Abstract
Background
Streptococcus mutans surface-protein antigen (SpaP, PAc, or antigen I/II) has been well known to play an important role in initial attachment to tooth surfaces. However, strains with weak SpaP-expression were recently reported to be found in natural populations of S. mutans. The S. mutans gbpC-negative strain Z1, which we previously isolated from saliva and plaque samples, apparently expresses relatively low levels of SpaP protein compared to S. mutans strains MT8148 or UA159.
Objective
To elucidate the mechanism for weak SpaP-expression in this strain, the spaP gene region in strain Z1 was amplified by polymerase chain reaction (PCR) and analyzed.
Methods
Allelic exchange mutants between strains Z1 and UA159 involving the spaP gene region were constructed. The SpaP protein expressed in the mutants was detected with Coomasie Brilliant Blue (CBB)-staining and Western blot analysis following SDS-PAGE.
Results
The 4689 bp spaP gene coding sequence for Z1 appeared to be intact. In contrast, a 20 bp nucleotide sequence appeared to be deleted from the region immediately upstream from the Z1 spaP gene when compared to the same region in UA159. The 216 bp and 237 bp intergenic fragments upstream from the spaP gene, respectively, from Z1 and UA159 were isolated, modified, and transformed into the other strain by allelic replacement. The resultant UA159-promoter region-mutant exhibited extremely weak SpaP-expression similar to that of strain Z1 and the Z1 complemented mutant expressed Spa protein levels like that of strain UA159.
Conclusion
These results suggest that weak SpaP-expression in strain Z1 resulted from a 20 bp-deletion in the spaP gene promoter region.
Keywords: antigen I/II, viridans streptococci, glucan-binding proteins, adhesins, mutant chimeric proteins
Streptococcus mutans is the primary etiologic agent of human dental caries (1). Two major virulence factors, cell surface protein antigen (SpaP/PAc) and glucosyltransferases, have primarily been investigated for their association with cariogenicity of this organism (2). In addition, S. mutans has also been studied in relation to infective endocarditis since this organism has been occasionally isolated from the blood of endocarditis patients (3). Several candidates for virulence factors as well as related molecules of this organism for the disease have been suggested, e.g. fibronectin-binding protein (4), enolase (5), or collagen adhesin (6), and so on (7, 8). Nakano et al. (9) reported that a PAc-defective mutant showed the lowest rate of phagocytosis and suggested that S. mutans clinical isolates with this phenotype may be high-risk strains for the development of bacteremia. Therefore, the SpaP protein may be a candidate for a virulence-related factor in infective endocarditis. In addition, Nakano et al. reported that 5 strains with extremely weak SpaP-expression were found from 45 randomly selected oral strains composed of S. mutans serotypes c, e, and f (15 strains each) in their laboratory collection and that serotype k strains with this phenotype were detected at higher frequency (7/11) (10).
We have identified a collagen adhesin gene cnm from S. mutans strain Z1 selected from our laboratory collection. This strain also exhibited extremely weak SpaP-expression. In the present study, we describe the analysis of the spaP gene regions and elucidate a likely explanation for weak SpaP-expression by constructing mutants harboring chimeric combinations of the spaP genes and their promoter regions.
Materials and methods
Bacterial strains
The S. mutans strains used were Z1 (6), which is serotype f, gbpC-negative, and cnm-positive strain previously isolated from a saliva sample at Tokyo Dental College (6, 11), MT8148 (12), and UA159 (13). The streptococci were maintained on Todd-Hewitt (TH) broth/agar plates. Escherichia coli strain DH5α was routinely used as a host for plasmid construction.
Cell surface protein sample preparation, SDS-PAGE, and Western blot analysis
Preparation of cell surface protein extracts from S. mutans strains was carried out as described previously (14) with a slight modification. Briefly, approximately 20 mg wet weight of early stationary phase streptococcal cells from 10 ml TH cultures were collected, washed, and suspended in SDS-sample buffer (100 mg wet weight/ml), boiled for 5 min, and supernatants were obtained as sample extracts. These streptococcal extracts were subjected to protein assays with the RC DC Protein Assay Reagents (Bio-Rad Laboratories, Inc., Hercules, CA), and frozen until electrophoretic analysis using the Laemmli-gel system. Following SDS-PAGE, the separated proteins were detected by Western blot analysis as described previously (15) with anti-PAc (SpaP) (1/2000 dilution), kindly provided by Y. Yamashita (Kyushu University, Japan), and anti-Cnm sera (1/2500 dilution) (6).
PCR amplification, nucleotide sequencing, sequence analysis, and plasmid construction
The regions corresponding to the surface protein antigen gene spaP (pac) and the upstream regions in Z1 and MT8148 were amplified and directly sequenced as described previously (16). Sequence analyses were carried out with GENETYX-MAC (Genetyx corporation, Tokyo, Japan) program.
Two plasmids for constructing SpaP-expression reporter mutants of S. mutans UA159 and Z1 strains were prepared using plasmid pSY31 derived from pZ63 harboring the scrA::lacZ gene fusion previously described (17). Constructed mutants besides the reporter mutants, plasmids used to construct these mutants, and primers used for amplification of the fragments cloned into these plasmids are summarized in Table 1. Fragments upstream from the spaP genes in UA159 and in Z1 were amplified with primer sets spa56/spaFuR and spa56Z/spaFuR, respectively. These fragments containing spaP upstream regions and the initiation codons of the spaP genes were digested with HindIII and BamHI and inserted into pSY31 digested with the same restriction enzymes to establish spaP::lacZ fusions. The resultant plasmids pZEJ1 and pZEK7 harboring the spaP::lacZ fusion genes were confirmed with restriction digestion and expression of β-galactosidase activities and used to transform S. mutans UA159 and Z1 strains, respectively.
Table 1.
S. mutans Strains, Plasmids, and Primers
| Strain, plasmid, or primer | Description | Reference or source |
|---|---|---|
| Straina | ||
| UA159 | Serotype c | 13 |
| Z1 | Serotype f | 6 |
| EJUA2 | Emr, UA159 transformed with pZEJ1 | This study |
| UKZ4 | Emr, Z1 transformed with pZEK7 | This study |
| E9UA2 | Kanr, UA159 transformed with the fragment amplified from pZE91 with primer P7/P8 | This study |
| E9Z1 | Kanr, Z1 transformed with the fragment amplified from pZE91 with primer P7/P8 | This study |
| E4UA1 | Emr/kans, E9UA2 transformed with the fragment amplified from pZE43 with primer P7/P8 | This study |
| E5UA1 | Emr/kans, E9UA2 transformed with the fragment amplified from pZE55 with primer P7/P8 | This study |
| E4Z1 | Emr/kans, E9Z1 transformed with the fragment amplified from pZE43 with primer P7/P8 | This study |
| Plasmida | ||
| PZ61 | pUC-type Kmr vector containing scrA::lacZ fusion and Emr genes | 17 |
| PSY31 | plasmid to construct lacZ gene fusion with an inserted gene as a HindIII-BamHI fragment, derived from pZ61 | This study |
| PZEJ1 | HindIII-BamHI fragment containing spaP upstream region and the initiation codon of the spaP gene from UA159, cloned into pSY31, resulted in the spaP::lacZ gene fusion | This study |
| PZEK7 | HindIII-BamHI fragment containing spaP upstream region and the initiation codon of the spaP gene from Z1, cloned into pSY31, resulted in the spaP::lacZ gene fusion | This study |
| PZDX2 | pUC19 containing fragment p40f3U and kanr gene | This study |
| PZDZ1 | pUC19 containing fragment p40f3U and Emr gene | This study |
| PZE91 | pUC19 containing fragment p40f3U, kanr gene, and fragment spa5U | This study |
| PZE43 | pUC19 containing fragment p40f3U, Emr gene, and fragment IGspa5U | This study |
| pZE55 | pUC19 containing fragment p40f3U, Emr gene, and fragment IGspa5Z | This study |
| Primerb | ||
| Spa56 | 5′-ATAAGCTTCCAGCACTCAAGGCA-3′, forward primer to amplify spaP upstream region from UA159 | This study |
| Spa56Z | 5′-ATAAGCTTCCAGCAATCAAGACATAAA-3′, forward primer to amplify spaP upstream region from Z1 | This study |
| SpaFuR | 5′-TAGGATCCCATAAATCCTCCAAATCTGAATAAATCTT-3′, reverse primer to amplify spaP upstream regions and the initiation codons of the spaP genes from UA159 and Z1 | This study |
| Spa31 | 5′-TCAATCAATGATAATATAACGACGAATAC-3′, reverse primer to amplify 444 bp fragment p40f3U (–681 to –238 in Fig. 2) | This study |
| Spa53 | 5′-AAAACACTGTGTGGTGCTGTTCTA-3′, forward primer to amplify 475 bp fragment spa5U (96 to 570 in Fig. 2) | This study |
| Spa32 | 5′-TAATGAATTCAACCTCGGCTTTATGAGCT-3′, reverse primer to amplify 475 bp fragment spa5U (96 to 570 in Fig. 2), 807 bp fragment IGspa5U (–237 to 570 in Fig. 2), and 786 bp fragment IGspa5Z (–216 to 570 in Fig. 2) | This study |
| Spa52 | 5′-AGTAAAAAAGGTTAGGATGACAAAATCCT-3′, forward primer to amplify 807 bp fragment IGspa5U (−237 to 570 in Fig. 2) and 786 bp fragment IGspa5Z (−216 to 570 in Fig. 2) | This study |
| P7 | 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′, used as a forward primer to amplify fragments p40f3U::kanr::spa5U, p40f3U::Emr::IGspa5U, and p40f3U::Emr::IGspa5Z | Toyobo, Osaka, Japan |
| P8 | 5′-AGCGGATAACAATTTCACACAGGAAAC-3′ used as a reverse primer to amplify fragments p40f3U::kanr::spa5U, p40f3U::Emr::IGspa5U, and p40f3U::Emr::IGspa5Z | Toyobo, Osaka, Japan |
Emr, erythromycin-resistant phenotype; kanr/kans, kanamycin resistant/sensitive phenotype in S. mutans; Kmr, kanamycin resistant phenotype in E. coli
Underlines, restriction enzyme (HindIII/BamHI/EcoRI) recognition sequences.
Several other plasmids were constructed for subsequent transformation of S. mutans to construct chimera-chromosome mutants, following manipulations of plasmid pUC19 with a kanamycin resistant gene BamHI cassette (kanr), an erythromycin resistant gene BamHI cassette (Emr), the 3′-portion of the p40f (SMU.609) gene (18) from UA159 (fragment p40f3), and the 5′-end of the spaP gene with or without the intergenic regions from either UA159 and Z1.
The PCR amplification was performed with high fidelity DNA polymerase KOD-Plus (Toyobo, Osaka, Japan) throughout this study. A 444 bp fragment, p40f3U (−681 to −238 in Fig. 2), was amplified from UA159 genomic DNA using primers spa56 and spa31. A 475 bp fragment, spa5U (96 to 570 in Fig. 2), was amplified from UA159 genomic DNA using primers spa53 and spa32 (containing an EcoRI site at the 5′ end). This fragment corresponds to the 5′ region of the spaP gene devoid of first 15 codons. An 807 bp fragment, IGspa5U (−237 to 570 in Fig. 2) was amplified from UA159 genomic DNA using primers spa52 and spa32. A 786 bp fragment, IGspa5Z (−216 to 570 in Fig. 2), was amplified from Z1 genomic DNA using primers spa52 and spa32.
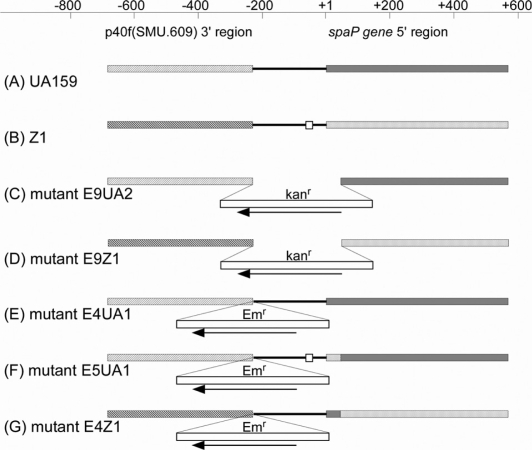
Fig. 2.
Gene arrangements of parental strains and their mutants. Chromosomal DNA regions corresponding to the DNA fragments used for transformation of S. mutans to construct the mutants are depicted in the figure and these regions in the five mutants are drawn with their background DNA, since we cannot identify the actual recombination points between the two homologous sequences. Positions and directions of the Kanr and Emr cassettes are indicated but their sizes are not drawn to scale. Open squares in (B) and (F) indicate 20 bp deletions, sparsely and densely doted small boxes in (F) and (G) indicate the first 15 identical codons of the spaP genes in strains Z1 and UA159.
HindIII digested p40f3U was initially cloned into HindIII/HincII digested pUC19 and then the kanr or Emr BamHI cassettes were inserted into the BamHI site of this plasmid. The resultant plasmids were designated as pZDX2 (p40f3U::kanr) and pZDZ1 (p40f3U::Emr), respectively. In addition, EcoRI-digested fragments spa5U, IGspa5U, and IGspa5Z were cloned into SmaI/EcoRI digested pUC19. The resultant plasmids were digested with HindIII/HincII, ligated with HindIII/SmaI p40f3U::kanr or p40f3U::Emr fragments excised from pZDX2 or pZDZ1, and screened with kanamycin- or erythromycin-resistant markers. As a result, three plasmids were finally constructed and designated as pZE91 (p40f3U::kanr::spa5U), pZE43 (p40f3U::Emr::IGspa5U), and pZE55 (p40f3U::Emr::IGspa5Z).
Construction of reporter mutants, mutants with chimeric chromosomes, and β-galactosidase assays
Plasmids pZEJ1 and pZEK7 were used to transform S. mutans strains UA159 and Z1, respectively, by procedures routinely carried out in this laboratory (19), and erythromycin-resistant colonies were selected as mutants EJUA2 and EKZ4 from which the chromosomal region corresponding to the spaP::lacZ fusion genes and upstream regions were amplified by PCR and sequenced to confirm the constructs.
Linear DNA fragments were amplified using primers P7 and P8 (Toyobo, Osaka, Japan) with plasmids pZE91, pZE43, and pZE55 as templates. The amplicons were used to transform S. mutans by procedures described above (19). An amplicon from pZE91 containing the p40f3U::kanr::spa5U fragment was transformed into S. mutans UA159 or Z1, and kanamycin-resistant colonies were selected as mutants E9UA2 and E9Z1. These mutants were further used as hosts for transformation with fragments p40f3U::Emr:: IGspa5U or p40f3U::Emr:: IGspa5Z amplified with pZE43 or pZE55, Emr/kans colonies were selected and mutants E4UA1, E5UA1, and E4Z1 were obtained. The chromosomal regions corresponding to the spaP genes and upstream regions were amplified by PCR and confirmed by sequencing with these constructs. Chromosomal rearrangements around the spaP5′ and upstream region in mutants are summarized in Fig. 2 with UA159 and Z1.
Reporter mutant cells were disrupted using Tissue Lyser (Qiagen, Hilden, Germany) with 0.2 mm Φ ceramics beads. Protein concentrations of cell free extracts were determined with DC Protein Assay Reagents (Bio-Rad Laboratories). Beta-galactosidase activities of these mutants were assayed according to the DyNA Quant Application Note 2 (GE Healthcare Global Headquarters, Pollards Wood, UK) using a DyNA Quant 200 fluorophotometer and 4-methylumbelliferyl-β-galactoside as a substrate.
Results
SpaP-expression and sequence analysis of spaP upstream regions
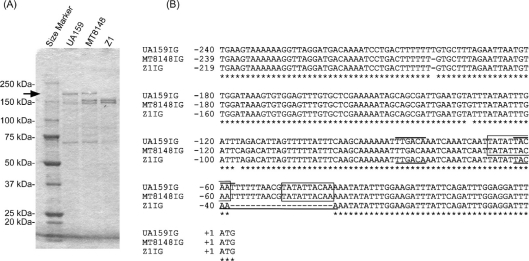
The S. mutans cell surface proteins were isolated from strains UA159, MT8148, and Z1 for analysis with SDS-PAGE as described under the Materials and methods section. A 190 kDa protein band corresponding to the SpaP protein was detected in UA159 and MT8148 but not observed in Z1 in Coomasie Brilliant Blue (CBB)-stained polyacrylamide gels (Fig. 1A). The entire spaP gene sequence of Z1 was determined (Acc#: AB516671) and an intact 4689 bp spaP gene coding sequence of Z1 was 99 and 97% identical to those of UA159 and MT8148, respectively. Therefore, weak SpaP-expression in Z1 as a cell wall protein was not the result of a truncation mutation within the coding sequence as reported for strain GS-5 (14). We subsequently determined the 216 bp and 236 bp (Acc#: AB548069) intergenic sequences upstream from the spaP genes, respectively, in both Z1 and MT8148. Multiple aligned sequence comparisons with UA159 were determined with the DDBJ ClustalW program (Fig. 1B). The region (−116 to −1) conserved between UA159 and MT8148 was distinct from that corresponding to Z1, where a 20 bp deletion was detected (open square in Fig. 2B).
Fig. 1.
CBB-stained polyacrylamide gel and intergenic sequence alignment. (A) SDS sample preparation was carried out as described under the Materials and methods. 0.72, 0.76, 0.86 µg protein of samples, respectively, from strains UA159, MT8148, and Z1 were applied to a 5–20% gradient SDS-polyacrylamide gel. The arrow indicates the positions of the 190 kDa bands corresponding to the SpaP proteins. (B) Depicted multiple aligned intergenic (IG) sequences start with the termination codons of the p40f (SMU.609) genes and end with the ATG initiation codons of the spaP genes in UA159IG, MT8148IG (AB548069), and Z1IG (AB516671) sequences. Each sequence is numbered +1 at the first nucleotide of the spaP ATG codon. Lines over the UA159IG and lines under the Z1IG sequences, respectively, indicate −35 and −10 regions of the putative promoter sequences. Boxes indicate direct repeat sequences.
SpaP-expression reporter mutants, EJUA2 and EKZ4, were used to quantify the difference in SpaP-expression between UA159 and Z1. Beta-galactosidase activities of mutants EJUA2 and EKZ4 were, respectively, 1.54±1.09 (n =6) and 0.0167±0.0079 (n =6). SpaP-expression in UA159 was deemed to be two orders of magnitude higher compared to that of Z1.
These results suggested that a sequence responsible for weak SpaP-expression in Z1 was contained within the 20 bp deleted region. In order to confirm this hypothesis, we constructed UA159- and Z1-mutants, respectively, harboring putative Z1 and UA159 spaP-promoters and examined the properties of these mutants.
Mutants with chimeric chromosomes and their SpaP-expression levels
In order to introduce the entire intergenic region between the p40f and spaP genes from Z1 into the corresponding region of UA159 by allelic exchange, we initially constructed a mutant E9UA2 (Fig. 2C), in which the intergenic region and the first 15 codons of the spaP gene were deleted from the chromosome following transformation with the fragment, p40f3U::kanr::spa5U. Using this mutant, the following mutants (E4UA1 and E5UA1, Fig. 2E and F) were constructed. A series of fragments used for these transformation procedures were amplified from the plasmids constructed as described under the Materials and methods section. Gene arrangements around the intergenic regions in these mutants as well as parental strains UA159 and Z1 are depicted in Fig. 2. Mutant E4UA1 was constructed as a control for E5UA1. Mutant E4Z1 was constructed from E9Z1 (Fig. 2D), which was constructed from Z1 with the same p40f3U::kanr::spa5U fragment used to construct E9UA2 from UA159. In order to minimize polar effects, the direction of transcription for the Emr gene was designed to be opposite that of the spaP gene in all mutants (Fig. 2). These arrangements should result in normalized SpaP-expression in these mutants even though it was possible that some promoter activity could still exist in the orientation opposite to that of the Emr gene promoter.
The N-terminal 190 SpaP amino acid sequence encoded by the fragment IGspa5U was completely identical to that encoded by the fragment IGspa5Z, although three nucleotide substitutions had occurred in the 570nt spaP 5′ coding sequences in both fragments. Therefore, E4UA1 and E5UA1 essentially harbor the same SpaP protein sequences as do UA159. The E4Z1 also expresses the same sequence of the SpaP protein as does strain Z1.
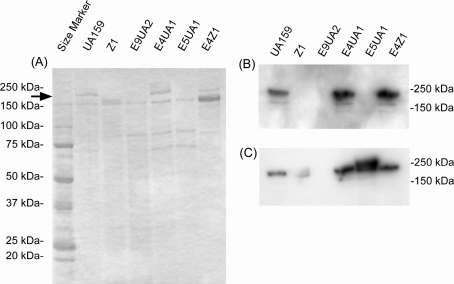
The SpaP-expression of these mutants and their parental strains was examined following CBB staining of polyacrylamide gels containing resolved cell surface protein extracts (Fig. 3A) and was determined by Western blotting analysis (Fig. 3B) with anti-PAc (SpaP) serum. A 190 kDa protein band corresponding to SpaP of E5UA1 was not detected in the CBB stained gel and Western blotting analysis similar to strain Z1 and the spaP-negative mutant E9UA2 (Fig. 3A). This weak expression did not result from insertion of the Emr gene cassette into a position immediately downstream of putative p40f gene because E4UA1, which has the same insertion arrangement of Emr gene cassette as E5UA1, expressed SpaP protein to the same extent as UA159. In addition, E4Z1 that harbors the intergenic region originating from UA159 expressed SpaP protein at similar levels as UA159. Since 190 kDa SpaP bands in E5UA1 and Z1 were not detected in the CBB stained gel and since the SpaP-expression level in UA159 presumed to be two orders of magnitude higher compared to that of Z1 was indicated by the β-galactosidase reporter assay, 12 ng protein (1/100 diluted) of samples from E4Z1, E4UA1, UA159, and 1.2 µg protein of samples from E5UA1 and Z1 were applied to the polyacrylamide gel for Western blotting. The analysis with anti-PAc (SpaP) serum (Fig. 3C) following SDS-PAGE indicated that anti-SpaP serum reactive bands of 1/100 diluted samples from E4Z1, E4UA1, and UA159 were almost the same extent as those of one fold amount of samples from E5UA1 and Z1. In addition to SpaP-expression, the chromosome background of E4Z1 was confirmed by Western blot analysis with anti-Cnm (collagen adhesin) serum as a strain-specific marker for Z1 (data not shown).
Fig. 3.
CBB-stained polyacrylamide gel (A) and Western blot analyses (B, C) of mutants and their parental strains. (A) Samples (1.2 µg protein) prepared as described under Materials and methods were applied to the 5–20% gradient SDS-polyacrylamide gel. (B) Diluted samples (12 ng protein) were applied to the gel for the following Western blotting analysis. Anti-PAc (SpaP) serum diluted 1:2000 was used as the first antibody. The ECL Plus Western blotting reagents (GE Healthcare, Buckinghamshire, UK) were used for detection. (C) Samples for Z1, E9UA2, and E5UA1 (1.2 µg) as well as for UA159, E4UA1, and E4Z1 (12 ng) were applied to the gel and determined by Western blotting analysis.
These results suggest that the intergenic region, specifically the 20 bp deletion within the upstream spaP region in Z1, affected SpaP-expression.
Discussion
The S. mutans SpaP protein has been well known to play a role in saliva-mediated aggregation and initial attachment to tooth surfaces since the late 1980s (20). SpaP protein is a predominant surface protein and is readily detected in CBB-stained polyacrylamide gels under a variety of growth conditions in several other strains examined in this laboratory besides UA159 and MT8148. A simple method to prepare cell surface protein samples was utilized in the present study. However, the weak SpaP-expression observed in strain Z1 and mutant E5UA1 does not result from a failure of cell wall protein extraction. This is suggested since we have previously reported that, in contrast to the UA159 fraction, a 190 kDa protein band corresponding to the SpaP protein of S. mutans was only faintly observed in a Z1 wall-enriched fraction isolated following N-acetylmuramidase digestion (11). In addition, S. mutans strains exhibiting the weak SpaP-expression phenotype were detected among those naturally occurring in the oral cavity with lower frequency in serotypes c, e, f, and higher in serotype k strains (10). Therefore, our finding that Z1 did not express this protein at levels comparable to strain UA159 or MT8148 is consistent with these earlier studies.
Our results suggest that low level SpaP-expression is ascribed to a 20 bp nucleotide deletion from the region immediately upstream of the spaP gene. The distinct SpaP phenotypes observed for E4UA1 and E5UA1 in a UA159 genome background as well as the E4Z1 phenotype also support our hypothesis. However, we cannot formally exclude another possible mechanism for this phenotype, i.e. that the 20 bp nucleotide deletion was not detected in the region of chromosome of the weak SpaP-expressing strains. In addition, it is unlikely that all of the previously reported low level SpaP-expression strains harbor the same 20 bp nucleotide deletion since S. mutans is known for its genetic diversity. Three of seven such serotype k strains have mutations within the spaP genes (10), whose positions were distinct from that in strain GS-5 (14). However, five other strains (2 serotype c, 3 serotype f) with phenotypes (GbpC–, Cnm+) very similar to Z1 have also been detected in approximately 100 of our stock culture samples and these harbored the same 20 bp deletion as Z1 (unpublished data).
The SpaP-reactive bands in strains Z1 and E5UA1 shown in Fig. 3C were not as distinct as with the other strains. A minor fraction of post-translationally unprocessed protein or chemically modified proteins may have been included in these SDS samples prepared using our procedure. The amounts of UA159, E4UA1, and E4Z1 applied as samples in the Western blot analysis were 1/100 diluted compared to those of Z1 and E5UA1. Therefore, minor fractions in the former samples may not have detected. Anti-SpaP serum reactive bands in the mutants were detected at slightly higher levels than those in their parental strains. This may have resulted from additional transcription from promoter activities within the Emr fragment, although the Emr gene is arranged in the opposite orientation relative to the spaP genes. However, further analyses are necessary to resolve these questions. The approximately 150 kDa bands in Fig. 3A corresponded to glucosyltransferases (Gtf) B and C according to our previous Western blotting results and banding pattern difference between UA159/E9UA2/E4UA1/E5UA1 and Z1/E4Z1 very likely resulted from a difference in the genomic backgrounds between strains UA159 and Z1. However, a reason for difference in 150 kDa band intensities between Z1 and E4Z1 is presently unknown, since Gtf expression of E4Z1 should not have been affected by the introduced mutation.
The SpaP molecules consist of several domains including the P-region (12, 21). Another report concerning SpaP-expression/regulation indicated that the spaP gene lacking its P-region nucleotides resulted in a complete absence of surface-localized SpaP protein, although the reason for this is still unknown (22). However, this is not a finding affecting our hypothesis because the Z1 spaP gene is intact like that of UA159.
The UA159 intergenic sequence corresponding to the 20 bp deletion (−58 to −39) overlapped with the presumed spaP-promoter (-86 TTGACA-N17-TACAAT -59, lines over the sequences in Fig. 1B). This deleted sequence was likely changed to a low activity promoter (-66 TTGACA-N17-TACAAA -39, lines under the sequences in Fig. 1B) resulting from the 20 bp deletion in Z1. Furthermore, 10 bp direct repeats were detected overlapping with the spaP promoter regions in UA159 and MT8148 (boxes in Fig. 1B). Unequal crossover during DNA replication or homologous recombination involving the 10 bp direct repeats may have occurred in Z1 under certain selective pressures in the past. However, further experiments are required to determine whether the single nucleotide substitution in the −10 region or other factors are involved in the change to a low activity promoter. This line of work is currently in progress by employing a site-directed mutagenesis.
Strain Z1 is cnm-positive and gbpC-negative as described previously (6) in addition to the SpaP phenotype. How Z1 could have survived in the oral cavity with the GbpC-negative and weak SpaP phenotype is an interesting question. Inactivation of the S. mutans spaP resulted in a decrease in gp340-mediated cell aggregation that contributes to the clearance of S. mutans cells from the oral cavity in vivo (20, 23). Therefore, cells with low SpaP levels may be able to escape from such clearance and can attach to teeth by glucosyltransferases-mediated adherence mediated by adhesive glucans as suggested by Lemos et al. (24). Extremely low level SpaP expressing strains have been isolated as described above. A PAc-defective mutant showed the lowest rate of phagocytosis and suggested that S. mutans clinical isolates with this phenotype may be high-risk strains for the development of bacteremia (9). One of the strains isolated from endocarditis patients was reported to be cnm-positive (25) and gbpC-defective (26), although the mutation positions in the gbpC genes in these two strains were distinct. Further analyses for virulent phenotypes or genotypes of strains involved in infective endocarditis are necessary to further delineate the role of these factors in virulence.
Acknowledgements
We thank Y. Yamashita (Kyushu University, Fukuoka, Japan) for providing anti-PAc serum, and H. K. Kuramitsu (State University of New York at Buffalo, NY) for critical reading of the manuscript.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors. This investigation was supported in part by the ‘High-Tech Research Center’ Project for Private Universities: matching fund subsidy from the Ministry of Education, Culture, Sports, Science and Technology, 2006–2011.
References
- 1.Loesche WJ. Role of Streptococcus mutans in human dental caries. Microbiol Rev. 1986;50:353–80. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banas JA. Virulence properties of Streptococcus mutans . Front Biosci. 2004;9:1267–77. doi: 10.2741/1305. [DOI] [PubMed] [Google Scholar]
- 3.Nemoto H, Nakano K, Nomura R, Ooshima T. Molecular characterization of Streptococcus mutans strains isolated from the heart valve of an infective endocarditis patient. J Med Microbiol. 2008;57:891–5. doi: 10.1099/jmm.0.47836-0. [DOI] [PubMed] [Google Scholar]
- 4.Jung CJ, Zheng QH, Shieh YH, Lin CS, Chia JS. Streptococcus mutans autolysin AtlA is a fibronectin-binding protein and contributes to bacterial survival in the bloodstream and virulence for infective endocarditis. Mol Microbiol. 2009;74:888–902. doi: 10.1111/j.1365-2958.2009.06903.x. [DOI] [PubMed] [Google Scholar]
- 5.Jones MN, Holt RG. Cloning and characterization of an α-enolase of the oral pathogen Streptococcus mutans that binds human plasminogen. Biochem Biophys Res Commun. 2007;364:924–9. doi: 10.1016/j.bbrc.2007.10.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato Y, Okamoto K, Kagami A, Yamamoto Y, Igarashi T, Kizaki H. Streptococcus mutans strains harboring collagen-binding adhesin. J Dent Res. 2004;83:534–9. doi: 10.1177/154405910408300705. [DOI] [PubMed] [Google Scholar]
- 7.Han TK, Zhang C, Dao ML. Identification and characterization of collagen-binding activity in Streptococcus mutans wall-associated protein: a possible implication in dental root caries and endocarditis. Biochem Biophys Res Commun. 2006;343:787–92. doi: 10.1016/j.bbrc.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 8.Nagata E, Okayama H, Ito HO, Yamashita Y, Inoue M, Oho T. Serotype-specific polysaccharide of Streptococcus mutans contributes to infectivity in endocarditis. Oral Microbiol Immunol. 2006;21:420–3. doi: 10.1111/j.1399-302X.2006.00317.x. [DOI] [PubMed] [Google Scholar]
- 9.Nakano K, Tsuji M, Nishimura K, Nomura R, Ooshima T. Contribution of cell surface protein antigen PAc of Streptococcus mutans to bacteremia. Microbes Infect. 2006;8:114–21. doi: 10.1016/j.micinf.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Nakano K, Nomura R, Nemoto H, Lapirattanakul J, Taniguchi N, Gronroos L, et al. Protein antigen in serotype k Streptococcus mutans clinical isolates. J Dent Res. 2008;87:964–8. doi: 10.1177/154405910808701001. [DOI] [PubMed] [Google Scholar]
- 11.Sato Y, Okamoto K, Kagami A, Yamamoto Y, Ohta K, Igarashi T, et al. Application of in vitro mutagenesis to identify the gene responsible for cold agglutination phenotype of Streptococcus mutans . Microbiol Immunol. 2004;48:449–56. doi: 10.1111/j.1348-0421.2004.tb03535.x. [DOI] [PubMed] [Google Scholar]
- 12.Okahashi N, Sasakawa C, Yoshikawa M, Hamada S, Koga T. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans, implicated in dental caries. Mol Microbiol. 1989;3:673–8. doi: 10.1111/j.1365-2958.1989.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 13.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99:14434–9. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami Y, Nakano Y, Yamashita Y, Koga T. Identification of a frameshift mutation resulting in premature termination and loss of cell wall anchoring of the PAc antigen of Streptococcus mutans GS-5. Infect Immun. 1997;65:794–7. doi: 10.1128/iai.65.2.794-797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto-Shibayama K, Sato Y, Yamamoto Y, Ohta K, Kizaki H. Identification of a glucan-binding protein C gene homologue in Streptococcus macacae. Oral Microbiol Immunol. 2006;21:32–41. doi: 10.1111/j.1399-302X.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- 16.Sato Y, Okamoto-Shibayama K, Takada K, Igarashi T, Hirasawa M. Genes responsible for dextran-dependent aggregation of Streptococcus sobrinus strain 6715. Oral Microbiol Immunol. 2009;24:224–30. doi: 10.1111/j.1399-302X.2008.00499.x. [DOI] [PubMed] [Google Scholar]
- 17.Sato Y, Yamamoto Y, Suzuki R, Kizaki H, Kuramitsu HK. Construction of scrA::lacZ gene fusions to investigate regulation of the sucrose PTS of Streptococcus mutans . FEMS Microbiol Lett. 1991;79:339–45. doi: 10.1111/j.1574-6968.1991.tb04552.x. Available from: http://onlinelibrary.wiley.com/doi/10.1111/fml.1991.79.issue-2-3/issuetoc. [DOI] [PubMed] [Google Scholar]
- 18.Catt DM, Gregory RL. Streptococcus mutans murein hydrolase. J Bacteriol. 2005;187:7863–5. doi: 10.1128/JB.187.22.7863-7865.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry D, Kuramitsu HK. Genetic transformation of Streptococcus mutans . Infect Immun. 1981;32:1295–7. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SF, Progulske-Fox A, Erdos GW, Piacentini DA, Ayakawa GY, Crowley PJ, et al. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II) Infect Immun. 1989;57:3306–13. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly C, Evans P, Bergmeier L, Lee SF, Progulske-Fox A, Harris AC, et al. Sequence analysis of the cloned streptococcal cell surface antigen I/II. FEBS Lett. 1989;258:127–32. doi: 10.1016/0014-5793(89)81632-1. [DOI] [PubMed] [Google Scholar]
- 22.Brady LJ, Cvitkovitch DG, Geric CM, Addison MN, Joyce JC, Crowley PJ, et al. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect Immun. 1998;66:4274–82. doi: 10.1128/iai.66.9.4274-4282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koga T, Okahashi N, Takahashi I, Kanamoto T, Asakawa H, Iwaki M. Surface hydrophobicity, adherence, and aggregation of cell surface protein antigen mutants of Streptococcus mutans serotype c. Infect Immun. 1990;58:289–96. doi: 10.1128/iai.58.2.289-296.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr Issues Mol Biol. 2005;7:95–107. [PubMed] [Google Scholar]
- 25.Nakano K, Nomura R, Taniguchi N, Lapirattanakul J, Kojima A, Naka S, et al. Molecular characterization of Streptococcus mutans strains containing the cnm gene encoding a collagen-binding adhesin. Arch Oral Biol. 2010;55:34–9. doi: 10.1016/j.archoralbio.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Nakano K, Matsumura M, Kawaguchi M, Fujiwara T, Sobue S, Nakagawa I, et al. Attenuation of glucan-binding protein C reduces the cariogenicity of Streptococcus mutans: analysis of strains isolated from human blood. J Dent Res. 2002;81:376–9. doi: 10.1177/0810376. [DOI] [PubMed] [Google Scholar]