Abstract
Archaea, organisms that make up the third domain of cellular life are members of the human oral microflora. They are strikingly less diverse than oral bacteria and appear to be relatively rare with respect to their numerical abundance. Since they have been exclusively found in association with oral infections such as periodontitis and apical periodontitis and given their unique physiology and energy metabolism, it is highly plausible that they are more than just secondary colonizers of infected areas, but instead are actively involved in the overall poly-microbial infection process. Conversely, it is a highly challenging task to clearly demonstrate their possible active participation – mostly due to the difficulty to grow them in routine microbiology laboratories. This current review points out the importance for understanding the medical impact of methanogens and aims at devising strategies for elucidating the true function of archaea in the oral ecosystem.
Keywords: methanogenic archaea, human microbial ecosystems, oral infections, interspecies hydrogen transfer
Archaea – one of the three domains of cellular life (besides Bacteria and Eukarya) consist of the four major divisions Euryarchaeota, Crenarchaeota, Korarchaeota, and Nanoarchaeota. While the human oral microbiota is primarily composed of organisms from the domain Bacteria, members of the domain Archaea are conspicuously underrepresented. This is surprising, since Archaea are widespread in nature and are capable of occupying almost any ecological niche, even those exhibiting extreme environmental conditions. Putting in numbers, approximately 700 bacterial phylotypes (1, 2) are ‘confronted’ with less than a handful of methane-producing Archaea (methanogenic archaea). In light of the overwhelmingly high diversity and dominance of bacteria, the question arises whether few archaeal species matter at all in oral microbiology. Or, in other words: Are archaea an important or worthwhile research object for promoting our understanding of the role of the human microbiota in health and disease? Owing to their unique physiology and energy metabolism, their explicit occurrence at sites of infection with reasonably high proportions and prevalence, the answer is clearly yes. This current review details the life style that distinguishes methanogenic archaea from bacteria, discusses the most recent findings underscoring the importance of oral archaea and suggests strategies on how to get to the bottom of the true ecological functions of archaea in humans.
Detection of archaea in the oral cavity
Methanogenic archaea have been detected in the oral cavity by both cultivation-based methods and molecular methods previously (e.g. 3, 4). However, due to the difficulty in cultivating them, evidence of their existence in this human ecosystem has primarily been provided by PCR-based techniques targeting either the 16S rRNA gene and/or the functional gene mcrA encoding for the Methyl-Coenzym M reductase, a key enzyme involved in methanogenesis. In the essence, only one distinct cultivated representative of methanogenic Archaea, namely Methanobrevibacter oralis (3), is currently known and its pre-dominance has been verified by several PCR-based studies. The genus Methanobrevibacter consists of several species of which many appear to be specialized to the intestinal tract systems of animals (e.g. M. smithii in the human gut systems). Phylogenetically, this genus groups within the order Methanobacteriales – one of the five known orders of methanogens, all of which belong to Euryarchaeota (Fig. 1). Recently an additional as yet uncultivated Methanobrevibacter phylotype in the oral cavity has been reported by PCR-based methods (5). Furthermore, first evidence of the presence of Thermoplasma-like organisms (also belonging to Euryarchaeota) in the oral cavity exists as well (6). Two aspects are worth notifying here: first, all of the above named organisms have been identified in samples from periodontal plaque and in some cases even in association with apical periodontitis, suggesting medical relevance (see later in this article). Second, it is plausible to assume that the full diversity of oral (methanogenic) archaea has not yet been fully explored for reasons also outlined later in this article.
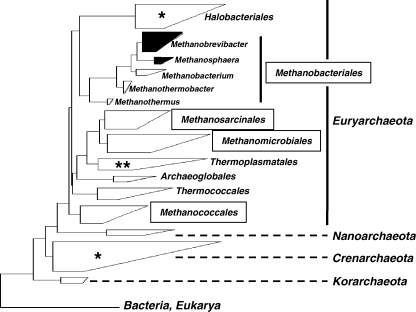
Fig. 1.
Dendrogramm depicting major taxonomic groups of Archaea. The dendrogramm is based on representative 16S rRNA gene sequences (clustered in boxes) calculated with the ARB software package (www.arb-home.de). The size and shape of the boxes roughly reflect the proportions of known phylotypes and phylogenetic depth. Note, that the tree is not a rigorous phylogenetic analysis, but an attempt to convey sequence relationship among archaeal organisms. The names in frames indicate four major orders of methanogens. Another fifth order of methanogens that is represented by only one species, namely, Methanopyrus kandleri is not included in the tree. The boxes that contain recognized human methanogens (i.e. Methanobrevibacter oralis as primary colonizer of the oral cavity, M. smithii and Methanosphaera stadtmanae as primary colonizer of the gut system) are highlighted in black. The stars within light boxes indicate sporadic detection of 16S rRNA genes (single star: detection in the human gut system, double star: detection in the gut system and oral cavity). For further information see main text.
What makes M. oralis (or other methanogenic Archaea) so unique compared to oral bacteria?
Methanogens and interspecies hydrogen transfer
Methanogenic archaea (in the following referred to as ‘methanogens’), while ubiquitous in anaerobic environments, such as wetlands, rice fields, biological treatment systems, or the gastro-intestinal tract system of ruminants, are the only biological source of methane (CH4) on Earth. And since CH4 is a prominent greenhouse gas (with a global warming potential 21 times higher than CO2) (7), methanogens have been extensively studied in different natural habitats. From these studies it has become evident that methanogens closely interact with a variety of bacterial species, in that they collaboratively degrade organic matter under anaerobic conditions. Important is that this cross-feeding behavior, or syntrophic growth on substrates, is obligatory for these partners as none of them would be able to utilize the required substrates alone due to thermo-dynamical constraints. The substrates in question are volatile fatty acids (VFAs) such as acetate, propionate, and butyrate (8). Although being important intermediate metabolites, they are unfavorable substrates for anaerobes, since their oxidation to H2 and CO2 is endergonic under standard conditions [that is the changes in the Gibbs free energy are positive, (8)]. Oxidation is thermodynamically only feasible when the H2 partial pressure is kept low. This introduces the key-role of methanogens that use such ‘simple’ molecules like H2 and CO2 as substrates to form CH4, a considerably exergonic reaction. It has been demonstrated that the resulting free energy is not only sufficient for growth of the methanogenic populations but also for those bacterial populations that transformed the VFA thereby generating H2 (8, 9). This syntrophic growth on VFA has received a general term in literature, namely, interspecies hydrogen transfer (9). It is now recognized as a mutually beneficial, unidirectional process that plays a central role in the anaerobic fermentation of organic matter in natural environments. It is important to understand that VFA fermentation is not inhibited by mere accumulation of H2 (i.e. end-product inhibition). If so, simple, non-enzymatic removal of H2 out of the ecological system (for instance diffusion) would make the biochemical reaction run autonomously. Instead, the energy that becomes available by the anaerobic oxidation of H2 is eminent, making metabolic H2-consumtion essential for anaerobic use of VFAs. This implies that methanogens and syntrophic bacterial partners have to be physically, very closely connected for efficient interspecies hydrogen transfer.
Possible medical importance of methanogens
If we apply this knowledge to methanogens present exclusively at sites of poly-microbial anaerobic biofilms in the oral cavity (e.g. periodontal pockets), it is plausible to assume that they support the growth of fermenting bacteria, which themselves are opportunistic pathogens. Hence, methanogens – if active – are involved in the overall infectious process with interspecies hydrogen transfer being an indirect mechanism of virulence. But are methanogens really in a viable state in dental plaque? As a matter of fact this question has already been answered by relatively old studies that have provided evidence of CH4 formation from isolated methanogenic species derived from primate but also human plaque samples (10–12). However, it is not simply the question of activity (why should a given microorganism not be active at least for some time in a given habitat, anyway?). The question is rather, what precisely is the nature of the activity within periodontal plaque and what are the microbial interactions? In this context it is important to note that methanogens can form methane independently from H2 by using other electron donors, such as methanol, methylamine, acetate, ethanol, or formiate (8). In fact, oral methanogens are able to use some of these compounds besides H2 and CO2 (12). But only in the case of H2 used as substrate, a support of fermenting pathogens by methanogens (through interspecies hydrogen transfer) is possible. So, what we need to know is, whether H2-consumption linked with syntrophic growth on VFA really occurs within the complex oral plaque consortium. Before finding answers to this, it is furthermore notable that there exists also a direct potential of methanogens for harming the host: The mechanism under discussion is the capability of methanogens to effectively transform heavy metals or metalloids into volatile methylated derivatives that are known to be more toxic than the original compounds (13). This feature is shared with some bacteria, but interestingly, methanogens isolated from the human gut have been shown to possess a much higher potential for metal(loid) derivatization (e.g. bismuth, selenium, tellurium, and mercury) in vitro compared to bacterial gut isolates (14). The immediate consequences of such transformation for human health have to be elucidated. However, as an example, use of bismuth containing compounds in pharmaceutical products have been linked with poisoning during prolonged medical therapy with the consequences of renal failure and mental disorders as described by Michalke et al. (13). It is likely that frequent exposure with such heavy metals through use of cosmetics or pharmaceutical products increases the possibility of its methylation by methanogens followed by increased toxicity. Whether or not methanogens in the oral cavity also have the potential or opportunity for such toxic transformations has not been investigated so far, but given the high number and diversity of dental materials potentially releasing heavy metals (crowns, bridges, amalgam, composites) this could be of great clinical importance as well.
Possible clinical relevance of methanogens in the oral cavity
Methanobrevibacter oralis has long been identified in periodontal pockets and its proportional increase with severity of disease has meanwhile been confirmed by various groups. The prevalence of detection using PCR-based methods varied greatly not only due to methodological differences but probably also due to different geographic/ethnic distribution of M. oralis [i.e. 22% for Japanese patients (15), 36% for US patients (4), 43% for German patients (16), and even up to 73% in Chinese patients (6)]. Notably, M. oralis was never detected at healthy sites; hence, this organism exhibits a positive predictive value for periodontitis of almost 100%, which is to our knowledge not reached by any bacterial species involved in this disease.
Mean relative proportions of these archaeal species varied depending on the study but also on the severity of periodontal disease [e.g. ranging from approximately 0.5 up to 18% compared to the entire prokaryotic load (16, 4, respectively)]. To our knowledge for no other periodonto-pathogenic bacterium, a mean proportional level as high as 18% has been described. But even values below 1% are not negligible, since particular members of the genus Methanobrevibacter are known to consume H2 almost completely if present at such low abundance (e.g. shown for mesophilic bioprocesses treating several types of wastewater) (17). Evidence of clinical significance of M. oralis has also been given by immunological findings. Yamabe et al. (15) investigated its distribution in Japanese patients with periodontitis and examined the serum IgG responses to archaeal components. Western immunoblotting detected IgG antibodies against M. oralis in sera from 8 of 11 tested patients suggesting antigenic potential. Furthermore, in a follow-up study Yamabe et al. (18) identified one of the antigenic molecules as subunit of the group II chaperonins (also known as thermosomes in Archaea and CCT in Eukarya). The authors (18) also demonstrated cross-reactivity with the human chaperonin CCT applying western immunoblotting. This is especially important given our knowledge regarding cross-reactivity of bacterial chaperonins with human molecules. For instance, members of the bacterial Hsp60 group (group I Chaperonin also known as GroEL in bacteria) are highly antigenic molecules (19, 20) and present in many pathogenic bacteria. Furthermore, several immune disorders such as rheumatoid arthritis or rheumatoid fever are thought to be triggered by these molecules. Although further investigation is needed for definite conclusions, the data indicate that antigenic molecules of M. oralis could have the potential to act as modifier or even initiator of inflammatory processes such as periodontitis.
Interestingly, further evidence that the human immune system recognizes and responds to Archaea comes from older studies that investigated the potential of archaeal cell membrane lipids (incorporated into liposomes and generally referred to as ‘archaeosomes’) as self-adjuvanting antigen-delivery vehicles. Such archaeal lipid structures derived from a range of different environmental archaea but also from members of the genus Methanobrevibacter exhibited higher adjuvant activity than conventional adjuvants such as bacterial liposomes [described in more detail in (21, 22)]. For instance, vaccination of mice with archaeosome-entrapped listeriolysin leads to rapid and prolonged specific immunity against Listeria monocytogenes infection and is superior to several other antigen-delivery vaccine strategies. This implies that the distinct structures of archaeal lipids (branched phytonyl chains attached to glycerol backbones via ether bonds, as opposed to the ester-linked fatty-acyl chains in Bacteria and Eukarya) not only confer considerable stability to liposomes but also that the mammalian immune system seems to be genetically programmed to ‘consider’ archaea as potential pathogens.
The identification of M. oralis at primary sterile localizations provides more evidence of medical relevance, although the question about cause and effect (23) remains unanswered so far. M. oralis was identified in 25% of samples retrieved from patients with endodontic infections previously (24). Further studies on US and Chinese patients have confirmed the presence of M. oralis in infected root canals, though with varying prevalence (25, 26). The latter study reported three additional interesting observations worth notifying. First, the combined presence of archaea (i.e. M. oralis) and bacteria was associated with a significantly higher prevalence of clinical symptoms (e.g. pain) compared to the number of cases with sole presence of bacteria. Second, archaea were identified also in persisting or secondary endodontic infections; that is, in cases of failed root canal treatment. This is especially interesting, since the response of methanogens to classical endodontic disinfectants such as sodium hypochloride or chlorhexidine has not been investigated so far. Lastly, Jiang et al. (26) identified archaea using an rRNA-based approach. This demonstrates that M. oralis was in fact present in a viable state in infected root canals and reduces speculations that the former DNA-based studies only detected the stable DNA released from cells already dead or damaged.
The significance of these findings have yet to be elucidated; however, it is conceivable that methanogens are more than just secondary colonizers of infected areas. Their occurrence in endodontic environments also implies the existence of strategies to invade and survive primary sterile compartments. This in turn leads to the intriguing question how likely the involvement of M. oralis or other archaea in infectious processes of otherwise sterile sites is, such as brain abscesses, peritonitis, or endocarditis? This question is warranted since distinct members of the endogenous microflora (from the oral cavity) while harmless in their natural location can cause severe life-threatening infections. Syntrophic interactions of these species with methanogens could be a driving factor for such kinds of polymicrobial diseases, provided that methanogens also get access to the primary sterile sites. However, our initial attempts to look for methanogens in extra-oral, ‘real’ clinical samples were unsuccessful so far. Non-detection of methanogens was accompanied with cross-reaction of archaeal PCR primers with human DNA. So, even if present, it is likely that methanogens are overlooked due to the unfavorable ratio of human DNA versus microbial DNA in those clinical samples.
Does M. oralis serve as keystone species in H2-consumption of oral ecosystems?
As mentioned above, the overwhelmingly high number of oral bacteria gives rise to the question if few archaeal species do matter at all. They do if we look at a well-recognized principle in ecology, namely, the concept of keystone species. This concept has been developed by ecological and conservation biologists over 40 years ago. According to that concept, keystone species (traditionally referred to animals) are rare members of a complex community and are usually only noticed when they are removed or they disappear from an ecosystem, resulting in dramatic changes to the rest of the community (27). In other words, a keystone species is one whose impact on its community or ecosystem is large – in any case greater than would be expected from its relative abundance or total biomass. This phenomenon is well accepted among ecologists and has been observed in a wide range of ecosystems. While providing a powerful model for understanding the forces that organize ecological communities, the term keystone species has also found its way into the literature of microbial ecologists (e.g. 28).
Just consider that microbial communities are composed of members who can carry out the same or similar biochemical reactions (functional redundancy) and of members exhibiting unique physiological traits (keystone species). As a consequence, the functional diversity of complex microbial communities is much lower than their phylogenetic diversity (29). In the case of the anaerobic degradation of organic matter, this means that most biochemical conversions can be performed by more than one bacterial species. This should especially be true for human microbial ecosystems since the host depends on a more or less stable supply with metabolic end-products of his microbial symbionts [‘top-to-down selection’30]. If a species goes extinct, other species (with the same functions) will replace it, thereby maintaining the microbial homeostasis. By contrast, this is not true for keystone species, which are niche specialists with unique physiologies that cannot or only hardly be replaced. If keystone species within macro-organisms fail for some reason, the effect will be either beneficial or harmful to the host depending on their ecological role for a physiological or pathogenic microbial community in the first place. In the case of methanogens at sites of oral infections, one could think of a beneficial outcome upon their elimination for reasons given earlier. It should be noted though that methanogens can theoretically be replaced by two different functional groups from the domain Bacteria, namely, dissimilatory sulfate reducers (SRB) and reductive acetogens, both of which can grow on H2 with the resulting end-products H2S and acetate, respectively. In fact, all three hydrogenotrophic functional groups were identified in sub-gingival plaque samples previously (16). However, an association with disease could be established only for methanogens and SRB, as reductive acetogens were also detected in healthy persons and their levels (unlike methanogens and SRB) even decreased with severity of the periodontal conditions. Furthermore, methanogens and SRB were found to the exclusion of each other in 46% of patients, although multiple plaque samples per patient were pooled prior to analysis (16). This indicates an apparent lack of one group in the oral cavity – probably due to host-specific factors. By contrast, both H2-consuming microbial groups were found together in only about 20% of patients (16). Hence, for a substantial number of periodontitis patients, H2-consumption seems to be a process performed by only one type (keystone) species because functional replacement is not possible due to the absence of an appropriate alternative syntrophic partner.
Future strategies
Lessons from human gut microbiota studies
Syntrophic archaeal-bacterial interactions
The fact that both the human intestinal tract system and the oral cavity share surprisingly few bacterial species (31, 32) applies also to archaea since it is not M. oralis but a related species, namely, M. smithii as the major archaeal player in the human gut system. The disorders in which M. smithii is probably involved are inflammatory bowel disease (or Crohn's disease), irritable bowel syndrome, colorectal cancer, diverticulosis, or obesity and current knowledge has been well discussed in the review articles by Nakamura et al. (33) and Roccarina et al. (34). Although different human disorders may be linked with different microbial members of either compartment, it is likely that the nature of archaeal interactions with gut bacteria – in particular the role as H2-consumer and supporter of the fermenting microbiota – has meaningful parallels to the situation in the oral cavity. Since much more attention has been devoted to the role of gut archaea, the most striking novel insights and possible implications for oral archaea are addressed subsequently. Based on a gnotobiotic mouse model, a link between M. smithii and host energy balance could be established (35). The colon of germ-free mice was colonized with a polysaccharide-degrading bacterium (Bacteroides thetaiotaomicron), a sulfate-reducing bacterium (Desulfovibrio piger), and M. smithii in different combinations (35). B. thetaiotaomicron degraded polyfructose-containing glycans more efficiently in the presence of M. smithii. In addition, mice colonized with these two microbes showed increased fermentation products and an increase in adiposity (increased storage of energy in fat cells). These effects were not observed after co-colonization with the pair B. thetaiotaomicron and D. piger (as alternative H2-consumer), emphasizing the critical role of M. smithii in promoting polysaccharide degradation, followed by absorption of the fatty acids that lead to liver lipogenesis and formation of fat deposits. These experiments substantiate the hypothesis that by providing the final step in energy extraction from degradation of organic compounds, methanogens have an enormous potential for altering the whole gut physiology with profound consequences for the host. As such they could also be an interesting target for therapeutical manipulation of obesity. In fact, further evidence has recently been provided by Zhang et al. (36). Using real-time PCR, they detected significantly higher numbers of H2-utilizing methanogens in three obese individuals than in three normal-weight or three post-gastric-bypass individuals. The numbers of the H2-producing Prevotellaceae were also highly enriched in the obese individuals supporting the hypothesis that interspecies hydrogen transfer may take place between these bacterial species and M. smithii. It is currently discussed that exactly this bacterial-archaeal interaction is an important mechanism for increasing energy uptake by the human large intestine in obese persons. Likewise, since many species of Prevotellacaea (55 distinct Prevotella taxons according to the HOMD database [http://www.homd.org/]) are common members in periodontal pockets, analogous relationships could exist with M. oralis.
A good indication about the nature of the syntrophically used substrate in humans does also exist. Abell et al. (37) found a negative correlation between mean fecal butyrate concentration and methanogen abundance by testing eight individuals weekly over a 12-week period and using molecular methods. Their findings suggest on the one hand an indirect association of methanogens with colorectal cancer (considering that butyrate is the primary energy source for immunologically active cells lining the colon, e.g. enhancement of cell cycle arrest and apoptosis) (38, 39). On the other hand it is likely that the methanogens live in syntrophy with butyrate degrading organisms via interspecies hydrogen transfer. Hence, butyrate producing and butyrate fermenting bacteria may also be interesting candidates to look at when searching for syntrophic partners of M. oralis in the oral cavity as well.
Detection targets to reveal diversity
Although this review focuses on oral archaea, novel insights regarding the diversity of archaea in the gut system are given here, because it is likely that M. oralis shares its niches in the oral cavity with so far undetected methanogens. Scanlan et al. (40) and Mihajlovski et al. (41) have identified mcrA gene sequences only distantly related to cultured methanogens from the five recognized orders (i.e. Methanomicrobiales, Methanopyrales, Methanobacteriales, Methanococcales, and Methanosarcinales, Fig. 1). McrA encodes one of the key enzymes (Methyl-Coenzyme M reductase, see later) involved in H2-consumption and is specific to methanogens. The authors hypothesized that this novel mcrA gene type corresponds to an uncultured phylotype that makes up a putative sixth methanogenic order (41). In a subsequent extended study, by using newly designed PCR primers specific to the novel group, Mihajlovski et al. (42) identified further mcrA gene types also grouping within the same clade of this putative sixth methanogenic order. In addition, these sequence types shared close to moderate similarity with a number of clone sequences recovered from animal studies subjecting cattle and pigs (42), further supporting the possibility of a novel group of methanogens adapted to the animal intestinal tract. Besides finding a correlation between that novel mcrA-gene type and age of the tested individuals, Mihajlovski et al. (42) detected also 16S rRNA gene sequences grouping within the Thermoplasmatales but with no cultured relatives (Fig. 1). The 16S rRNA gene sequences were only found in the samples that were also positive for the novel mcrA gene type. It is, therefore, likely that a novel sister group of Thermoplasmatales probably capable of methanogenesis exists in the human gut system. Since Thermoplasma-like sequences have also been identified in samples from periodontal pockets (6), it would be highly interesting to test the newly designed specific mcrA-primers in order to elucidate whether a novel as yet undetected form of methanogens there also exists.
Knowledge regarding archaeal diversity has been further advanced by a study of Oxley et al. (43). In their study of the microbiota in colonic mucosal biopsies from 8 out of 39 patients with inflammatory bowel disease, they found 16S rDNA sequences representing a phylogenetically rich diversity of halophilic archaea (15 different phylotypes) from the Halobacteriaceae (haloarchaea) including those with no directly related cultured representatives. This means that archaeal diversity within humans is not limited to the coherent group of methanogens (Fig. 1). Furthermore, aerobic enrichment cultures prepared from a patient biopsy at low salinity (2.5% NaCl) yielded haloarchaeal sequence types and subsequent microscopic observation after fluorescence in situ hybridization provided evidence of the presence of viable archaeal cells in these cultures. These results prove the survival of haloarchaea in the human digestive system and suggest that they may be members of the mucosal microbiota. Whether they constitute regular colonizers and whether they occur in abundance comparative to methanogenic archaea is unknown so far. In addition since most intestinal microbes initially enter through the oral cavity, an intriguing question is whether distinct niches in the oral cavity also provide appropriate conditions for colonization by this novel group of Archaea.
Sample processing with higher DNA extraction efficiency and primer design strategies for PCR analysis
As stated above, recent findings have enlarged the known diversity of Archaea in humans now including further phylotypes of Methanobrevibacter but even an entire novel order of methanogens, as well as members of Thermoplasmatales and Halobacteriacaea. The question arises here why their presence has largely eluded detection in many studies before. One apparent answer is that the primers used to amplify their DNA were not specific and/or sensitive enough. From our experiences, most primers designed in older, environmental studies for characterization of naturally occurring archaea are not suitable for detection of human-associated archaea. In many cases cross-reaction with human DNA was observed in our laboratory, in an extent that we rarely observed with Bacteria-specific primers. The reasons may be the unfavorable ratio between archaeal-to-human DNA (consider the relative low abundance of archaea compared to bacteria), but also a higher degree of sequence similarity between archaea-specific primers and human gene sequences. Hence, careful design of novel primers as well as the use of multiple molecular targets (16S rDNA and mcrA) is highly important in recovering a wider range of human methanogens. An alternative approach may be the separation of human and microbial DNA prior to PCR amplification. Two different approaches have been developed (44), which might also be useful for metagenomic studies of the human microbiome (45). The fact that the choice of the molecular approach greatly influences our perception of archaeal diversity and prevalence in humans can be best illustrated in another most recent study, again focused on the human gut flora. Dridi et al. (46) developed a new protocol for the extraction and PCR-based detection of M. smithii and Methanosphaera stadtmanae DNA in human stool samples. The protocol included a mechanical lysis step with glass beads that was applied twice, combined with an overnight incubation with proteinase K. The PCR-based detection included newly designed primers targeting the 16S rRNA gene but also the rpoB gene, which encodes the β subunit of RNA polymerase, one of the core genes shared by Bacteria and Archaea. By testing fecal samples from 700 (!) volunteers by real-time quantitative PCR, they found M. smithii in virtually all individuals and M. stadtmanae in almost 30% of cases. Double treatment with glass beads was performed because gut methanogens have been shown to possess a proteinase K resistant cell wall (47) – and apparently the extensive mechanical action was decisive in the efficiency of DNA extraction. Similarly Salonen et al. (48) found an increased prevalence of methanogenic Archaea in human fecal samples by repeated mechanical disruption steps, compared to other extraction protocols. These findings revise our perspective that methanogens colonize the gut of only about half of the human population. Instead, M. smithii appears as an almost ubiquitous inhabitant of the intestinal microbiome. As such, it sheds additional light on the paramount role that methanogenic species may have in the overall microbial ecology of the digestion process. Given the effect of differential DNA extraction protocols on the recovery of archaeal DNA as indicated by the above studies, the true prevalence and proportions of oral archaea has also to be re-considered.
All of the above mentioned Archaea belong phylogenetically to the major archaeal phylum Euryarchaeota. Whether or not Archaea other than Euryarchaeota (namely from the phyla Crenarchaeota, Korarchaeota, or Nanoarchaeota) are as yet undetected colonizers of the oral cavity is currently speculative (Fig. 1). However, members belonging to Crenarchaeota have been detected in the human gut system previously with PCR-based methods (49). In order to reproduce this intriguing result, we tested the same primers and PCR conditions in our laboratory. No 16S rRNA genes of Crenarchaeota were obtained in our study, neither from five selected fecal nor from five oral samples, so that the findings of Rieu-Lesme et al. (49) are preliminary for now and require further investigation. In addition, when testing further Crenarchaeota specific-primers (50) we only amplified and detected the 16S rRNA gene of M. oralis in oral samples, indicating an apparent lack of Crenarchaeota and an abundance of M. oralis sufficiently high enough to overcome mismatches to the crenarchaeota primers. In another first attempt, we tested published primers specifically designed for the detection of Korarchaeota (51) and Nanoarchaeota (52) with oral samples. In both cases we observed cross-reaction with human DNA, even under highly stringent PCR conditions. Again, new primer systems and/or efficient strategies for removal of human DNA are required to find clear answers.
Genome, metagenome, pangenome analysis
A major step toward a better understanding of the function and dynamics of human archaea will be done by results originating from the Human Microbiome Project (45). Whole genome analysis of several M. smithii strains in comparison with close relatives from environmental systems has already been performed and shown that M. smithii is highly adapted to the human gut system (53). This includes its capability of (1) producting surface glycans resembling those found in the gut mucosa, (2) expressing of adhesion-like proteins, (3) consuming of a high variety of fermentation products by saccharolytic bacteria, and (4) effectively competing for nitrogenous nutrient sources (53). A genome comparison with M. oralis (to our knowledge there has so far been no strain subjected to whole genome sequence analysis) will lead to the discovery of those genes responsible for successful colonization of the human oral cavity versus gastrointestinal tract. However, genome analysis of a few isolated strains may not be sufficient. Given the recognized plasticity of the genomes of most opportunistic pathogenic bacteria (e.g. acquisition of virulence factors or antibiotic resistance mechanisms), it is important to assess the ‘pangenome’ of M. smithii and M. oralis as well by testing a high number of individuals. Since isolation of methanogens is a daunting task and even impossible for most laboratories the clue for gaining more genomic insights will come from metagenomic analysis, once the ever decreasing sequencing costs have reached a level making the testing of a high number of individuals affordable. Such metagenomic analyses for establishing a microbial gene catalogue of the human gut or oral cavity will not only reveal whether or not horizontal gene transfer is common in M. oralis and/or M. smithii but also – once a sufficiently high number of individuals is tested – if certain human physiological disorders are potentially linked with a given archaeal strain (or phylotype).
Phenotype identification strategies
Monitoring interspecies hydrogen transfer via stable isotope probing
The possibility that interspecies hydrogen transfer based on methanogenesis is a virulence mechanism at sites of periodontal disease is unproven so far. If occurring, a close physical contact between methanogens and the complementary syntrophic partner (co-aggregation) is required (54). Such close physical contact has been shown in the case of natural environments where biomass is tightly aggregated, for instance in granular sludge from upflow anaerobic sludge blankets (55). Although the milieu of periodontal pockets provides different and varying conditions, close physical contact in the biofilm should be possible since archaeal-bacterial co-aggregation along with interspecies hydrogen transfer has even been demonstrated in the case of liquid media (54). Crucial for co-aggregation and interspecies hydrogen transfer in the liquid media were the type of substrate (i.e. VFA) and the identity and activity of the syntrophic bacterial partner. Hence, if we wish to find out whether such interactions also occur in the oral cavity, we should find the syntrophic partner(s) and the collaboratively used VFAs. A clue to this could be stable isotope probing (SIP) of different organic compounds such as butyrate or propionate, which has become a popular method to investigate syntrophic methanogen-bacterial interactions in various habitats (e.g. 56, 57). The advantage of SIP is that it provides a link between the phylogeny and function of microbes in a complex community without the need for species isolation. The idea is that the physiologically active subset of organisms within environmental samples (here clinical samples, i.e. periodontal plaque samples), when offered substrates labeled with a stable isotope (typically 13C), would incorporate the heavier C-source into cell components including DNA (Fig. 2). Stable isotopes are the non-radioactive isotopes of elements (e.g. 15N, 18O, 2H, 13C) that do not emit ionizing radiation and thus have the advantage over radioisotope techniques of not affecting the viability of some organisms. After an appropriate time of incubation with the labeled substrate, DNA is extracted and the ‘light’ and ‘heavy’ fractions are physically separated by density-gradient (isopycnic) ultracentrifugation (58). The ‘heavy’ DNA is subjected to PCR amplification of ribosomal RNA and functional genes (e.g. mcrA of methanogens) to assess the identity of those microorganisms that feed on the labeled VFA. If mcrA was identified in the ‘heavy’ DNA, it could be concluded that methanogens were actively involved in the degradation process of VFA and have assimilated the 13C-source into their DNA. In addition, using universal PCR targeting the 16S rRNA gene, those organisms that oxidized the VFA could also be identified. A more advanced approach would even be to look at labeled rRNA (59) instead of DNA because of its higher turn-over rates and because it takes considerable time until DNA is sufficiently labeled up to detectable levels given that DNA-labeling requires cell proliferation, and methanogens are slow-growing organisms.
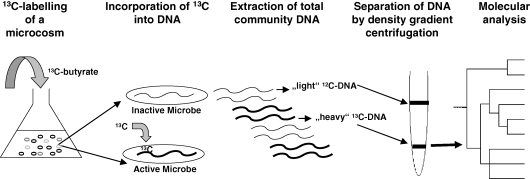
Fig. 2.
Schematic representation of a SIP experiment. Clinical samples (e.g. from periodontal plaque) are incubated anaerobically in a liquid medium with addition of 13C-labelled butyrate as substrate. If syntrophic growth occurs between a butyrate-degrading bacterium and methanogens (via interspecies hydrogen transfer), the labeled 13C will ultimately be assimilated and incorporated into their DNA. After some time of incubation, total DNA is extracted and the ‘heavy’ and ‘light’ fractions are physically separated. Analysis of ‘heavy’ DNA by PCR, cloning, and comparative sequence analysis will then allow identification of the active microbial populations in the microcosm. For further information see main text.
Besides adaptation of this technique from protocols from environmental microbiologists – a major challenge is to achieve in vivo conditions as close as possible when incubating sub-gingival plaque samples in the laboratory. Clearly, sub-gingival plaque samples do not fully resemble the periodontal environment and a laboratory-based study of microbial activity in plaque does not necessarily reflect the true microbial activity at site of infection.
In worst case, SIP experiments might result in changes in the metabolic activity of the flora over time and thus not reflect the natural process at sites of infection. Nonetheless, once methanogenic activities and putative syntrophic partners were assessed through SIP, it shows proof-of-principle and the results could be verified by follow-up experiments. These could be SIP experiments with different types and/or concentration of VFAs and varying incubation times. Furthermore, real-time quantitative PCR directly from DNA from sub-gingival plaque of the same patients could be performed to specifically trace those microorganisms identified via SIP. Such experiments would not only lead to the identification of interacting bacterial partners but also – once established – enable monitoring the dynamics of such partnerships over time through repeated sampling and testing. Using SIP for linking structure and function of human microbial ecosystems is still in its infancy. However, such techniques have been launched for studying the impact of the gut microflora on inflammatory bowel disease (60). It might be worthwhile to consult respective literature regarding technical challenges and possible drawbacks prior to setting up own experiments. Furthermore, isotopically labeled 15N has recently been successfully used to demonstrate that denitrification is an important microbial process taking place in dental plaque, a finding highly encouraging for the experiments proposed here (61).
Monitoring interspecies hydrogen transfer via inhibition of methanogenesis
Another interesting strategy to identify metabolic activity of methanogens in sub-gingival plaque could be its biochemical inhibition. As stated earlier, the end-product of methanogens (CH4) contributes substantially to global warming. In addition CH4 production in ruminants is believed to cause a substantial loss (2–15%) of gross energy intake thereby decreasing productivity (62). Therefore, in order to mitigate the formation of CH4 in anaerobic environments and in order to understand the ecology of its formation, a wide range of inhibiting compounds have been investigated (e.g. 63–65). Clearly, in our case methanogenesis per se is not what we need to prevent in periodontal plaque but H2-consumption. Both reactions however, although at opposite ends of a complex biochemical cascade within methanogens, are tightly linked such that once the final step of CH4 formation is inhibited, the entire process is blocked. For example, 2-bromoethane sulfonate (BES), which is a structural analogue of coenzyme M, is often used to specifically inhibit methanogenesis (63). Coenzyme M is the C1-carrier, required for the final methyl-transfer reactions in the metabolism of methanogens. BES stops CH4 formation and as a consequence should also impede interspecies hydrogen transfer, if existing. If we now consider a SIP experiment as described in the previous paragraph (see also Fig. 2), methanogenesis and degradation of VFAs could be monitored with and without inhibitors. After incubation it could then be tested for the presence of mcrA in the ‘heavy’ DNA. Lack of mcrA in ‘heavy’ DNA versus presence of mcrA in positive controls (i.e. incubated samples without inhibition with BES) would not only demonstrate successful inhibition of methanogenesis but also that H2-consuming methanogenesis has actually taken place in the non-inhibited plaque samples. Such experiments would also provide the framework for the major next step that would have to follow as outlined below.
Final remarks
The above experiments should give substantial confidence/evidence regarding the occurrence of interspecies hydrogen transfer in periodontal plaque samples with M. oralis. As such it could provide an important avenue for novel strategies in the treatment of poly-microbial oral infections. Future research could then be directed at controlling the activity or levels of methanogens at the site of disease. An alluring possibility would be the search for compounds suitable for specifically inhibiting methanogenesis within patients. The intriguing advantage would be that, once methanogenesis is inhibited, this may negatively affect the associated pathogenic flora while simultaneously leaving the physiological flora unaffected. However, unlike in the case of laboratory demonstration or outdoor experiments, use of such inhibitors for therapeutic purposes will of course require substances to be entirely harmless to the host. This means that traditionally used substances like BES are unlikely candidates. However, a better strategy might be to look for further (natural) products, e.g. those produced by plants. It is known, that plants produce a diverse array of plant secondary metabolites (PSM) to protect against microbial and insects attacks. There even exists a number of PSM that are specifically active against methanogenesis. These compounds already hold great promise as natural, safe feed additives alternative to chemical additives to inhibit enteric methanogenesis in ruminants [reviewed in (66)]. These include saponins, tannins, and flavonoids. Since there have been described up to 200,000 different PSM worldwide (67), a biocompatible and effective substance may eventually also be found for interfering with CH4-formation in the oral cavity. However, before we have reached the position to ‘make the case’ against methanogens, more experimental data are needed to definitely understand their role in health and disease.
Conflict of interest and funding
This work was supported by the START program of the Faculty of Medicine, RWTH Aachen University Hospital, Germany.
References
- 1.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrari A, Brusa T, Rutili A, Canzi E, Biavati B. Isolation and characterization of Methanobrevibacter oralis sp. nov. Curr Microbiol. 1994;29:7–12. [Google Scholar]
- 4.Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. Methanogenic archaea and human periodontal disease. Proc Natl Acad Sci USA. 2004;101:6176–81. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vianna ME, Conrads G, Gomes BPFA, Horz HP. T-RFLP-based mcrA gene analysis of methanogenic archaea in association with oral infections and evidence of a novel Methanobrevibacter phylotype. Oral Microbiol Immunol. 2009;24:417–22. doi: 10.1111/j.1399-302X.2009.00539.x. [DOI] [PubMed] [Google Scholar]
- 6.Li CL, Liu DL, Jiang YT, Zhou YB, Zhang MZ, Jiang W, et al. Prevalence and molecular diversity of archaea in subgingival pockets of periodontitis patients. Oral Microbiol Immunol. 2009;24:343–6. doi: 10.1111/j.1399-302X.2009.00514.x. [DOI] [PubMed] [Google Scholar]
- 7.IPCC. Summary for policymakers. In: Solomon S., Qin D., Manning M., Chen Z., Marquis M., Averyt K.B., Tignor M., Miller HL, editors. Climate Change 2007: The Physical Science Basis. Cambridge, UK and New York, NY: Cambridge University Press; 2007. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. [Google Scholar]
- 8.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–91. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 9.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–80. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemp CW, Curtis MA, Robrish SA, Bowen WH. Biogenesis of methane in primate dental plaque. FEBS Lett. 1983;155:61–4. doi: 10.1016/0014-5793(83)80209-9. [DOI] [PubMed] [Google Scholar]
- 11.Robrish SA. Biotechnology and ecological studies on the oral cavity. Microb Ecol. 1986;12:53–64. doi: 10.1007/BF02153222. [DOI] [PubMed] [Google Scholar]
- 12.Robichaux M, Howell M, Boopathy R. Methanogenic activity in human periodontal pocket. Cur Microbiol. 2003;46:53–8. doi: 10.1007/s00284-002-3807-5. [DOI] [PubMed] [Google Scholar]
- 13.Michalke K, Schmidt A, Huber B, Meyer J, Sulkowski M, Hirner MA, et al. Role of intestinal microbiota in transformation of bismuth and other metals and metalloids into volatile methyl and hydride derivatives in humans and mice. Appl Environ Microbiol. 2008;74:3069–75. doi: 10.1128/AEM.02933-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer J, Michalke K, Kouril T, Hensel R. Volatilisation of metals and metalloids: an inherent feature of methanoarchaea? Syst Appl Microbiol. 2008;31:81–7. doi: 10.1016/j.syapm.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Yamabe K, Maeda H, Kokeguchi S, Tanimoto I, Sonoi N, Asakawa S, et al. Distribution of Archaea in Japanese patients with periodontitis and humoral immune response to the components. FEMS Microbiol Lett. 2008;287:69–75. doi: 10.1111/j.1574-6968.2008.01304.x. [DOI] [PubMed] [Google Scholar]
- 16.Vianna ME, Holtgraewe S, Seyfarth I, Conrads G, Horz HP. Quantitative analysis of three hydrogenotrophic microbial groups, methanogenic archaea, sulfate-reducing bacteria, and acetogenic bacteria, within plaque biofilms associated with human periodontal disease. J Bacteriol. 2008;190:3779–85. doi: 10.1128/JB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narihiro T, Terada T, Ohashi A, Wu JH, Liu WT, Araki N, et al. Quantitative detection of culturable methanogenic archaea abundance in anaerobic treatment systems using the sequence-specific rRNA cleavage method. ISME J. 2009;3:522–35. doi: 10.1038/ismej.2009.4. [DOI] [PubMed] [Google Scholar]
- 18.Yamabe K, Maeda H, Kokeguchi S, Soga Y, Meguro M, Naruishi K, et al. Antigenic group II chaperonin in Methanobrevibacter oralis may cross-react with human chaperonin CCT. Mol Oral Microbiol. 2010;25:112–22. doi: 10.1111/j.2041-1014.2009.00548.x. [DOI] [PubMed] [Google Scholar]
- 19.Zugel U, Kaufmann SHE. Immune response against heat shock proteins in infectious diseases. Immunobiol. 1999;201:22–35. doi: 10.1016/s0171-2985(99)80044-8. [DOI] [PubMed] [Google Scholar]
- 20.Zugel U, Kaufmann SHE. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin Microbiol Rev. 1999;12:19–39. doi: 10.1128/cmr.12.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckburg PB, Lepp PW, Relman DA. Archaea and their potential role in human disease. Infect Immun. 2003;71:591–6. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavicchioli R, Curmi PMG, Saunders N, Thomas T. Pathogenic archaea: do they exist? Bioessays. 2003;25:1119–28. doi: 10.1002/bies.10354. [DOI] [PubMed] [Google Scholar]
- 23.Editorial. Cause and effect. Nat Rev Microbiol. 2006;4:414. doi: 10.1038/nrmicro1437. [DOI] [PubMed] [Google Scholar]
- 24.Vianna ME, Conrads G, Gomes BP, Horz HP. Identification and quantification of archaea involved in primary endodontic infections. J Clin Microbiol. 2006;44:1274–82. doi: 10.1128/JCM.44.4.1274-1282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickerman MM, Brossard KA, Funk DB, Jesionowski AM, Gill SR. Phylogenetic analysis of bacterial and archaeal species in symptomatic and asymptomatic endodontic infections. J Med Microbiol. 2007;56:110–8. doi: 10.1099/jmm.0.46835-0. [DOI] [PubMed] [Google Scholar]
- 26.Jiang YT, Xia WW, Li CL, Jiang W, Liang JP. Preliminary study of the presence and association of bacteria and archaea in teeth with apical periodontitis. Int Endod J. 2009;42:1096–103. doi: 10.1111/j.1365-2591.2009.01639.x. [DOI] [PubMed] [Google Scholar]
- 27.Mills LS, Soule ME, Doak DF. The keystone-species concept in ecology and conservation. BioScience. 1993;43:219–24. [Google Scholar]
- 28.Allen EE, Banfield JF. Community genomics in microbial ecology and evolution. Nat Rev Microbiol. 2005;3:489–98. doi: 10.1038/nrmicro1157. [DOI] [PubMed] [Google Scholar]
- 29.Rousk J, Brookes PC, Baath E. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl Environ Microbiol. 2009;75:1589–96. doi: 10.1128/AEM.02775-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ley RE, Daniel A, Peterson DA, Gordon GI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Maukonen J, Motto J, Suihko ML, Saarela M. Intra-individual diversity and similarity of salivary and faecal microbiota. J Med Microbiol. 2008;57:1560–8. doi: 10.1099/jmm.0.47352-0. [DOI] [PubMed] [Google Scholar]
- 32.Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–36. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura N, Lin HC, McSweeney CS, Mackie RI, Gaskins HR. Mechanisms of microbial hydrogen disposal in the human colon and implications for health and disease. Ann Rev Food Science Technol. 2010;1:363–95. doi: 10.1146/annurev.food.102308.124101. [DOI] [PubMed] [Google Scholar]
- 34.Roccarina D, Lauritano EC, Gabrielli M, Franceschi F, Ojetti V, Gasbarrini A. The role of methane in intestinal diseases. Am J Gastroenterol. 2010;105:1250–6. doi: 10.1038/ajg.2009.744. [DOI] [PubMed] [Google Scholar]
- 35.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA. 2006;103:10011–6. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang HS, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu YS, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–70. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abell GCJ, Conlon MA, McOrist AL. Methanogenic archaea in adult human faecal samples are inversely related to butyrate concentration. Microb Ecol Health Dis. 2006;18:154–60. [Google Scholar]
- 38.Avivi-Green C, Polak-Charcon S, Madar Z, Schwartz B. Apoptosis cascade proteins are regulated in vivo by high intracolonic butyrate concentration: correlation with colon cancer inhibition. Oncol Res. 2000;12:83–95. doi: 10.3727/096504001108747558. [DOI] [PubMed] [Google Scholar]
- 39.Zgouras D, Stein J. Butyrate impairs intestinal tumor cell induced angiogenesis by inhibiting HIF-1 alpha nuclear trans-location. Biochem Biophys Res Commun. 2003;300:832–8. doi: 10.1016/s0006-291x(02)02916-9. [DOI] [PubMed] [Google Scholar]
- 40.Scanlan PD, Shanahan F, Marchesi JR. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol. 2008;8 doi: 10.1186/1471-2180-8-79. Article-no. 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mihajlovski A, Alric M, Brugere JF. A putative new order of methanogenic archaea inhabiting the human gut, as revealed by molecular analyses of the mcrA gene. Res Microbiol. 2008;159:516–21. doi: 10.1016/j.resmic.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 42.Mihajlovski A, Dore J, Levenez F, Alric M, Brugere JF. Molecular evaluation of the human gut methanogenic archaeal microbiota reveals an age-associated increase of the diversity. Environ Microbiol Rep. 2010;2:272–80. doi: 10.1111/j.1758-2229.2009.00116.x. [DOI] [PubMed] [Google Scholar]
- 43.Oxley APA, Lanfranconi MP, Würdemann D, Ott S, Schreiber S, McGenity TJ, et al. Halophilic archaea in the human intestinal mucosa. Environ Microbiol. 2010;12:2398–410. doi: 10.1111/j.1462-2920.2010.02212.x. [DOI] [PubMed] [Google Scholar]
- 44.Horz HP, Scheer S, Huenger F, Vianna ME, Conrads G. Selective isolation of bacterial DNA from human clinical specimens. J Microbiol Methods. 2008;72:98–102. doi: 10.1016/j.mimet.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The Human Microbiome Project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dridi B, Henry M, Khechine AE, Raoult D, Drancourt M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS ONE 2009. 4 doi: 10.1371/journal.pone.0007063. Article-ID e7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kubota K, Imachi H, Kawakami S, Nakamura K, Harada H, Ohashi A. Evaluation of enzymatic cell treatments for application of CARD-FISH to methanogens. J Microbiol Methods. 2008;72:54–9. doi: 10.1016/j.mimet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Salonen A, Nikkilä J, Jalanka-Tuovinen J, Immonen O, Rajilić-Stojanović M, Kekkonen RA, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–34. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Rieu-Lesme F, Delbes C, Sollelis L. Recovery of partial 16S rDNA sequences suggests the presence of Crenarchaeota in the human digestive ecosystem. Curr Microbiol. 2005;51:317–21. doi: 10.1007/s00284-005-0036-8. [DOI] [PubMed] [Google Scholar]
- 50.Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol. 2003;5:787–97. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- 51.Burggraf S, Heyder P, Eis N. A pivotal archaea group. Nature. 1997;385:780. doi: 10.1038/385780a0. [DOI] [PubMed] [Google Scholar]
- 52.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–7. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 53.Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA. 2007;104:10643–8. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishii S, Kosaka T, Hori K, Hotta Y, Watanabe K. Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus . Appl Environ Microbiol. 2006;71:7838–45. doi: 10.1128/AEM.71.12.7838-7845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Bok FAM, Plugge CM, Stams AJM. Interspecies electron transfer in methanogenic propionate degrading consortia. Water Res. 2004;38:1368–75. doi: 10.1016/j.watres.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 56.Hatamoto M, Imachi H, Yashiro Y, Ohashi A, Harada H. Detection of active butyrate-degrading microorganisms in methanogenic sludges by RNA-based stable isotope probing. Appl Environ Microbiol. 2008;74:3610–4. doi: 10.1128/AEM.00045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lueders T, Pommerenke B, Friedrich MW. Stable-isotope probing of microorganisms thriving at thermodynamic limits: syntrophic propionate oxidation in flooded soil. Appl Environ Microbiol. 2004;70:5778–86. doi: 10.1128/AEM.70.10.5778-5786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Radajewski S, McDonald IR, Murrell JC. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr Opin Biotechnol. 2003;14:296–302. doi: 10.1016/s0958-1669(03)00064-8. [DOI] [PubMed] [Google Scholar]
- 59.Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol. 2002;68:5367–73. doi: 10.1128/AEM.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barclay AR, Morrison DJ, Weaver LT. What is the role of the metabolic activity of the gut microbiota in inflammatory bowel disease? Probing for answers with stable isotopes. J Pediatr Gastroenterol Nutr. 2008;46:486–95. doi: 10.1097/MPG.0b013e3181615b3a. [DOI] [PubMed] [Google Scholar]
- 61.Schreiber F, Stief P, Gieseke A, Heisterkamp IM, Verstraete W, de Beer D, et al. Denitrification in human dental plaque. BMC Biology. 2010;8 doi: 10.1186/1741-7007-8-24. Article-no. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson KA, Johnson DE. Methane emissions from cattle. J Anim Sci. 1995;73:2483–92. doi: 10.2527/1995.7382483x. [DOI] [PubMed] [Google Scholar]
- 63.Conrad R, Klose M. Selective inhibition of reactions involved in methanogenesis and fatty acid production on rice roots. FEMS Microbiol Ecol. 2007;34:27–34. doi: 10.1111/j.1574-6941.2000.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 64.Ungerfeld EM, Rust SR, Boone DR, Liu Y. Effects of several inhibitors on pure cultures of ruminal methanogens. J Appl Microbiol. 2004;97:520–6. doi: 10.1111/j.1365-2672.2004.02330.x. [DOI] [PubMed] [Google Scholar]
- 65.Lenz M, Janzen N, Lens PNL. Selenium oxyanion inhibition of hydrogenotrophic and acetoclastic methanogenesis. Chemosphere. 2003;73:383–8. doi: 10.1016/j.chemosphere.2008.05.059. [DOI] [PubMed] [Google Scholar]
- 66.Patra AK, Saxena J. A new perspective on the use of plant secondary metabolites to inhibit methanogenesis in the rumen. Phytochemistry. 2010;71:1198–222. doi: 10.1016/j.phytochem.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 67.Hartmann T. From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry. 2007;68:2831–46. doi: 10.1016/j.phytochem.2007.09.017. [DOI] [PubMed] [Google Scholar]