convergence-extension movements seen in harpy mutants (Fig. 1B, E) could reflect sluggish cell movement or weakened signaling required for planar cell polarity.
The lack of cell division itself also likely perturbs some aspects of patterning and morphogenesis. In wild-type embryos, for example, cell divisions are oriented non-randomly during late gastrulation and may contribute somewhat to convergence and extension (Kane et al., 1992; Kane and Kimmel, 1993; Kimmel et al., 1994; Concha and Adams, 1998); such forces would be absent in the mutant. Cell division in the wild-type neural tube often separates daughter cells into right and left hemisegments where they enter equivalent cohorts of cells and often adopt similar fates (Kimmel et al., 1994; Geldmacher-Voss et al., 2003). Disruption of this process in harpy mutants possibly accounts for the asymmetric deficiencies in Rohon-Beard sensory neurons and primary motoneurons (Fig. 4I–K). This phenomenon is not limited to primary neural cell types, as there was a strong general tendency of mutant clones in wild-type hosts to remain isolated on one side of the neural tube (Fig. 8E,F). Development of many organ systems relies on maintaining a pool of stem cells or cycling progenitors. In the nervous system, for example, ongoing division of progenitors generates successive layers with distinct cell fates. The loss of proliferative centers in the brain of harpy mutants is reflected by the near absence of notch1b expression (Fig. 5D) and is likely responsible for a severe deficiency of many neural cell types (Figs. 4D, 5L,N). Similarly, the pancreas normally undergoes dramatic mitotic expansion but in the mutant it remains very small (Fig. 4H).
The relationship between endoreduplication and differentiation
BrdU incorporation shows that in mutants endoreduplication continues in a large fraction of cells past 24 hours but gradually ceases by 48 hours or soon thereafter. A recent study by Rhodes et al. (2009) confirms that cells in harpy hypomorphic mutants also continue to undergo eudoreduplication. Similar results have been obtained with cultured mammalian cells, in which depletion of Rca1/emi1 leads to limited endoreduplication with accumulation of greater than 4N DNA content (Di Fiore and Pines, 2007; Machida and Dutta, 2007). Endoreduplication eventually stops in cultured cells due to accumulation of DNA damage, which activates checkpoint kinases that halt further DNA synthesis (Verschuren et al., 2007). A similar process of checkpoint-mediated senescence could account for cessation of DNA synthesis in at least some cells in harpy mutants. However, the complex pattern of BrdU incorporation revealed dramatic variation in the time when different cells cease endoreduplication, suggesting that cells are not regulated in a uniform manner and that some other process influences endoreduplication. Indeed, our findings suggest that differentiation accounts for much of this variation, allowing some cells to withdraw early from endoreduplication. Specifically, analysis of early-differentiating cell types in harpy mutants showed that these cells no longer incorporate BrdU after 12 hours (Fig. 7, Table 2), similar to their wild-type counterparts. Conversely, cell types that proliferate extensively in wild-type embryos, such as blood cells and periderm, grow to an especially large size in harpy mutants (Fig. 3H,I), suggesting they undergo additional rounds of endoreduplication in lieu of cell division. Other developing tissues that continued to show sporadic BrdU-incorporation include the pronephros, otic placodes and the midbrain-hindbrain border. In each of these cases expression of pax2a was used to mark early stages in the development of these organ primordia rather than formation of terminally differentiated cell types. Thus, escape from endoreduplication in harpy mutant cells correlates with terminal differentiation, but not earlier steps in cell fate specification of lineage restriction.
The precise relationship between differentiation and cell division is only partially understood. The maternal-zygotic transition in zebrafish begins at the tenth division, after which cell cycles lengthen and become asynchronous as cells begin to take on different fates (Kane et al., 1992; Kane and Kimmel, 1993). Cells fated to become primary neurons become post-mitotic as early as cycle 16 whereas secondary neurons are born in cycle 17 or later (Kimmel et al., 1994). Differentiating cells can modulate the rate of cell division by changing expression levels of cell cycle inhibitors, such as Rb and Cip1/Kip1/p21, which repress E2F-dependent expression of Cyclins (reviewed by Robinson et al., 2002; De Falco et al., 2006; Bicknell et al., 2007; and Besson et al., 2008). This includes Cyclin E, which is required for entry into S-phase as well as endoreduplication (Knoblich et al., 1994; Geng et al., 2003; Parisi et al., 2003). Hence, the process of differentiation could block endoreduplication in harpy mutants by p21/Rb-mediated repression of Cyclin E.
Consequences of elevated Anaphase Promoting Complex/Cyclosome (APC/C) activity in harpy mutants
APC/C is an E3 ubiquitin ligase that normally targets mitotic Cyclins and Geminin for destruction, cyclical regulation of which is necessary for coordinating DNA synthesis and cell-cycle progression (Reimann et al., 2001a; Miller et al., 2006; Di Fiore and Pines, 2007; Machida and Dutta, 2007). We infer that APC/C remains constitutively active in harpy mutants, as has been shown in Drosophila, Xenopus and mammalian tissue culture following disruption of Rca1/emi1 (Reimann et al., 2001a; Grosskortenhaus and Sprenger, 2002; Machida and Dutta, 2007). This model accounts for the cell-autonomous block to mitosis in harpy mutants (Fig. 8H) as well as ongoing endoreduplication. This model also accounts for the high frequency of cells that escape the mitotic block in the hypomorphic allele of harpy in the study by Rhodes et al. (2009).
Constitutive APC/C activity could explain some of the developmental defects seen in these mutants. APC/C is known to target regulatory proteins required for axon growth and synaptic function (Juo and Kaplan, 2004; Konishi et al., 2004; van Roessel et al., 2004; Stegmuller et al.). Although some aspects of axonogenesis in mutant neurons were rescued by development within a wild-type host (Fig. 8C), rescue was neither uniform nor complete, indicating that at least some cell-autonomous defects contribute to axonal defects. APC/C also targets Id-related HLH proteins, which normally inhibit differentiation by antagonizing neurogenic and myogenic bHLH proteins (Lasorella et al., 2006). While in principle rapid clearing of Id proteins in harpy mutants could accelerate differentiation, we detected no instances of precocious expression of cell type markers. On the other hand, we did observe that an early pattern of expression of fgf8 in the hindbrain is abnormally retained through at least 24 hours in harpy mutants (Fig. 5F), possibly reflecting failure in the normal progression of interactions between diverse cell types.
Our short pulse BrdU incorporation studies (Fig. 2 I–N) suggested that endoreduplication is occurring in harpy mutants. What with the strong labeling of a fraction of the cells with only few intermediately labeled cells, cells seem to be either in S-phase or not, much like S-phase in cells in wild-type embryos. Nevertheless, our work only partially clarifies the dynamics of S-phase in the mutants: although we refer to multiple rounds of DNA synthesis as endoreduplication, this term is often used to imply that cells undergo definite S-phases separated by a G-phase. In principle, we do not know if there is a G-phase because we have not measured the S-phases delimiting this hypothetical G-phase. Second, when Rca1/emi1 is ablated, the APC/C could target geminin for degradation and it would become possible to allow re-initiation of DNA synthesis, termed re-replication (Sivaprasad et al., 2007; Porter, 2008; van Leuken et al., 2008). Because we see only slightly higher numbers of positive nuclei with long pulses, it is possible that a large subset of the cell population may be in re-replication. Clearly, further experiments will be needed to answer these questions.
EXPERIMENTAL PROCEDURES
Fish Strains, Staging and Genotyping
Wild-type zebrafish strains were derived from the AB line (Eugene, OR) or the tupLongfin line (Tübingen, Germany). Embryos were developed in fish water containing methylene blue at 28.5°C and staged according to standard protocols (Kimmel et al., 1995). hrpti245 was recovered in a large-scale ENU-mutagenesis screen (Haffter et al., 1996). hrpx1 was induced by post-meiotic ENU-mutagenesis (Riley and Grunwald, 1995) and recovered in a small-scale screen.
Lineage Tracing and Time Lapse Analysis
Cell labeling was adapted from protocols previously described in Warga and Nüsslein-Volhard (1999) and in Warga et al. (2009). Briefly, an individual blastomere was labeled at the 1K-cell stages with a 5% solution of rhodamine-dextran (10,000 MW; Invitrogen, formerly Molecular Probes). Immediately after labeling, embryos were mounted in a 3% solution of methylcellulose (Sigma) and recorded in multiplane as described in Warga and Kane (2003). Generally between 8 to 10 embryos were recorded simultaneously for up to 12 hours. Afterwards, the recordings were analyzed using Cytos Software, as described in Kane et al. (2005) and in McFarland et al. (2005).
Cell Transplantation
Transplantation was carried out between doming and 30% epiboly stage as described in Ho and Kane (1990). In some of our experiments we included the fixable tracer biotinylated-Dextran (Invitrogen). In such experiments, hosts were fixed in 4% paraformaldehyde, and the clone was visualized as described in Warga and Kane (2007) with the following modification: Diaminobenzidine (Sigma) was used as the enzyme substrate following protocols previously described in Warga and Nüsslein-Volhard (1999).
Analysis of gene expression
Whole-mount in situ hybridization was carried out as described previously in Thisse and Thisse (1993) and Phillips et al. (2006). Wholemount immunostaining was performed as described in Riley et al. (1999) and Phillips et al. (2006) using antibodies purchased from the Developmental Studies Hybridoma Bank directed against Grasp (zn8,1:100), Islet1/2 (39.4D5 1:100), Pax7 (1:150), 3A10 (1:20), or anti-HuC (ZIRC, Eugene OR, 1:100), or commercial antibodies against Pax2 (Covance, 1:100), phospho-histone H3 (Upstate 06-570, 1:350) or acetylated tubulin (Sigma, 1:2000).
BrdU incorporation
BrdU pulse labeling was performed as described by Phillips et al. (2006) using anti-BrdU (Beckton-Dickinson, 1:250).
DAPI-staining
Embryos were fixed and stained simultaneously for 15–20 minutes in a 4% paraformaldehyde solution in PBS containing 0.01% DAPI and either photographed immediately or disassociated by gently crushing between coverslips before photographing.
Dissociation of embryos
To obtain cell-counts, dechorionated embryos were incubated for 5–10 minutes at 37°C in modified Holtfretter’s buffer (60 mM NaCl, 0.6 mM KCl, 0.9 mM CaCl, 5 mM HEPES pH 7.4) containing collagenase (Sigma C9891) at a concentration of 20 mg/ml (embryos at 12 hours or older) or 7 mg/ml (embryos at 6 hours). Embryos were monitored while gently triturating with a glass pipette until dissociation was complete. Dissociated cells were suspended in Hotfretter’s buffer and counted with a hemocytometer.
ACKNOWLEDGEMENTS
We thank Sumi Himangshu and April Kraus for spirited help on the harpy project through the years, and Robert Ho, Yi-Lin Yan, Benjamin Geiger, Yun-Jin Jiang, and Sharon Amacher for probes and advice. This work was supported by a National Institutes of Health grant NIDCD DC003806 to BBR and a Pew Fellowship to DAK.
List of abbreviations
- bmn
branchio-motor nuclei
- dc
diencephalon
- ep
epiphysis
- ed
endoderm
- edc
endothelial cell
- fp
floor plate
- hb
hindbrain
- hbn
hindbrain neurons
- hc
hypochord
- hgl
hatching gland
- ls
lens
- mhb
midbrain hindbrain border
- mp
muscle pioneers
- Mth
Mauthner neurons
- mus
muscle
- nc
notochord
- np
nose placode
- oc
otocyst
- op
otic placode
- os
optic stalk
- ov
otic vesicle
- pcr
pancreas
- pdm
periderm
- pmn
primary motor neurons
- pp
pharyngeal pouch
- prd
pronephric duct
- prn
pronephric neck
- prt
pronephric tubule
- prt
pronephros
- px
pharynx
- r4
4th rhombomere
- rb
Rohon-Beard sensory neuron
- rbc
red blood cell
- scn
spinal cord neurons
- sin
spinal interneurons
- tc
telencephalon
- tg
trigeminal ganglia
- vb
ventral brain
REFERENCES
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. Journal of Neuroscience. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel B, Eisen JS. Regulation of neuronal specification in the zebrafish spinal cord by Delta function. Development. 1998;125:371–380. doi: 10.1242/dev.125.3.371. [DOI] [PubMed] [Google Scholar]
- Baker NE. Patterning signals and proliferation in Drosophila imaginal discs. Curr Opin Genet Dev. 2007;17:287–293. doi: 10.1016/j.gde.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Bicknell KA, Coxon CH, Brooks G. Can the cardiomyocyte cell cycle be reprogrammed? J Mol Cell Cardiol. 2007;42:706–721. doi: 10.1016/j.yjmcc.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Bierkamp C, Campos-Ortega JA. A zebrafish homologue of the Drosophila neurogenic gene Notch and its pattern of transcription during early embryogenesis. Mechanisms of Development. 1993;43:87–100. doi: 10.1016/0925-4773(93)90027-u. [DOI] [PubMed] [Google Scholar]
- Caron SJ, Prober D, Choy M, Schier AF. In vivo birthdating by BAPTISM reveals that trigeminal sensory neuron diversity depends on early neurogenesis. Development. 2008;135:3259–3269. doi: 10.1242/dev.023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Adams RJ. Oriented cell divisions and cellular morphogenesis in the zebrafish gastrula and neurula: a time-lapse analysis. Development. 1998;125:983–994. doi: 10.1242/dev.125.6.983. [DOI] [PubMed] [Google Scholar]
- De Falco G, Comes F, Simone C. pRb: master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene. 2006;25:5244–5249. doi: 10.1038/sj.onc.1209623. [DOI] [PubMed] [Google Scholar]
- Di Fiore B, Pines J. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J Cell Biol. 2007;177:425–437. doi: 10.1083/jcb.200611166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Zavitz KH, Thomas BJ, Lin M, Campbell S, Zipursky SL. Control of G1 in the developing Drosophila eye: rca1 regulates Cyclin A. Genes Dev. 1997;11:94–105. doi: 10.1101/gad.11.1.94. [DOI] [PubMed] [Google Scholar]
- Geldmacher-Voss B, Reugels AM, Pauls S, Campos-Ortega JA. A 90-degree rotation of the mitotic spindle changes the orientation of mitoses of zebrafish neuroepithelial cells. Development. 2003;130:3767–3780. doi: 10.1242/dev.00603. [DOI] [PubMed] [Google Scholar]
- Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Sprenger F. Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev Cell. 2002;2:29–40. doi: 10.1016/s1534-5807(01)00104-6. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Lemaire P, Kato K. Community effects and related phenomena in development. Cell. 1993;75:831–834. doi: 10.1016/0092-8674(93)90526-v. [DOI] [PubMed] [Google Scholar]
- Haddon C, Smithers L, Schneider-Maunoury S, Coche T, Henrique D, Lewis J. Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development. 1998;125:359–370. doi: 10.1242/dev.125.3.359. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nüsslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Harris WA, Hartenstein V. Neuronal determination without cell division in Xenopus embryos. Neuron. 1991;6:499–515. doi: 10.1016/0896-6273(91)90053-3. [DOI] [PubMed] [Google Scholar]
- Hatta K, Bremiller R, Westerfield M, Kimmel CB. Diversity of expression of engrailed-like antigens in zebrafish. Development. 1991;112:821–832. doi: 10.1242/dev.112.3.821. [DOI] [PubMed] [Google Scholar]
- Hirsinger E, Stellabotte F, Devoto SH, Westerfield M. Hedgehog signaling is required for commitment but not initial induction of slow muscle precursors. Developmental Biology. 2004;275:143–157. doi: 10.1016/j.ydbio.2004.07.030. [DOI] [PubMed] [Google Scholar]
- Ho RK, Kane DA. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–730. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- Horikawa K, Ishimatsu K, Yoshimoto E, Kondo S, Takeda H. Noise-resistant and synchronized oscillation of the segmentation clock. Nature. 2006;441:719–723. doi: 10.1038/nature04861. [DOI] [PubMed] [Google Scholar]
- Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Developmental Dynamics. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- Juo P, Kaplan JM. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr Biol. 2004;14:2057–2062. doi: 10.1016/j.cub.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kane DA, Maischein HM, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kelsh RN, Mullins MC, Odenthal J, Warga RM, Nüsslein-Volhard C. The zebrafish early arrest mutants. Development. 1996;123:57–66. doi: 10.1242/dev.123.1.57. [DOI] [PubMed] [Google Scholar]
- Kane DA, McFarland KN, Warga RM. Mutations in half baked/E-cadherin block cell behaviors that are necessary for teleost epiboly. Development. 2005;132:1105–1116. doi: 10.1242/dev.01668. [DOI] [PubMed] [Google Scholar]
- Kane DA, Warga RM, Kimmel CB. Mitotic domains in the early embryo of the zebrafish. Nature. 1992;360:735–737. doi: 10.1038/360735a0. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Kane DA. Cell cycles and clonal strings during formation of the zebrafish central nervous system. Development. 1994;120:265–276. doi: 10.1242/dev.120.2.265. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Korzh V, Edlund T, Thor S. Zebrafish primary neurons initiate expression of the LIM homeodomain protein Isl-1 at the end of gastrulation. Development. 1993;118:417–425. doi: 10.1242/dev.118.2.417. [DOI] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Fjose A. Expression of the zebrafish paired box gene pax<zf-b> during early neurogenesis. Development. 1991a;113:1193. doi: 10.1242/dev.113.4.1193. [DOI] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Moens U, Ericson JU, Fjose A. Zebrafish pax zf-a—A paired box-containing gene expressed in the neural tube. EMBO J. 1991b;10:3609–3619. doi: 10.1002/j.1460-2075.1991.tb04927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak SJ, Phillips BT, Heck R, Riley BB. An expanded domain of fgf3 expression in the hindbrain of zebrafish valentino mutants results in mis-patterning of the otic vesicle. Development. 2002;129:5279–5287. doi: 10.1242/dev.129.22.5279. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- Leger S, Brand M. Fgf8 and Fgf3 are required for zebrafish ear placode induction, maintenance and inner ear patterning. Mech Dev. 2002;119:91–108. doi: 10.1016/s0925-4773(02)00343-x. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–194. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani FV, Ahn CP, Martin GR. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–405. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland KN, Warga RM, Kane DA. Genetic locus half baked is necessary for morphogenesis of the ectoderm. Developmental Dynamics. 2005;233:390–406. doi: 10.1002/dvdy.20325. [DOI] [PubMed] [Google Scholar]
- Mendelson B. Development of Reticulospinal Neurons of the Zebrafish. I. Time of Origin. Journal of Comparative Neurology. 1986;251:160–171. doi: 10.1002/cne.902510203. [DOI] [PubMed] [Google Scholar]
- Metcalfe WK, Myers PZ, Trevarrow B, Bass MB, Kimmel CB. Primary neurons that express the L2/HNK-1 carbohydrate during early development in the zebrafish. Development. 1990;110:491–504. doi: 10.1242/dev.110.2.491. [DOI] [PubMed] [Google Scholar]
- Mikkola I, Fjose A, Kuwada JY, Wilson S, Guddal PH, Krauss S. The paired domain-containing nuclear factor pax[b] is expressed in specific commissural interneurons in zebrafish embryos. J Neurobiol. 1992;23:933–946. doi: 10.1002/neu.480230802. [DOI] [PubMed] [Google Scholar]
- Miller JJ, Summers MK, Hansen DV, Nachury MV, Lehman NL, Loktev A, Jackson PK. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 2006;20:2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millimaki BB, Sweet EM, Dhason MS, Riley BB. Zebrafish atoh1 genes: classic proneural activity in the inner ear and regulation by Fgf and Notch. Development. 2007;134:295–305. doi: 10.1242/dev.02734. [DOI] [PubMed] [Google Scholar]
- Myers PZ, Eisen JS, Westerfield M. Development and axonal outgrowth of identified motoneurons in the zebrafish. J. Neurosci. 1986;6:2278–2289. doi: 10.1523/JNEUROSCI.06-08-02278.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogare DE, Arguello A, Sazer S, Lane ME. Zebrafish cdc25a is expressed during early development and limiting for post-blastoderm cell cycle progression. Dev Dyn. 2007;236:3427–3435. doi: 10.1002/dvdy.21363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenthal J, Nüsslein-Volhard C. fork head domain genes in zebrafish. Development Genes & Evolution. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cellcycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Parisi T, Beck AR, Rougier N, McNeil T, Lucian L, Werb Z, Amati B. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. Embo J. 2003;22:4794–4803. doi: 10.1093/emboj/cdg482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BT, Kwon HJ, Melton C, Houghtaling P, Fritz A, Riley BB. Zebrafish msxB, msxC and msxE function together to refine the neural-nonneural border and regulate cranial placodes and neural crest development. Developmental Biology. 2006;294:376–390. doi: 10.1016/j.ydbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Porter AC. Preventing DNA over-replication: a Cdk perspective. Cell Div. 2008;3:3. doi: 10.1186/1747-1028-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001a;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Reimann JD, Gardner BE, Margottin-Goguet F, Jackson PK. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev. 2001b;15:3278–3285. doi: 10.1101/gad.945701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Amsterdam A, Sanda T, Moreau LA, McKenna K, Heinrichs S, Ganem NJ, Ho KW, Neuberg DS, Johnston A, Ahn Y, Kutok JL, Hromas R, Wray J, Lee C, Murphy C, Radtke I, Downing JR, Fleming MD, MacConaill LE, Amatruda JF, Gutierrez A, Galinsky I, Stone RM, Ross EA, Pellman DS, Kanki JP, Look AT. Emi1 maintains genomic integrity during zebrafish embryogenesis and cooperates with p53 in tumor suppression. Mol Cell Biol. 2009;29:5911–5922. doi: 10.1128/MCB.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BB, Chiang M, Farmer L, Heck R. The deltaA gene of zebrafish mediates lateral inhibition of hair cells in the inner ear and is regulated by pax2.1. Development Supplement. 1999;126:5669–5678. doi: 10.1242/dev.126.24.5669. [DOI] [PubMed] [Google Scholar]
- Riley BB, Grunwald DJ. Efficient induction of point mutations allowing recovery of specific locus mutations in zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5997–6001. doi: 10.1073/pnas.92.13.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson IM, Ranjan R, Schwarz TL. Synaptotagmins I and IV promote transmitter release independently of Ca(2+) binding in the C(2)A domain. Nature. 2002;418:336–340. doi: 10.1038/nature00915. [DOI] [PubMed] [Google Scholar]
- Rollins MB, Andrews MT. Morphogenesis and regulated gene activity are independent of DNA replication in Xenopus embryos. Development. 1991;112:559–569. doi: 10.1242/dev.112.2.559. [DOI] [PubMed] [Google Scholar]
- Rossi CC, Kaji T, Artinger KB. Transcriptional control of Rohon-Beard sensory neuron development at the neural plate border. Dev Dyn. 2009;238:931–943. doi: 10.1002/dvdy.21915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Prince V, Ingham PW. Plasticity in zebrafish hox expression in the hindbrain and cranial neural crest. Developmental Biology. 2001;231:201–216. doi: 10.1006/dbio.2000.9997. [DOI] [PubMed] [Google Scholar]
- Sivaprasad U, Machida YJ, Dutta A. APC/C--the master controller of origin licensing? Cell Div. 2007;2:8. doi: 10.1186/1747-1028-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmuller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 2006;50:389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Strähle U, Blader P, Henrigue D, Ingham PW. axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes and Development. 1993;7:1436–1446. doi: 10.1101/gad.7.7b.1436. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- Ulloa F, Briscoe J. Morphogens and the control of cell proliferation and patterning in the spinal cord. Cell Cycle. 2007;6:2640–2649. doi: 10.4161/cc.6.21.4822. [DOI] [PubMed] [Google Scholar]
- van Leuken R, Clijsters L, Wolthuis R. To cell cycle, swing the APC/C. Biochim Biophys Acta. 2008;1786:49–59. doi: 10.1016/j.bbcan.2008.05.002. [DOI] [PubMed] [Google Scholar]
- van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell. 2004;119:707–718. doi: 10.1016/j.cell.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Verschuren EW, Ban KH, Masek MA, Lehman NL, Jackson PK. Loss of Emi1-dependent anaphase-promoting complex/cyclosome inhibition deregulates E2F target expression and elicits DNA damage-induced senescence. Mol Cell Biol. 2007;27:7955–7965. doi: 10.1128/MCB.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warga RM, Kane DA. One-eyed pinhead regulates cell motility independent of Squint/Cyclops signaling. Developmental Biology. 2003;261:391–411. doi: 10.1016/s0012-1606(03)00328-2. [DOI] [PubMed] [Google Scholar]
- Warga RM, Kane DA. A role for N-cadherin in mesodermal morphogenesis during gastrulation. Developmental Biology. 2007;310:211–225. doi: 10.1016/j.ydbio.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Warga RM, Kane DA, Ho RK. Fate mapping embryonic blood in zebrafish: multi- and unipotential lineages are segregated at gastrulation. Dev Cell. 2009;16:744–755. doi: 10.1016/j.devcel.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warga RM, Nüsslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–838. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- Wiellette E, Grinblat Y, Austen M, Hirsinger E, Amsterdam A, Walker C, Westerfield M, Sive H. Combined haploid and insertional mutation screen in the zebrafish. Genesis: the Journal of Genetics & Development. 2004;40:231–240. doi: 10.1002/gene.20090. [DOI] [PubMed] [Google Scholar]
- Won M, Ro H, Park HC, Kim KE, Huh TL, Kim CH, Rhee M. Dynamic expression patterns of zebrafish 1G5 (1G5z), a calmodulin kinase-like gene in the developing nervous system. Dev Dyn. 2006;235:835–842. doi: 10.1002/dvdy.20693. [DOI] [PubMed] [Google Scholar]
- Zamir E, Kam Z, Yarden A. Transcription-dependent induction of G1 phase during the zebra fish midblastula transition. Molecular & Cellular Biology. 1997;17:529–536. doi: 10.1128/mcb.17.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kendrick C, Julich D, Holley SA. Cell cycle progression is required for zebrafish somite morphogenesis but not segmentation clock function. Development. 2008;135:2065–2070. doi: 10.1242/dev.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]