Abstract
Our goal was to gain a better understanding of the contribution of hydrophobic interactions to protein stability. We measured the change in conformational stability, Δ(ΔG), for hydrophobic mutants of four proteins: villin head piece subdomain (VHP) with 36 residues, a surface protein from Borrelia burgdorferi (VlsE) with 341 residues, and two proteins previously studied in our laboratory, ribonucleases Sa and T1. We compare our results with previous studies and reach the following conclusions. 1. Hydrophobic interactions contribute less to the stability of a small protein, VHP (0.6 ± 0.3 kcal/mole per –CH2– group), than to the stability of a large protein, VlsE (1.6 ± 0.3 kcal/mol per –CH2– group). 2. Hydrophobic interactions make the major contribution to the stability of VHP (40 kcal/mol) and the major contributors are (in kcal/mol): Phe 18 (3.9), Met 13 (3.1), Phe 7 (2.9), Phe 11 (2.7), and Leu 21 (2.7). 3. Based on Δ(ΔG) values for 148 hydrophobic mutants in 13 proteins, burying a –CH2– group on folding contributes, on average, 1.1 ± 0.5 kcal/mol to protein stability. 4. The experimental Δ(ΔG) values for aliphatic side chains (Ala, Val, Ile, and Leu) are in good agreement with their ΔGtr values from water to cyclohexane. 5. For 22 proteins with 36 to 534 residues, hydrophobic interactions contribute 60 ± 4% and hydrogen bonds 40 ± 4% to protein stability. 6. Conformational entropy contributes about 2.4 kcal/mol per residue to protein instability. The globular conformation of proteins is stabilized predominately by hydrophobic interactions.
Keywords: Hydrophobic interactions, hydrogen bonds, conformational entropy, protein stability, large proteins, small proteins
Introduction
By the late 1930’s, the structure of globular proteins was beginning to be understood and Bernal concluded: 1 “Ionic bonds are plainly out of the question ··· the hydrophobe groups of the proteins must hold it together. ··· the protein molecule in solution must have its hydrophobe groups out of contact with water, that is, in contact with each other, ··· In this way a force of association is provided which is not so much that of attraction between hydrophobe groups, which is always weak, but that of repulsion of the groups out of the water medium.” The importance of this suggestion was not widely appreciated until 1959 when Kauzmann’s influential review was published. 2 He presented convincing evidence that: “The hydrophobic bond is probably one of the more important factors involved in stabilizing the folded configuration in many native proteins.” This kindled Tanford’s long-term interest in the hydrophobic effect and he used the limited model compound data available in 1962 to show that: 3 “··· the stability of the native conformation in water can be explained ··· entirely on the basis of the hydrophobic interactions of the non-polar parts of the molecule.” Tanford has written an interesting history of the hydrophobic effect. 4
In 1971, Tanford and Nozaki published the first hydrophobicity scale, using ethanol as a model for the interior of the protein.5 Other hydrophobicity scales followed using octanol 6, 7, N-methylacetamide,8 and cyclohexane 9 as models of the protein interior. The applications and reliability of these scales have been discussed.10–13
In 1987, Yutani’s group was the first to gain a better understanding of the contribution of the hydrophobic effect to protein stability, by studying hydrophobic mutants of tryptophan synthase α subunit (Trp Syn α).14 This was followed by similar studies of other proteins: barnase (Ba) 15–17; staph nuclease (SN) 18, 19; gene V protein20; T4 lysozyme (T4 Lyso) 21–23; chymotrypsin inhibitor 2 (CI2) 24; FK506-binding protein (FKBP);25 human lysozyme (hLyso)26; fibronectin type III domains 27; ubiquitin 28; Sac7d and Sso7d 29; and apoflavodoxin (ApoFlavo) 30. These studies showed that the contribution of a buried nonpolar side chain to protein stability depends on two factors: first, the hydrophobicity of the side chain and the amount of side chain surface area removed from contact with water when the protein folds, and, second, the van der Waals interactions of the side chain when the protein is folded and unfolded. The importance of hydrophobicity had long been recognized 2, 31, but it became clear only later that van der Waals interactions may make an even larger contribution because of the tight packing in the interior of folded proteins. 19, 28, 32, 33
It seems likely that the stabilizing and destabilizing forces that contribute to protein stability will depend on protein size. As globular proteins become smaller, it will not be possible to completely bury hydrophobic side chains, and the charged side chains will be closer together and potentially in different environments than in a larger protein. In addition, the denatured state ensembles might depend on the size of the protein and this could influence protein stability.
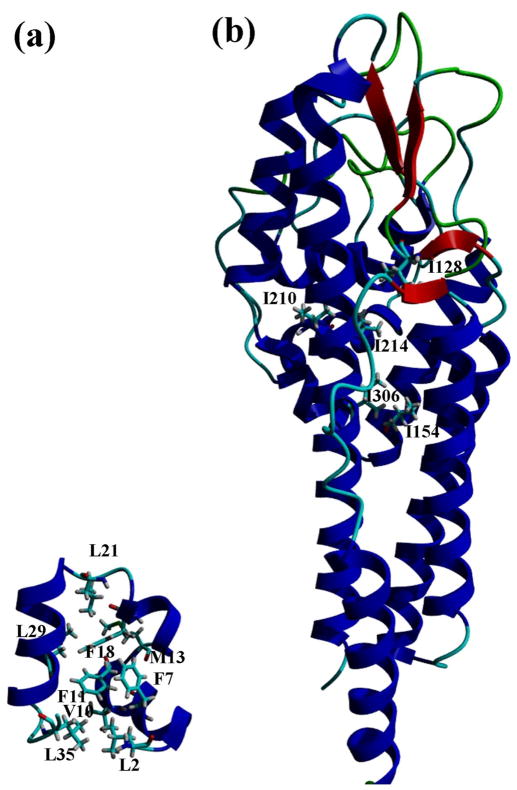
In this paper, we examine the contribution of hydrophobic interactions to the stability of a small protein, villin headpiece subdomain (VHP) with 36 residues34, 35, and of a large protein, Borrelia burgdorferi protein (VlsE) with 341 residues.36, 37 The mutants studied are superimposed on their structures in Fig. 1. In addition, we have studied the contribution of hydrophobic interactions to the stability of two proteins previously studied in our laboratory, ribonucleases Sa (RNase Sa) and T1 (RNase T1). Aided by previous results, these new data allow us to gain an improved understanding of the contribution of hydrophobic interactions to protein stability.
Fig. 1.
Ribbon diagrams of the VHP (a) and VlsE (b) structures showing the mutated residues. The figures were generated using the YASARA program103 and the crystal structures of VHP (PDB code: 1YRI) 35 and VlsE (PDB code: 1L8W) 37.
Results
VlsE
Jones and Wittung-Stafshede previously studied the denaturation of VlsE by urea and guanidine hydrochloride (GuHCl), and found it to be completely reversible, as do we for urea denaturation. 37 They followed the unfolding with both fluorescence and circular dichroism and got identical results which is consistent with a two-state folding mechanism. They also reported kinetic data that were consistent with a two-state mechanism. Based on their results and conclusions, we have assumed a two-state folding mechanism for VlsE for the analysis of our results.37
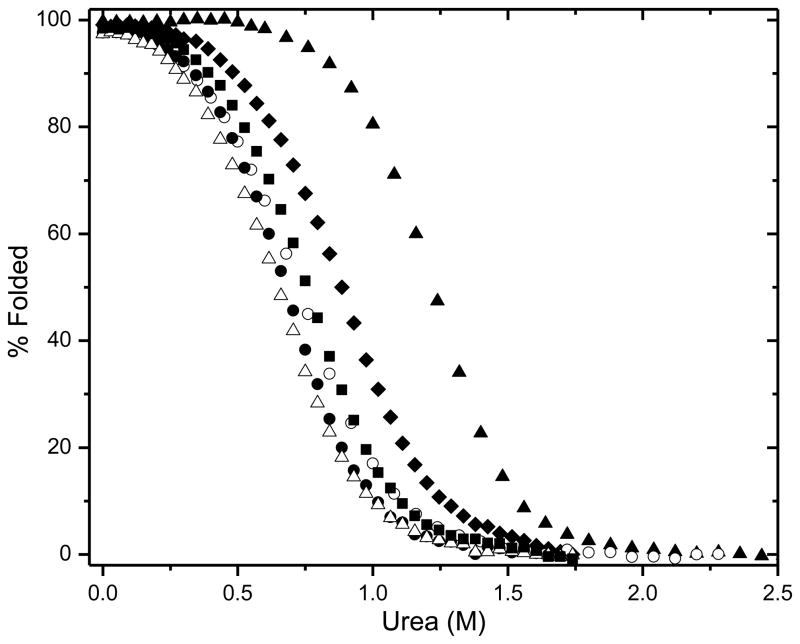
To study the contribution of the hydrophobic effect to the stability of VlsE, we prepared five Ile to Val mutants (Fig. 1b). All of these Ile residues are completely buried, and the mutants contain one less –CH2– group than wild-type. Urea denaturation curves were determined by measuring the circular dichroism at 220 nm as a function of urea concentration, and analyzed using the linear extrapolation method. Typical results are shown in the Supplementary Data. The unfolding curves for wild type VlsE and the five mutants are shown in Fig. 2 and the results are given in Table 1. The mutants are all less stable than wild type and the decreases in stability range from 1.1 to 1.9 kcal/mol with an average = 1.6 ± 0.3 kcal/mol.
Fig. 2.
Percent folded protein as a function of urea concentration for WT VlsE (▴) and the hydrophobic variants I128V (◆), I306V (■), I154V (○), I210V (●), and I214V (▵), in 5 mM Sodium Phosphate, pH 7.0, 25 °C.
Table 1.
Parameters characterizing the urea unfolding of VlsE and hydrophobic variants in 5 mM Sodium Phosphate, pH 7.0, 25 °C a
| Variant | Urea 1/2b (M) | Δ Urea 1/2c (M) | md (cal mol−1M−1) | ΔG (H2O)e (kcal mol−1) | ΔΔG f (kcal mol−1) |
|---|---|---|---|---|---|
| WT | 1.19 | - | 3865 | 4.6 | - |
| I128V | 0.88 | − 0.31 | 3360 | 3.0 | − 1.1 |
| I154V | 0.70 | − 0.49 | 3350 | 2.3 | − 1.8 |
| I210V | 0.67 | − 0.52 | 3770 | 2.5 | − 1.9 |
| I214V | 0.65 | − 0.54 | 3480 | 2.3 | − 1.9 |
| I306V | 0.77 | − 0.42 | 3700 | 2.8 | − 1.5 |
The urea denaturation curves were analyzed as described. 98
Midpoint of the unfolding curve. The error is ± 2 %.
Δ Urea 1/2 = Urea 1/2 (variant) − Urea 1/2 (WT).
The slope of plots of ΔG versus [Urea]. The error is ± 10%.
ΔG (H2O) = the intercept of plots of ΔG versus [Urea] at 0 M Urea.
From Δ Urea 1/2 × the average m value of WT and the variants (3590 cal mol−1M−1). The negative values indicate a decrease in stability.
VHP
McKnight et al. were the first to report studies of the thermal and GuHCl denaturation of the VHP subdomain. 34 Since then, many studies of different aspects of the folding of VHP and related variants have been published. (For recent examples, see Refs. 38 and 39). Our VHP has an N-terminal Met and is identical to the protein designated HP-36 by McKnight et al.34 We also find the thermal, urea, and GuHCl denaturation to be completely reversible and our Tm = 74.4 °C is in good agreement with their value of 72°C determined under different conditions. They find a stability of 3.3 ± 0.4 kcal/mol from GuHCl denaturation at 4°C and we find 2.7 kcal/mol from both urea and GuHCl denaturation at 25°C. 34
To study the contribution of the hydrophobic effect to the stability of VHP, we prepared the following nine mutants: L2A, F7A, V10A, F11A, M13A, F18A, L21A, L29A, and L35A (Fig. 1a). Urea denaturation curves were determined by measuring circular dichroism of VHP at 222 nm as a function of urea concentration, and analyzed using the linear extrapolation method. Typical results are shown in the Supplementary Data. The results from studies of five mutants using urea denaturation are summarized in Table 2. The stabilities of all nine mutants were measured using thermal denaturation and the results are given in Table 3. The Δ(ΔG) values from urea and thermal denaturation curves are in reasonable agreement and we use the average of the two for the mutants where both are available.
Table 2.
Parameters characterizing the urea unfolding of VHP and hydrophobic variants in 50 mM Sodium Phosphate, pH 7.0, 25 °C a
| Variant | Urea 1/2b (M) | Δ Urea 1/2c (M) | md (cal mol−1M−1) | ΔG (H2O) e (kcal mol−1) | ΔΔG f (kcal mol−1) |
|---|---|---|---|---|---|
| WT | 6.17 | - | 450 | 2.7 | - |
| L2A | 3.22 | − 2.95 | 470 | 1.5 | − 1.4 |
| V10A | 4.61 | − 1.56 | 460 | 2.1 | − 0.7 |
| F11A | 1.30 | − 4.87 | 494 | 0.6 | − 2.3 |
| L29A | 2.76 | − 3.41 | 450 | 1.2 | − 1.6 |
| L35A | 5.19 | − 0.98 | 470 | 2.4 | − 0.5 |
The urea denaturation curves were analyzed as described. 98
Midpoint of the unfolding curve. The error is ± 1%.
Δ Urea 1/2 = Urea 1/2 (variant) − Urea 1/2 (WT).
The slope of plots of ΔG versus [Urea]. The error is ± 10%.
ΔG (H2O) = the intercept of plots of ΔG versus [Urea] at 0 M Urea.
From Δ Urea 1/2 × the average m value of WT and the variants (466 cal mol−1M−1).
Table 3.
Parameters characterizing the thermal unfolding of VHP and hydrophobic variants in 50 mM sodium phosphate, pH 7.0 a
| Variant | Δ Hmb (kcal mol−1) | Δ Smc (cal mol−1 K−1) | Tmd (°C) | Δ Tme (°C) | Δ(ΔG) f (kcal mol −1) |
|---|---|---|---|---|---|
| WT | 30 | 88 | 74.4 | - | - |
| L2A | 23 | 70 | 60.4 | − 14.0 | − 1.0 |
| F7A | 18 | 56 | 41.7 | − 32.7 | − 2.3 |
| V10A | 30 | 89 | 67.0 | − 7.4 | − 0.5 |
| F11A | 22 | 70 | 48.8 | − 25.6 | − 1.8 |
| M13A | 15 | 47 | 34.7 | − 39.7 | − 2.8 |
| F18A | 20 | 65 | 28.3 | − 46.1 | − 3.3 |
| L21A | 22 | 68 | 45.3 | − 29.1 | − 2.1 |
| L29A | 24 | 74 | 55.3 | − 19.1 | − 1.4 |
| L35A | 29 | 86 | 67.2 | − 7.2 | − 0.5 |
The thermal denaturation curves were analyzed as described. 98
Enthalpy of unfolding at Tm. The error is ± 2 kcal mol−1.
Δ Hm/Tm. The error is ± 5 cal mol−1 K−1.
Midpoint of the unfolding curve. The error is ± 0.5 °C.
Δ Tm = Tm (variant) − Tm (WT).
Δ(ΔG) = ΔTm × the average Δ Sm value of WT and the variants (71 cal mol−1 K−1).
RNase Sa
We have previously used RNase Sa to study many different aspects of protein stability and folding. 40–45 To study the contribution of the hydrophobic effect to the stability of RNase Sa, we prepared 5 Ile to Val and 5 Leu to Ala mutants. Three thermal denaturation curves were determined for each mutant and the average results are summarized in Table 4.
Table 4.
Parameters characterizing the thermal unfolding of RNase Sa and hydrophobic variants in 30 mM MOPS, pH 7.0 a
| Variant | ΔHmb (kcal mol−1) | Δ Smc (cal mol−1 K−1) | ΔTmd (°C) | Δ(ΔG) e (kcal mol −1) |
|---|---|---|---|---|
| WT | 92 | 286 | - | - |
| L8A | 89 | 283 | − 5.0 | − 1.34 |
| L11A | 78 | 252 | − 11.9 | − 3.08 |
| L19A | 73 | 236 | − 13.8 | − 3.65 |
| L21A | 85 | 265 | − 1.6 | − 0.41 |
| I22V | 74 | 234 | − 5.7 | − 1.48 |
| I58V | 89 | 277 | 0.11 | 0.00 |
| I70V | 87 | 274 | − 3.4 | − .94 |
| I71V | 80 | 249 | − 0.3 | − 0.13 |
| L91A | 91 | 288 | − 3.2 | − 0.84 |
| I92V | 84 | 266 | − 3.9 | − 1.00 |
The thermal denaturation curves were analyzed as described. 98 The data in the table is the average value of three experiments.
Enthalpy of unfolding at Tm. The error is ± 5 kcal mol−1.
Δ Hm/Tm. The error is ± 15 cal mol−1 K−1.
Δ Tm = Tm (variant) − Tm (WT). The Tm for WT is 48.4 °C.40
Δ(ΔG) = ΔTm × the average Δ Sm value of WT and the variants (264 cal mol−1 K−1).
RNase T1
Like RNase Sa, RNase T1 is a member of the microbial ribonuclease family that we have used in many previous studies. 46–48 To investigate the contribution of the hydrophobic effect to the stability of RNase T1, we prepared 2 Ile to Val, 2 Val to Ala, and 1 Leu to Ala mutants. Two urea denaturation curves were determined for each of the mutants and average results are summarized in Table 5. Devos et al. 49 previously studied the V16A and V78A mutants of RNase T1 and determined their structures. Our Δ(ΔG) values in Table 5 are in excellent agreement with their values of 2.49 kcal/mol for V16A and 4.08 kcal/mol for V78A which were also obtained from an analysis of urea denaturation experiments. The thermodynamics of folding of these hydrophobic mutants of RNase T1 were also studied using differential scanning calorimetry and the results are given in the Supplementary Data.
Table 5.
Parameters characterizing the urea unfolding of RNase T1 and hydrophobic variants a
| Variant | Urea 1/2b (M) | Δ Urea 1/2c (M) | md (cal mol−1M−1) | Δ G (H2O)e (kcal mol−1) | ΔΔG f (kcal mol−1) |
|---|---|---|---|---|---|
| pH 7.0, 25 °C, 30 mM MOPS Buffer | |||||
| WT | 5.30 | - | 1210 | 6.4 | - |
| V16A | 3.12 | − 2.19 | 1265 | 3.9 | − 2.7 |
| I61V | 3.64 | − 1.66 | 1255 | 4.6 | − 2.1 |
| V78A | 2.05 | − 3.25 | 1280 | 2.6 | − 4.0 |
| L86A | 1.70 | − 3.61 | 1211 | 2.1 | − 4.4 |
| I90V | 4.49 | − 0.81 | 1150 | 5.2 | − 1.0 |
| pH 5.0, 25 ° C, 30mM Formate or Acetate Buffer | |||||
| WT | 6.89 | - | 1384 | 9.5 | - |
| V78A | 3.78 | − 3.12 | 1321 | 5.0 | − 4.2 |
The urea denaturation curves were analyzed as previously described. 98 The data in the table is the average value of two experiments.
Midpoint of the unfolding curve. The error is ± 1%.
Δ Urea 1/2 = Urea 1/2 (variant) − Urea 1/2 (WT).
The slope of plots of ΔG versus [Urea]. The error is ± 10%.
ΔG (H2O) = the intercept of plots of ΔG versus [Urea] at 0 M Urea.
From Δ Urea 1/2 × the average m value of WT and the variants at pH 7 (1230 cal mol−1M−1) and pH 3 (1342 cal mol−1M−1).
Other Proteins
We have also compiled Δ(ΔG) values for 138 hydrophobic mutants in 11 proteins and these are available in the Supplementary Data. The compilation includes 33 I → V, 39 V → A, 26 I → A, and 40 L → A mutants.
Discussion
Previous studies have shown that the Δ(ΔG) values for hydrophobic mutants are determined mainly by two factors: first, a constant term that depends on the difference in hydrophobicity between the wild-type and mutant side chains and, second, a variable term that depends on the difference in van der Waals interactions of the side chains. It is the second term that leads to the range of values (See Table 7 below) that are observed for mutants of the same type in different proteins and at different sites in the same protein. It is determined by the distance to neighboring atoms and their polarizability, so the local environment is important. 19, 28, 32, 33 In mutants of known structure, a cavity is generally observed in the mutant and Matthew’s group showed that larger cavities result in larger Δ(ΔG) values because the loss of van der Waals interactions is greater. 22, 23 A similar observation was made by Bowie’s group for the membrane protein bacterial rhodopsin (b Rhod).50 These studies and others suggest that the van der Waals interactions of the nonpolar side chains can make a larger contribution to the Δ(ΔG) values than the differences in hydrophobicity. 19, 28, 32, 33 When the Δ(ΔG) values from these two studies are extrapolated to zero cavity volume, the Δ(ΔG) values for the T4 lysozyme are greater than for rhodopsin.22, 23, 50 This probably results because the rhodopsin side chains are in a more nonpolar environment in the unfolded protein so the difference in hydrophobicity is smaller than for T4 lysozyme. We will make use of these ideas below in interpreting our results.
Table 7.
Average Δ(ΔG) values for 138 hydrophobic mutants from 11 proteins
| Substitution | Volume a (Å3) | Δ Gtr to Octanol a | Δ Gtr to Cylohexane a | Range | Δ(ΔG)b Average | Per –CH2– |
|---|---|---|---|---|---|---|
| Ile → Val (33) | 25.8 | − 0.80 | − 0.88 | − 0.1 to 2.3 | 1.1 ± 0.6 | 1.1 ± 0.6 |
| Val → Ala (39) | 49.0 | − 1.24 | − 2.23 | 0 to 4.9 | 2.4 ± 1.1 | 1.2 ± 0.5 |
| Ile → Ala (26) | 74.8 | − 2.04 | − 3.11 | 0.7 to 5.4 | 3.1 ± 1.2 | 1.0 ± 0.4 |
| Leu → Ala (40) | 74.5 | − 1.90 | − 3.11 | 0.2 to 6.2 | 3.2 ± 1.2 | 1.1 ± 0.4 |
From Table 1 in Ref. (52).
From the Supplementary Data.
Hydrophobic interactions contribute more to the stability of a large protein than a small protein
The results for VlsE, VHP, RNase Sa, and RNase T1 from Tables 1 to 5 are combined and summarized in Table 6. For VHP, none of the side chains are 100% buried, but we studied all of the hydrophobic side chains that are over 70% buried. In contrast, over 40% of the aliphatic side chains are 100% buried in VlsE because of its larger size. The hydrophobicity term discussed above depends on the accessible hydrophobic surface area that is removed from contact with water.2, 23 Consequently, this term will be reduced for side chains that are not completely buried in the folded protein. An approximate correction can be made by dividing the observed Δ(ΔG) values by the fraction buried for the side chain, so that the Δ(ΔG) values are compared at the same accessibility, 100% buried.51, 52 The observed and corrected Δ(ΔG) values are given in Table 6. This correction will have no effect on the van der Waals term that also contributes to the Δ(ΔG) values. The same correction was made for all of the mutations analyzed in Table 7 when the wild type side chain was less than 100% buried. The average corrected Δ(ΔG) values are never over 6% greater than the observed values so the corrections will not change any of the conclusions we reach.
Table 6.
Observed and accessibility corrected Δ(Δ)G values for the hydrophobic mutants of VlsE, VHP, RNase Sa, and RNase T1
| Protein | Variant | Side Chain % Buried a | Δ(ΔG) Obs b | Δ(ΔG) Acc. Corrected c | per –CH2– d |
|---|---|---|---|---|---|
| VlsE | I128V | 100 | 1.1 | 1.1 | 1.1 |
| I154V | 100 | 1.8 | 1.8 | 1.8 | |
| I210V | 100 | 1.9 | 1.9 | 1.9 | |
| I214V | 100 | 1.9 | 1.9 | 1.9 | |
| I306V | 100 | 1.5 | 1.5 | 1.5 | |
| VHP | V10A | 70 | 0.6 | 0.9 | 0.4 |
| L2A | 70 | 1.2 | 1.7 | 0.6 | |
| L21A | 74 | 2.1 | 2.8 | 0.9 | |
| L29A | 73 | 1.5 | 2.1 | 0.7 | |
| L35A | 79 | 0.5 | 0.6 | 0.2 | |
| F7A | 98 | 2.3 | 2.3 | - | |
| F11A | 98 | 2.1 | 2.1 | - | |
| F18A | 98 | 3.3 | 3.4 | - | |
| M13A | 72 | 2.8 | 3.9 | - | |
| RNase Sa | I22V | 81 | 1.5 | 1.8 | 1.9 |
| I58V | 50 | 0 | 0 | 0 | |
| I70V | 100 | 0.9 | 0.9 | 0.9 | |
| I71V | 100 | 0.1 | 0.1 | 0.1 | |
| I92V | 100 | 1.0 | 1.0 | 1.0 | |
| L8A | 84 | 1.3 | 1.6 | 0.5 | |
| L11A | 97 | 3.1 | 3.2 | 1.1 | |
| L19A | 95 | 3.7 | 3.8 | 1.3 | |
| L21A | 67 | 0.4 | 0.6 | 0.2 | |
| L91A | 64 | 0.8 | 1.3 | 0.4 | |
| RNase T1 | I61V | 97 | 2.1 | 2.2 | 2.2 |
| I90V | 100 | 1.0 | 1.0 | 1.0 | |
| V16A | 98 | 2.7 | 2.8 | 1.4 | |
| V78A | 100 | 4.1 | 4.1 | 2.1 | |
| L86A | 74 | 4.4 | 5.9 | 2.0 |
The % buried for the side chain in the wild-type protein was calculated using PFIS . 99
The VlsE results are from Table 1. The VHP results are based on the data in Tables 2 and 3 and the average was used when two Δ(ΔG) values were available. The RNase Sa results are the average of three values for each mutant and are from Table 4. The RNase T1 results are the average of two values for each mutant and are from Table 5.
Δ(ΔG) accessibility corrected = Δ(ΔG) observed/(side chain fraction buried).
the per –CH2– values = Δ(ΔG) accessibility corrected values for the I to V mutants, divided by 2 for the V to A mutants and divided by 3 for the I to A and L to A mutants.
One goal of our studies was to see if the burial of nonpolar groups makes the same contribution to the stability of a small protein, VHP (36 residues) as it does to the stability of a large protein, VlsE (341 residues). To consider this, we analyzed the results of previous studies of the contribution of hydrophobic interactions to protein stability. For the mutants considered, a larger aliphatic side chain is replaced by a smaller to minimize steric effects. The proteins ranged in size from 64 residues (chymotrypsin inhibitor 2) to 168 residues (apoflavodoxin). (Table 3 in the Supplementary Data gives the proteins and the mutations used in the analysis, and Tables 4 to 7 list the results for individual proteins for 33 Ile to Val, 39 Val to Ala, 26 Ile to Ala, and 40 Leu to Ala variants. Only side chains that were at least 70% buried in the wild type protein were included. The results for RNases Sa and T1 reported here were included in the analysis.) The analysis included 138 mutations that removed a total of 309 –CH2– groups in 11 proteins. The range and average of the Δ(ΔG) values for each type of mutation are given in Table 7. In addition, the differences in side-chain size are given along with the Δ(ΔG) values expected when octanol or cyclohexane are used to model the interior of the protein.
The range of Δ(ΔG) values is large, and this gives rise to a large standard deviations from the average values. This shows that individual side chains are in quite different environments in folded proteins. The average experimental Δ(ΔG) values are in good agreement with the Δ(ΔG) values based on cyclohexane. We will make use of this observation below. For the four types of mutants in Table 7, the Δ(ΔG) values per –CH2– group are similar suggesting that the average of the van der Waals contacts lost are similar even though the potential differences in volume between the side chains is large.
The results for VHP and VlsE are compared with the other proteins in Table 8. The average Δ(ΔG) value for VHP is lower than that for any other protein. This was expected. None of the hydrophobic side chains in VHP are completely buried so they will have less favorable van der Waals interactions than a completely buried side chain. Since VHP is small, it should be possible for theoreticians to use these results to achieve a better understanding of the contribution of the van der Waals interactions of the hydrophobic side chains to the stability of globular proteins.
Table 8.
Number of Residues and average Δ(ΔG) values per –CH2– group for 148 mutations in 13 proteins
| Protein (# Mutations) | # Residues a | Δ(ΔG) (kcal/mol) b |
|---|---|---|
| VlsE (5) | 341 | 1.6 ± 0.3 |
| ApoFlavo (14) | 168 | 0.9 ± 0.4 |
| T4 Lyso (25) | 164 | 1.0 ± 0.4 |
| SN (28) | 141 | 1.2 ± 0.5 |
| hLyso (11) | 130 | 0.7 ± 0.3 |
| Ba (15) | 110 | 1.2 ± 0.4 |
| FKBP (15) | 107 | 1.2 ± 0.3 |
| RNase T1 (5) | 104 | 1.7 ± 0.5 |
| RNase Sa (7) | 96 | 1.0 ± 0.5 |
| AcCoABP (5) | 86 | 0.9 ± 0.5 |
| Ubiquitin (4) | 76 | 1.3 ± 0.4 |
| CI2 (9) | 64 | 1.1 ± 0.7 |
| VHP (5) | 36 | 0.6 ± 0.3 |
| Average | 125 | 1.1 ± 0.5 |
This is the number of residues in the protein.
These are the average Δ(ΔG) values for all of the aliphatic mutants studied for these proteins.
The individual Δ(ΔG) values for all of the proteins are given in the Supplementary Data.
The average Δ(ΔG) values for RNase T1 and VlsE are considerably higher than for any of the other proteins. The results for VlsE are interesting and allow us to draw some conclusions. In 2000, Fleming and Richards examined the packing density in a large sample of proteins and concluded: 53 “One would expect increased overall packing in large proteins, proteins with a high percentage of helix and proteins with a high percentage of aromatic and small residues and a low percentage of aliphatic residues.” They also found that some proteins had packing densities that differed significantly from the average. In contrast, Liang and Dill examined the packing density in the interiors of globular proteins, and concluded, in part,54 “We find that larger proteins are packed more loosely than smaller proteins. And we find that the enthalpies of folding (per amino acid) are independent of the packing density of a protein, indicating that van der Waals interactions are not a dominant component of the folding forces.” Both of these studies and others have pointed out that the packing of protein interiors is heterogeneous and this is one of the reasons for the differences between proteins seen in Table 8. 55–57 VlsE was not included in these analyses, but our results suggest that it may be more tightly packed than the other proteins that have been studied, all of which are considerably smaller than VlsE.
It seemed possible that the denatured state ensembles (DSE) of larger proteins might have more structure under physiological conditions than those of smaller proteins because the percentage of nonpolar residues increases with size.58 If so, the hydrophobic effect might make a smaller contribution to the stability of proteins as they become larger. The results with VlsE suggest that this is not the case. The results for RNase T1 are interesting. The average Δ(ΔG) is larger than for any other protein. This suggests that the cavities left in the RNase T1 mutants might be larger than in the other proteins. This is supported by the results of De Vos et al.49 who determined the structures of the V16A and V78A mutants and concluded: “Little or no adaptation of the immediately surrounding residues is observed that reduces the volume of the induced cavity ···” The atomic temperature factors of the wild-type protein also suggest that the mutations are in a rigid part of the molecule. This might result because this protein has two disulfide bonds so that it is more difficult for the structure to relax and fill the cavity. However, this is not consistent with the results for human lysozyme. After VHP, human lysozyme has the lowest average Δ(ΔG) per –CH2– group. This suggests that this protein does not have large cavities in the mutants, despite the fact that this protein has four disulfide bonds, more than any of the other proteins in Table 8.
Forces contributing to protein stability
The average observed Δ(ΔG) values in Table 7 are in good agreement with the values predicted from model compound studies using cyclohexane (CH) as a model for the protein interior. This agreement probably results because, on average, CH is more polarizable than the interior of the protein but is not as tightly packed and the two effects compensate.9, 52 This suggests that the ΔGtr values from water to CH can be used to estimate the average contribution of the hydrophobic effect to protein stability. The aliphatic residues and the –CH2– groups from the polar and charged side chains make the largest contribution to the hydrophobic effect, ranging from 78% for VHP to 96% for VlsE with an average of 86 ± 5%. For the nonpolar side chains that can also form hydrogen bonds, Trp, Tyr and Cys, it is preferable to use the ΔGtr values for octanol so that hydrogen bonding does not contribute.6, 52 In Table 9, we estimate the contribution of the hydrophobic effect to the stability of 22 proteins with the number of residues ranging from 36 to 534. These are all single-domain proteins and one, bacterial rhodopsin, is a membrane protein. 50, 59, 60 The details of how these calculations are done and the ΔGtr values used are given in the footnotes to Table 9.
Table 9.
Forces contributing to the conformational stability of 22 proteins of increasing size
| Protein a | pdbb | #Res c | S-S d | HB e | HB/Res f | HΦLBg | HΦUBg | HΦ/Res h | Total Stability i | CE (Calculated) j | Pred Stability (CE = 2.41) k | % Diff (Total vs. Pred) l |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VHP | 1yrf | 35 | 0 | 28 | 0.8 | 34.45 | 44.86 | 1.28 | 72.86 | 2.08 | 84.35 | 13.62 |
| Protein G | 1pgb | 56 | 0 | 54 | 0.96 | 51.24 | 66.72 | 1.19 | 120.72 | 2.16 | 134.96 | 10.55 |
| Ubiquitin | 1ubq | 76 | 0 | 69 | 0.91 | 92.63 | 120.63 | 1.59 | 189.63 | 2.50 | 183.16 | −3.53 |
| Barstar | 1bta | 89 | 0 | 69 | 0.78 | 115.35 | 150.21 | 1.69 | 219.21 | 2.46 | 214.49 | −2.20 |
| RNase Sa | 1rgg | 96 | 5 (1) | 90 | 0.94 | 83.92 | 109.29 | 1.14 | 204.29 | 2.13 | 231.36 | 11.70 |
| RNase T1 | 9rnt | 104 | 7 (2) | 99 | 0.95 | 77.05 | 100.33 | 0.96 | 206.33 | 1.98 | 250.64 | 17.68 |
| Barnase | 1bni | 108 | 0 | 108 | 1.00 | 105.31 | 137.14 | 1.27 | 245.14 | 2.27 | 260.28 | 5.82 |
| RNase A | 9rsa | 124 | 15 (4) | 110 | 0.89 | 109.42 | 142.48 | 1.15 | 267.48 | 2.16 | 298.84 | 10.49 |
| Che Y | 3chy | 128 | 0 | 122 | 0.95 | 154.72 | 201.48 | 1.57 | 323.48 | 2.53 | 308.48 | −4.86 |
| hu Lyso | 1lz1 | 130 | 18.2 (4) | 137 | 1.05 | 139.89 | 182.17 | 1.40 | 337.37 | 2.60 | 313.30 | −7.68 |
| Staph Nuc | 1ey0 | 136 | 0 | 120 | 0.88 | 154.66 | 201.40 | 1.48 | 321.40 | 2.36 | 327.76 | 1.94 |
| T4 Lyso | 1l63 | 162 | 0 | 188 | 1.16 | 186.02 | 242.24 | 1.50 | 430.24 | 2.66 | 390.42 | −10.20 |
| Adn Kinase | 3adk | 195 | 0 | 160 | 0.82 | 219.71 | 286.12 | 1.47 | 446.12 | 2.29 | 469.95 | 5.07 |
| b Rhod | 1c3w | 222 | 0 | 243 | 1.09 | 269.12 | 350.46 | 1.58 | 593.46 | 2.67 | 535.02 | −10.92 |
| Carb Anhy | 1cah | 258 | 0 | 238 | 0.92 | 288.43 | 375.60 | 1.46 | 613.60 | 2.38 | 621.78 | 1.32 |
| Trp Syn α | 1wq5 | 258 | 0 | 225 | 0.87 | 290.95 | 378.89 | 1.47 | 603.89 | 2.34 | 621.78 | 2.88 |
| VlsE | 1l8w | 271 | 0 | 251 | 0.93 | 290.07 | 377.74 | 1.39 | 628.74 | 2.32 | 653.11 | 3.73 |
| b Lipase | 1cvl | 316 | 4.1 (1) | 330 | 1.04 | 333.61 | 434.43 | 1.37 | 768.53 | 2.43 | 761.56 | −0.92 |
| Pepsinogen | 3psg | 365 | 7.3 (3) | 319 | 0.87 | 388.89 | 506.41 | 1.39 | 832.71 | 2.28 | 879.65 | 5.34 |
| MBP | 1omp | 370 | 0 | 337 | 0.91 | 423.53 | 551.53 | 1.49 | 888.53 | 2.40 | 891.70 | 0.36 |
| Glu Amyl | 1glm | 470 | 10 (3) | 503 | 1.07 | 505.75 | 658.59 | 1.40 | 1171.59 | 2.49 | 1132.70 | −3.43 |
| CR Lipase | 1crl | 534 | 5.9 (2) | 548 | 1.03 | 568.97 | 740.93 | 1.39 | 1294.83 | 2.42 | 1286.94 | −0.61 |
The abbreviations not defined in the text are: Adn Kinase = adenylate kinase; Carb Anhy = carbonic anhydrase; MBP = maltose binding protein; Glu Amyl =glucoamylase; CR Lipase = Candida rugosa lipase.
http://www.pdb.org/ and Ref. (100).
# Res = number of residues observed in the crystal structure. For some of the proteins, this is less than the number of residues in the protein: VHP = 36; Barnase = 110; Staph Nuc = 149; T4 Lyso = 164; Carb Anhy = 259; Trp Syn α = 268; VlsE = 341; Lipase = 319; Pepsinogen = 370.
Eight of the proteins contain disulfide bonds and the number is shown in parentheses. The contribution of the disulfide bonds to the stability is given in kcal/mol by the number in the column. The values for RNase Sa 40 and RNase T1101 are measured values and the values for the other proteins were calculated as described. 101
This column gives the number of hydrogen bonds in the protein and is also an estimate of the contribution of hydrogen bonds to the stability in kcal/mol since we assume each intramolecular hydrogen bond contributes 1 kcal/mol to the stability.41, 68 The number of hydrogen bonds was calculated with PFIS. The criteria used to establish a hydrogen bond is the same as that used in previous studies 75, 102 except we count hydrogen bonds when the distance between the two electronegative atoms is 3.6 Å or less.
This gives the hydrogen bonding contribution per residue in kcal/mol.
This gives the hydrophobic interaction contribution in kcal/mol when the denatured state is modeled using the lower boundary model (Column 7) or the upper boundary model (Column 8) of Creamer et al. 61 For five proteins ranging from 110 to 275 residues, they found that the denatured state ASA of the upper boundary model was 17.8 ± 0.6 % less and the lower boundary model was 36.9 ± 0.5 % less than the ASA based on the Gly-X-Gly tripeptide model.
This gives the hydrophobic interaction contribution per residue in kcal/mol when the denatured state is modeled using the upper boundary model of Creamer et al. 61 (Column 8).
Total Stability = S-S +HB + HΦUB
CE (Calculated) = (Total Stability)/(# Residues). CE is the conformational entropy contribution per residue in kcal/mol needed to give an instability equal to the Total Stability.
Pred Stability = 2.41*(# Residues). When Total Stability is plotted vs. the # residues, the least-squares slope is 2.41 kcal/mol per residue. This is the average conformational entropy contribution per residue that gives the best agreement with Total Stability.
This is the % difference between the Total Stability (Column 10) and Pred Stability (Column 12).
The hydrophobic interaction contribution depends on how many nonpolar groups are buried on folding and this depends on the model used for the denatured state ensemble (DSE). Table 9 gives results for two different DSE models. For the least accessible DSE, the lower boundary model of Creamer et al.61 is used which models the DSE using 17 residue fragments of the proteins with the same conformation they have in the folded proteins. This gives hydrophobic interaction contributions of 34 kcal/mol for VHP and 569 kcal/mol for CR lipase (Column 7). These estimates are probably too low. For our analysis, we will use the upper bound model of Creamer et al.61 which models the proteins in an extended β-sheet conformation. Now the contribution of hydrophobic interactions varies from 45 for VHP to 741 for CR lipase (Column 8). For the individual proteins, the contribution of hydrophobic interactions per residue is lowest for RNase T1 at 1.0 kcal/mol and highest for barstar at 1.7 kcal/mol with an average of 1.4 ± 0.2 kcal/mol per residue (Column 9). In comparison, the average is 1.1 ± 0.1 for the lower bound model. The upper bound model gives the estimate expected if the unfolded protein is completely accessible to water. More likely, the correct extent of exposure is intermediate between the upper and lower bound models. 62, 63
Until 1990, the prevailing idea was that intramolecular hydrogen bonds were necessary for maintaining the structure of a protein but made only a small contribution to protein stability.64, 65 More recent evidence shows that hydrogen bonds make a large contribution to protein stability.41, 46, 60, 66–71 For 35 Tyr to Phe mutations with the Tyr –OH group hydrogen bonded, Δ(ΔG) = −1.4 ± 0.9 kcal/mol, and for 17 mutations with the –OH group not hydrogen bonded, Δ(ΔG) = − 0.2 ± 0.4 kcal/mol.41, 68 Bowie has shown that the difference between these two Δ(ΔG) values, − 1.2 kcal/mol, is similar to the contribution of a hydrogen bond to protein stability determined by double mutant cycle analyses.72 A similar analysis of Thr to Val mutations leads to an estimate of 0.9 kcal/mol per hydrogen bond.68 A study of Asn to Ala mutants in which the Asn amide group is hydrogen bonded to a peptide group gives an estimate of 1.1 ± 0.6 kcal/mol per hydrogen bond. 69, 70, 73, 74 Finally, studies of amide to ester mutations of backbone peptide groups suggest that these hydrogen bonds contribute 1.1 ± 1.6 kcal/mol per hydrogen bond to protein stability. These results cover hydrogen bonds between side chains, between backbone groups, and side chain – backbone hydrogen bonds. Consequently, we will make the conservative assumption that each intramolecular hydrogen bond in a folded protein contributes 1 kcal/mol to the stability.
In Table 9, the number of hydrogen bonds formed by each protein is shown and this provides an estimate of the amount they contribute to the stability. The contribution from hydrogen bonds ranges from 28 kcal/mol for VHP to 548 for CR lipase. When the hydrogen bond contribution per residue is considered, they range from 0.8 kcal/mol for barstar to 1.2 kcal/mol for T4 lysozyme. The average number of hydrogen bonds formed in these proteins is 0.95 ± 0.10 per residue, with 65% between peptide groups, 23% between peptide groups and side chains, and 12% between side chains. In a larger sample of proteins, the Rose group found an average of 1.1 hydrogen bonds per residue. 75
The total stability is the sum of the contribution of disulfide bonds, hydrogen bonds, and hydrophobic interactions (Column 10). It ranges from 83 kcal/mol for VHP to 1456 kcal/mol for the CR lipase. There is a good correlation between the stability estimates and the number of residues: HB = 0.98 ± 0.02 (#Residues) with a correlation coefficient of 0.989; HΦ = 1.72 ± 0.02 (#Residues) with a correlation coefficient of 0.994; and Total Stability = 2.41 ± 0.02 (#Residues) with a correlation coefficient of 0.997. (These plots are shown in Figure 2 in the Supplementary Data.) This analysis suggests that both hydrogen bonds and hydrophobic interactions make large contributions to the conformational stability of globular proteins.
In a previous study, a different approach suggested that polar group burial contributes more to protein stability than nonpolar group burial.74 This was based, in part, on a paper by Harpaz et al.76 that analyzed the completely buried surface in a sample of 108 proteins. They showed that the total volume of the buried residues was 298,100 Å3. Of this, 123,653 Å3 came from 9 nonpolar side chains (Table 9), and 14,242 Å3 from the –CH2– groups of the polar and charged side chains, for a total of 137,895 Å3. (Peptide groups contribute 92,000 Å3.) Based on the data available at that time, the contribution of hydrophobic group burial was estimated using 49 cal/mol per Å3 of nonpolar volume buried. Using the more recent data presented here, the contribution is 44 ± 3 cal/mol per Å3, and this leads to (137,895 Å3 × .044 kcal/mol/Å3) = 6067 kcal/mol as the contribution of the hydrophobic effect to the stability of these proteins. If the approach used in Table 9 is applied to the same data, it leads to a 5954 kcal/mol estimate of the contribution the hydrophobic effect. It is reassuring that these two estimates differ by less than 2%. In the same paper, we estimated that burying the 92,000 Å3 of peptide groups contributed 7200 kcal/mol to the stability.
The major destabilizing force in protein folding is conformational entropy. Studies by the Freire77 and Record78 groups suggested that 1.7 kcal/mol per residue can be used to estimate the contribution of conformational entropy to protein stability. However, this estimate is too small to compensate for the total stabilities shown in Table 9. This suggests either that these conformational entropy estimates are too low, or that our estimates of the stabilizing forces are too large, or that there are other forces that make a major contribution to protein instability. We consider these topics next.
Forces contributing to protein instability
When the prevailing thought was that hydrogen bonds make only a small contribution to stability, it was assumed that polar groups that were not hydrogen bonded would make a large unfavorable contribution to the stability because they would form hydrogen bonds to water when the protein unfolds. This was first analyzed in a paper from Finney’s group. 79 They showed for a large sample of proteins that there is an excellent correlation (0.99) between “lost hydrogen bonds” and three stabilizing forces: hydrophobic effect, disulfide bonds, and ion pairs. (The hydrophobic effect contributed over 95% to their stabilizing forces.) They found that over 90% of the polar atoms form intramolecular hydrogen bonds or hydrogen bonds to water molecules. However, about 24% of the carbonyl oxygen atoms and 6% of the amide hydrogens of the peptide bonds in proteins do not form hydrogen bonds, and this is the major source of the lost hydrogen bonds.
More recently, analyses by Fleming and Rose 80 led them to conclude: “Unsatisfied backbone polar groups are energetically expensive, to the degree that they almost never occur.” For Tyr to Phe mutations discussed above, Δ(ΔG) = − 0.2 ± 0.4 kcal/mol for the Tyr residues that were not hydrogen bonded.68 This suggests that side chains that are not hydrogen bonded do not make a large unfavorable contribution to the stability. Even for 40 Val to Thr mutations, Δ(ΔG) = 1.8 ± 1.1 kcal/mol,41, 68 showing that when a polar –OH group is placed at a site designed for a –CH3 group, the penalty is far less than the cost of 5 to 6 kcal/mol proposed by Fleming and Rose.80 Consequently, we doubt that non hydrogen bonded polar groups make a large contribution to protein instability.
Another interesting suggestion was made by Kajander et al.58 They analyzed the structures of a large sample of proteins of different molecular weights and showed that 65% more charged groups are buried in proteins containing 700 amino acids than in proteins containing 100 amino acids. From this they concluded: “Nature may use charge burial to reduce protein stability; not all buried charges are fully stabilized by a prearranged protein environment.” If so, removing them may be a way of increasing the stability of proteins. The Garcia-Moreno group has shown that burying a charge in a nonpolar environment in staph nuclease can decrease the stability by over 5 kcal/mol,81 and we have observed that removing a naturally occurring buried charge in RNase Sa can increase the stability by over 3 kcal/mol.42 However, if buried charged groups are hydrogen bonded, they can make a large favorable contribution to protein stability.82
Kajander et al.58 included the backbone atoms in their analysis so that the peptide C = O and NH groups were classed as polar uncharged and the α-carbon was classed as aliphatic. For a different perspective, we have analyzed the proteins in Table 9 and considered just the side chains. The fraction burial of nonpolar side chains (Ala, Val, Ile, Leu, Met, Phe, Tyr, Trp, Cys), polar uncharged side chains (Asn, Gln, Ser, Thr, His) and polar charged side chains (Asp, Glu, Lys, Arg) was determined as a function of the number of residues. (The plots are shown in Figure 3 in the Supplementary Data.) For each group, the fraction of buried side chains increases linearly with the number of residues: for nonpolar, fraction buried = (# residues) (0.363 ± 0.007) with a correlation coefficient of 0.99; for polar uncharged, fraction buried = (# residues) (0.155 ± 0.009) with a correlation coefficient of 0.93; and for polar charged, fraction buried = (# residues) (0.117 ± .005) with a correlation coefficient of 0.95. For the polar charged side chains, if we consider only the O atoms for Asp and Glu, the N for Lys, and the two outermost N for Arg, the fraction buried is similar: fraction buried = 0.112 ± 0.004 (# residues) with a correlation coefficient of 0.95. The % of the charged groups that are buried is lowest for barstar (22%) and greatest for glucose amylase (70%). For the small and large proteins studied here, 28% of the charged groups are buried in VHP, and 50% are buried in VlsE. So, we see a trend similar to that observed by Kajander.58 This subject deserves further study.
The destabilizing force of most interest is conformational entropy. As noted above, two different approaches suggest that the entropic cost of folding a protein is about 1.7 kcal/mol per residue.77, 78 We have used this estimate in previous papers. 40, 83 We now think this estimate is too low. The estimates of the conformational entropy needed to balance the total stability for each protein are given in Table 9. The smallest estimate is 2.0 kcal/mol per residue for RNase T1 and the largest estimates are 2.7 for T4 lysozyme and rhodopsin. A value of 2.41 ± 0.02 kcal/mol per residue gives the best fit for all 22 proteins. Using this value gives the destabilizing estimates (Column 12) shown in Table 9. In only two cases do the stabilizing and destabilizing estimates differ by over 12%. The predicted stabilities are 17% too low for RNase T1 and 14% too low for VHP. This reflects the fact that the contribution of hydrophobic interactions is exceptionally low for RNase T1, and the contribution of hydrogen bonds is exceptionally low for VHP. For Rhodopsin, the predicted stability is 11% too high. This is expected since rhodopsin is a membrane protein. The measured values for the contribution of hydrogen bonds and hydrophobic interactions to rhodopsin are both lower than the values found with soluble proteins. 50, 60 This results because the unfolded protein will be in a more nonpolar environment and this will tend to lower the contribution that both hydrogen bonds and hydrophobic interactions make to the stability. The stability estimates used in Table 9 are based on nonmembrane proteins and they are expected to lead to an overestimate of the stability of a membrane protein like rhodopsin.
On the basis of experimental data, Makhatadze and Privalov have pointed out why previous conformational entropy estimates are too low and suggest an average value of 3.6 kcal/mol per residue.84 Using an extended all-atom simulation of the folding of a three-helix bundle protein, Boczko and Brooks85 estimated a conformational entropy cost of 2.6 kcal/mol per residue. These articles and others were reviewed by Brady and Sharp86 who suggested that the conformational entropy cost would be between 2.4 and 3.7 kcal/mol per residue. Thus, our estimate of 2.41 kcal/mol per residue is in line with other experimental and theoretical estimates.
Our estimates of the total stability of the proteins given in Table 9 should be considered a lower limit for the following reasons. First, the ΔGtr values for CH used to calculate the contribution of the burial of the nine nonpolar side chains and the –CH2– groups to protein stability are more likely too small than too large. Second, the contribution of individual hydrogen bonds to stability is more likely to be greater than 1 kcal/mol than less. 67 Third, other forces that contribute to protein stability such as charge – charge interactions, ion pairs, and π – cation interactions will generally make a small favorable contribution.87, 88 If the estimates of the total stability were higher, the estimates for the contribution of conformational entropy would also be higher, but they would still be in the range suggested by Brady and Sharp.86
Stability of VHP
Because of its small size, the thermodynamics and kinetics of folding of VHP are currently being studied by a variety of experimental35, 39, 89 and theoretical 90–92 approaches. Consequently, there is considerable interest in determining which residues make important contributions to the stability and the mechanism of folding of VHP. In 2002, Frank et al.93 replaced Met 13 and the three Phe residues with Leu and were able to conclude that Phe18 makes the largest contribution to the stability. Our results support this conclusion. Raleigh’s group has shown that the double mutant with Ala at position 28 and Met at residue 30 raises the Tm by over 20°C.38 They have also shown that a π - cation interaction between Phe 7 and Arg 15 contributes to the stability.38 The Kelly lab has studied five backbone and two side chain hydrogen bonds in VHP. 39 They estimate that the backbone hydrogen bonds contribute 1.5 kcal/mol per hydrogen bond and the two side chain hydrogen bonds contribute about 1 kcal/mol per hydrogen bond. The nine VHP mutants studied here (Table 6) have not been studied previously. We next consider what these mutants reveal about the stability of VHP.
The results in Table 6 show that the aromatic rings of Phe 7, 11, and 18 contribute 2.3, 2.1, and 3.3 kcal/mol for a total contribution of 7.7 kcal/mol to the stability of VHP. In addition, the 14 –CH2– group equivalents removed by the 5 aliphatic mutations contribute 5.9 kcal/mol and –CH2–S–CH3 group of Met 13 contributes 2.8 kcal/mol. This amounts to 16.4 kcal/mol of stability based on our measured Δ(ΔG) values. As we did in Table 9, we can estimate the contribution of the other six nonpolar side chains (Ala 9, 17, 19, Leu 23, Trp 24, and Phe 36) that are only partially buried as 5 kcal/mol. In addition, another 34 –CH2– groups are buried when the protein folds, and using 0.6 kcal/mol per –CH2– group, these would contribute another 20 kcal/mol to the stability. These are from the polar and charged side chains and from the groups not removed in the mutants in Table 8. Based on this, we estimate that the burial of nonpolar groups contributes about 41 kcal/mol to the stability of VHP. This compares with an estimate of 45 kcal/mol in Table 9 which is higher because 1 kcal/mol was used for the contribution of the –CH2– groups rather than the observed value for VHP of 0.6 kcal/mol. In comparison, VHP forms 28 hydrogen bonds on folding: 23 between peptide groups, 4 between peptide groups and side chains, and only one between two side chains. If we assign 1 kcal/mol per hydrogen bond, then hydrogen bonds contribute considerably less to the stability than hydrophobic interactions. But if we use 1.5 kcal/mol per peptide hydrogen bond, as found for five of the 23 backbone hydrogen bonds, 39 then hydrogen bonds (40 kcal/mol) and hydrophobic interactions (41 kcal/mol) make about the same contribution to the stability. Thus, even for a small protein where the nonpolar side chains cannot be completely buried, hydrophobic interactions contributes more to the stability than hydrogen bonds.
Concluding Remarks
For the 22 proteins in Table 9, the average contribution of hydrophobic interactions to the stability is 60 ± 4 %. The contribution is smallest for RNase T1 (54%) and greatest for Barstar (73%). The average contribution of hydrogen bonds to the stability is 40 ± 4%. The contribution is smallest for Barstar (27%) and largest for RNase T1 (43%). The largest contribution of disulfide bonds to the stability is for RNase A (5%). This analysis suggests that hydrophobic interactions make the dominant contribution to protein stability, always greater than 50%. Hydrogen bonds also make a large contribution to stability, but always less than hydrophobic interactions even for the smallest globular proteins. The loss in conformational entropy when a protein folds is the major destabilizing force, but the burial of non hydrogen bonded polar or charged groups may also contribute instability in fine-tuning protein stability.
Materials and Methods
All buffers and chemicals were of reagent grade. Urea was from Amresco or Nacalai Tesque (Kyoto, Japan), and was used without further purification. The plasmids for VHP, VlsE, and their variants were derived from pET vectors (Novagen) and have been described previously.94 The plasmids for RNase Sa, RNase T1, and their variants were derived from the pEH100 plasmid as described previously.40, 95 The expression hosts for all proteins and variants were either E. coli strains RY1988 (MQ), DS2000, or C41(DE3).94, 96 Oligonucleotide primers for mutagenesis were from Integrated DNA Technologies (Coralville, IA). Site-directed mutagenesis was performed using a QuikChange Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA). Mutant plasmids were sequenced by the Gene Technologies Laboratory, Texas A&M University.
VHP, VlsE, and their variants were expressed and purified as described previously.63, 94 RNases Sa, T1, and their variants were expressed and purified as described previously.82, 95, 96 The purity of all proteins was confirmed by SDS PAGE and MALDI-TOF mass spectrometry.
Urea and thermal denaturation curves were determined using either an AVIV 62DS or 202SF spectropolarimeter (Aviv Instruments, Lakewood, NJ) to follow unfolding. The methods for VHP and VlsE have been described previously.63 The methods for RNases Sa, T1, and their variants have been previously described. 97, 98 The analysis of urea and thermal denaturation curves assumed a two-state unfolding model and was done as described.97, 98
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM 37039 and GM 52483, Robert A. Welch Foundation Grants BE-1060 and BE-1281, and the Tom and Jean McMullin Professorship. We thank George Makhatadze, Evan Powers, Jeff Kelly, and James Bowie for providing important information, and Jeff Myers and Doug Laurents for providing suggestions for the manuscript. We also thank Michael Perham and Pernilla Wittung-Stafshede for help with the expression of VlsE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bernal JD. Structure of proteins. Nature. 1939;143:663–667. [Google Scholar]
- 2.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 3.Tanford C. Contribution of hydrophobic interactions to the stability of the globular conformation of proteins. J Am Chem Soc. 1962;84:4240–4247. [Google Scholar]
- 4.Tanford C. How protein chemists learned about the hydrophobic factor. Protein Sci. 1997;6:1358–66. doi: 10.1002/pro.5560060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nozaki Y, Tanford C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions establishment of a hydrophobicity scale. J Biol Chem. 1971;246:2211–2217. [PubMed] [Google Scholar]
- 6.Fauchere JL, Pliska VE. Hydrophobic parameters of amino-acid side-chains from the partitioning of N-acetyl-amino-acid amides. Eur J Med Chem. 1983;18:369–375. [Google Scholar]
- 7.Wimley WC, Creamer TP, White SH. Solvation energies of amino acid side chains and backbone in a family of host-guest pentapeptides. Biochemistry. 1996;35:5109–24. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
- 8.Damodaran S, Song KB. The role of solvent polarity in the free energy of transfer of amino acid side chains from water to organic solvents. J Biol Chem. 1986;261:7220–2. [PubMed] [Google Scholar]
- 9.Radzicka A, Wolfenden R. Comparing the polarites of the amino acids; side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral squeous solution. Biochemistry. 1988;27:1644–1670. [Google Scholar]
- 10.Wesson L, Eisenberg D. Atomic solvation parameters applied to molecular dynamics of proteins in solution. Protein Sci. 1992;1:227–35. doi: 10.1002/pro.5560010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose GD, Wolfenden R. Hydrogen bonding, hydrophobicity, packing, and protein folding. Annu Rev Biophys Biomol Struct. 1993;22:381–415. doi: 10.1146/annurev.bb.22.060193.002121. [DOI] [PubMed] [Google Scholar]
- 12.Karplus PA. Hydrophobicity regained. Protein Sci. 1997;6:1302–7. doi: 10.1002/pro.5560060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan HS, Dill KA. Solvation: how to obtain microscopic energies from partitioning and solvation experiments. Annu Rev Biophys Biomol Struct. 1997;26:425–59. doi: 10.1146/annurev.biophys.26.1.425. [DOI] [PubMed] [Google Scholar]
- 14.Yutani K, Ogasahara K, Tsujita T, Sugino Y. Dependence of conformational stability on hydrophobicity of the amino acid residue in a series of variant proteins aubstituted at a unique position of tryptophan synthase alpha subunit. Proc Natl Acad Sci USA. 1987;84:4441–4444. doi: 10.1073/pnas.84.13.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellis JT, Nyberg K, Sail D, Fersht AR. Contribution of hydrophobic interactions to protein stability. Nature. 1988;333:784–786. doi: 10.1038/333784a0. [DOI] [PubMed] [Google Scholar]
- 16.Kellis JT, Nyberg K, Fersht AR. Energetics of complementary side chain packing in a protein hydrophobic core. Biochemistry. 1989;28:4914–4922. doi: 10.1021/bi00437a058. [DOI] [PubMed] [Google Scholar]
- 17.Serrano L, Kellis JT, Cann P, Matouschek A, Fersht AR. The folding of an enzyme: II. Substructure of barnase and the contribution of different interactions to protein stability. J Mol Biol. 1992;224:783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- 18.Shortle D, Stites WE, Meeker AK. Contributions of the large hydrophobic amino acids to the stability of staphylococcal nuclease. Biochemistry. 1990;29:8033–8041. doi: 10.1021/bi00487a007. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Stites WE. Packing is a key selection factor in the evolution of protein hydrophobic cores. Biochemistry. 2001;40:15280–15289. doi: 10.1021/bi011776v. [DOI] [PubMed] [Google Scholar]
- 20.Sandberg WS, Terwilliger TC. Energetics of repacking a protein interior. Proc Natl Acad Sci USA. 1991;88:1706–1710. doi: 10.1073/pnas.88.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eriksson AE, Baase WA, Zhang XJ, Heinz DW, Blaber M, Baldwin EP, Matthews BW. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 22.Xu J, Baase WA, Baldwin E, Matthews BW. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998;7:158–177. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baase WA, Liu AD, Tronrud DE, Matthews BW. Lessons from the lysozyme of phage T4. Protein Sci. 2010;19:631–641. doi: 10.1002/pro.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson SE, Moracci M, elMasry N, Johnson CM, Fersht AR. Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2. Biochemistry. 1993;32:11259–69. doi: 10.1021/bi00093a001. [DOI] [PubMed] [Google Scholar]
- 25.Main ERG, Fulton KF, Jackson SE. Context-Dependent Nature of Destabilizing Mutations on the Stability of FKBP12. Biochemistry. 1998;37:6145–6153. doi: 10.1021/bi973111s. [DOI] [PubMed] [Google Scholar]
- 26.Takano K, Yamagata Y, Yutani K. A general rule for the relationship between hydrophobic effect and conformational stability of a protein: stability and structure of a series of hydrophobic mutants of human lysozyme. J Mol Biol. 1998;280:749–761. doi: 10.1006/jmbi.1998.1906. [DOI] [PubMed] [Google Scholar]
- 27.Cota E, Hamill SJ, Fowler SB, Clarke J. Two proteins with the same structure respond very differently to mutation: the role of plasticity in protein stability. J Mol Biol. 2000;302:713–725. doi: 10.1006/jmbi.2000.4053. [DOI] [PubMed] [Google Scholar]
- 28.Loladze VV, Ermolenko DN, Makhatadze GI. Thermodynamic consequences of burial of polar and non-polar amino acid residues in the protein interior. J Mol Biol. 2002;320:343–357. doi: 10.1016/S0022-2836(02)00465-5. [DOI] [PubMed] [Google Scholar]
- 29.Clark AT, McCrary BS, Edmondson SP, Shriver JW. Thermodynamics of Core Hydrophobicity and Packing in the Hyperthermophile Proteins Sac7d and Sso7d. Biochemistry. 2004;43:2840–2853. doi: 10.1021/bi0358263. [DOI] [PubMed] [Google Scholar]
- 30.Bueno M, Campos LA, Estrada J, Sancho J. Energetics of aliphatic deletions in protein cores. Protein Sci. 2006;15:1858–1872. doi: 10.1110/ps.062274906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 32.Ratnaparkhi GS, Varadarajan R. Thermodynamic and structural studies of cavity formation in proteins suggest that loss of packing interactions rather than the hydrophobic effect dominates the observed energetics. Biochemistry. 2000;39:12365–12374. doi: 10.1021/bi000775k. [DOI] [PubMed] [Google Scholar]
- 33.Prevost M, Wodak SJ, Tidor B, Karplus M. Contribution of the hydrophobic effect to protein stability: analysis based on simulations of the Ile-96----Ala mutation in barnase. Proc Natl Acad Sci U S A. 1991;88:10880–4. doi: 10.1073/pnas.88.23.10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKnight JC, Doering DS, Matsudaira PT, Kim PS. A Thermostable 35-Residue Subdomain within Villin Headpiece. J Mol Biol. 1996;260:126–134. doi: 10.1006/jmbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- 35.Chiu TK, Kubelka J, Herbst-Irmer R, Eaton WA, Hofrichter J, Davies DR. High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein. Proc Natl Acad Sci USA. 2005;102:7517–7522. doi: 10.1073/pnas.0502495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eicken C, Sharma V, Klabunde T, Lawrenz MB, Hardham JM, Norris SJ, Sacchettini JC. Crystal structure of lyme disease variable surface antigen VlsE of Borrelia burgdorferi. J Biol Chem. 2002;277:21691–21696. doi: 10.1074/jbc.M201547200. [DOI] [PubMed] [Google Scholar]
- 37.Jones K, Wittung-Stafshede P. The largest protein observed to fold by two-state kinetic mechanism does not obey contact-order correlation. J Am Chem Soc. 2003;125:9606–7. doi: 10.1021/ja0358807. [DOI] [PubMed] [Google Scholar]
- 38.Xiao S, Bi Y, Shan B, Raleigh DP. Analysis of core packing in a cooperatively folded miniature protein: the ultrafast folding villin headpiece helical subdomain. Biochemistry. 2009;48:4607–16. doi: 10.1021/bi8021763. [DOI] [PubMed] [Google Scholar]
- 39.Bunagan MR, Gao J, Kelly JW, Gai F. Probing the folding transition state structure of the villin headpiece subdomain via side chain and backbone mutagenesis. J Am Chem Soc. 2009;131:7470–6. doi: 10.1021/ja901860f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pace CN, Hebert EJ, Shaw KL, Schell D, Both V, Krajcikova D, Sevcik J, Wilson KS, Dauter Z, Hartley RW, Grimsley GR. Conformational stability and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa3. J Mol Biol. 1998;279:271–286. doi: 10.1006/jmbi.1998.1760. [DOI] [PubMed] [Google Scholar]
- 41.Takano K, Scholtz JM, Sacchettini JC, Pace CN. The Contribution of Polar Group Burial to Protein Stability Is Strongly Context-dependent. J Biol Chem. 2003;278:31790–31795. doi: 10.1074/jbc.M304177200. [DOI] [PubMed] [Google Scholar]
- 42.Trevino SR, Gokulan K, Newsom S, Thurlkill RL, Shaw KL, Mitkevich VA, Makarov AA, Sacchettini JC, Scholtz JM, Pace CN. Asp79 makes a large, unfavorable contribution to the stability of RNase Sa. J Mol Biol. 2007;354:967–978. doi: 10.1016/j.jmb.2005.09.091. [DOI] [PubMed] [Google Scholar]
- 43.Trevino SR, Schaefer S, Scholtz JM, Pace CN. Increasing protein conformational stability by optimizing [beta]-turn sequence. J Mol Biol. 2007;373:211–218. doi: 10.1016/j.jmb.2007.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu H, Grimsley GR, Razvi A, Scholtz JM, Pace CN. Increasing protein stability by improving beta-turns. Proteins. 2009;77:491–8. doi: 10.1002/prot.22509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu H, Grimsley GR, Scholtz JM, Pace CN. Increasing protein stability: Importance of Delta Cp and the denatured state. Protein Sci. 2010;19:1044–1052. doi: 10.1002/pro.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shirley BA, Stanssens P, Hahn U, Pace CN. Contribution of hydrogen bonding to the conformational stability of ribonuclease T1. Biochemistry. 1992;31:725–732. doi: 10.1021/bi00118a013. [DOI] [PubMed] [Google Scholar]
- 47.Myers JK, Pace CN, Scholtz JM. Helix propensities are identical in proteins and peptides. Biochemistry. 1997b;36:10923–10929. doi: 10.1021/bi9707180. [DOI] [PubMed] [Google Scholar]
- 48.Grimsley GR, Shaw KL, Fee LR, Alston RW, Huyghues-Despointes BMP, Thurlkill RL, Scholtz JM, Pace CN. Increasing protein stability by altering long-range coulombic interactions. Protein Sci. 1999;8:1843–1849. doi: 10.1110/ps.8.9.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Vos S, Backmann J, Prevost M, Steyaert J, Loris R. Hydrophobic Core Manipulations in Ribonuclease T1. Biochemistry. 2001;40:10140–10149. doi: 10.1021/bi010565n. [DOI] [PubMed] [Google Scholar]
- 50.Joh NH, Oberai A, Yang D, Whitelegge JP, Bowie JU. Similar energetic contributions of packing in the core of membrane and water-soluble proteins. J Am Chem Soc. 2009;131:10846–7. doi: 10.1021/ja904711k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pace CN. Contribution of the hydrophobic effect to globular protein stability. J Mol Biol. 1992;226:29–35. doi: 10.1016/0022-2836(92)90121-y. [DOI] [PubMed] [Google Scholar]
- 52.Pace CN. Evaluating contribution of hydrogen bonding and hydrophobic bonding to protein folding. Methods Enzymol. 1995;259:538–554. doi: 10.1016/0076-6879(95)59060-9. [DOI] [PubMed] [Google Scholar]
- 53.Fleming PJ, Richards FM. Protein packing: dependence on protein size, secondary structure and amino acid composition. J Mol Biol. 2000;299:487–98. doi: 10.1006/jmbi.2000.3750. [DOI] [PubMed] [Google Scholar]
- 54.Liang J, Dill KA. Are Proteins Well-Packed? Biophysical Journal. 2001;81:751–766. doi: 10.1016/S0006-3495(01)75739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuntz ID, Crippen GM. Protein densities. Int J Pept Protein Res. 1979;13:223–8. doi: 10.1111/j.1399-3011.1979.tb01872.x. [DOI] [PubMed] [Google Scholar]
- 56.Kurochkina N, Privalov G. Heterogeneity of packing: structural approach. Protein Sci. 1998;7:897–905. doi: 10.1002/pro.5560070407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schell D, Tsai J, Scholtz JM, Pace CN. Hydrogen bonding increases packing density in the protein interior. Proteins: Struct Funct Genet. 2006;63:278–282. doi: 10.1002/prot.20826. [DOI] [PubMed] [Google Scholar]
- 58.Kajander T, Kahn PC, Passila SH, Cohen DC, Lehtio L, Adolfsen W, Warwicker J, Schell U, Goldman A. Buried charged surface in proteins. Structure. 2000;8:1203–1214. doi: 10.1016/s0969-2126(00)00520-7. [DOI] [PubMed] [Google Scholar]
- 59.Faham S, Yang D, Bare E, Yohannan S, Whitelegge JP, Bowie JU. Side-chain contributions to membrane protein structure and stability. J Mol Biol. 2004;335:297–305. doi: 10.1016/j.jmb.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 60.Joh NH, Min A, Faham S, Whitelegge JP, Yang D, Woods VL, Bowie JU. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453:1266–70. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Creamer TP, Srinivasan R, Rose GD. Modeling Unfolded States of Proteins and Peptides. II Backbone Solvent Accessibility. Biochemistry. 1997;36:2832–2835. doi: 10.1021/bi962819o. [DOI] [PubMed] [Google Scholar]
- 62.Auton M, Holthauzen LMF, Bolen DW. Anatomy of energetic changes accompanying urea-induced protein denaturation. Proc Natl Acad Sci USA. 2007;104:15317–15322. doi: 10.1073/pnas.0706251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pace CN, Huyghues-Despointes BM, Fu H, Takano K, Scholtz JM, Grimsley GR. Urea denatured state ensembles contain extensive secondary structure that is increased in hydrophobic proteins. Protein Sci. 2010;19:929–943. doi: 10.1002/pro.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 65.Tanford C, Anfinsen CB, JJTEaFMR . Adv Protein Chem. Vol. 24. Academic Press; 1970. Protein Denaturation: Part C. Theoretical Models for The Mechanism of Denaturation; pp. 1–95. [PubMed] [Google Scholar]
- 66.Fersht AR. The hydrogen bond in molecular recogition. Trends Biochem Sci. 1987;12:301–304. [Google Scholar]
- 67.Myers JK, Pace CN. Hydrogen bonding stabilizes globular proteins. Biophys J. 1996;71:2033–2039. doi: 10.1016/S0006-3495(96)79401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pace CN, Trevino S, Prabhakaran E, Scholtz JM. Protein structure, stability, and solubility in water and other solvents. Philos Trans R Soc Lond B Biol Sci. 2004:359. doi: 10.1098/rstb.2004.1500. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Powers ET, Deechongkit S, Kelly JW. Backbone-backbone H-bonds make context-dependent contributions to protein folding kinetics and thermodynamics: lessons from amide-to-ester mutations. Adv Prot Chem. 2006;72:39–78. doi: 10.1016/S0065-3233(05)72002-7. [DOI] [PubMed] [Google Scholar]
- 70.Gao J, Bosco DA, Powers ET, Kelley JW. Localized thermodynamic coupling between hydrogen bonding and the microenvironment polarity sustantially stabilizes proteins. Nature. 2009 doi: 10.1038/nsmb.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pace CN. Energetics of protein hydrogen bonds. Nat Struct Mol Biol. 2009;16:681–2. doi: 10.1038/nsmb0709-681. [DOI] [PubMed] [Google Scholar]
- 72.Bowie JU. Membrane protein folding: how important are hydrogen bonds? Curr Opin Struct Biol. 2010 doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hebert EJ, Giletto A, Sevcik J, Urbanikova L, Wilson KS, Dauter Z, Pace CN. Contribution of a conserved asparagine to the conformational stability of ribonucleases Sa, Ba, and T1. Biochemistry. 1998;37:16192–16200. doi: 10.1021/bi9815243. [DOI] [PubMed] [Google Scholar]
- 74.Pace CN. Polar group burial contributes more to protein stability than nonpolar group burial. Biochemistry. 2001;40:310–3. doi: 10.1021/bi001574j. [DOI] [PubMed] [Google Scholar]
- 75.Stickle DF, Presta LG, Dill KA, Rose GD. Hydrogen bonding in globular proteins. J Mol Biol. 1992;226:1143–1159. doi: 10.1016/0022-2836(92)91058-w. [DOI] [PubMed] [Google Scholar]
- 76.Harpaz Y, Gerstein M, Chothia C. Volume changes on protein folding. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 77.D’Aquino JA, Gomez J, Hilser VJ, Lee KH, Amzel LM, Freire E. The magnitude of the backbone conformational entropy change in protein folding. Proteins. 1996;25:143–56. doi: 10.1002/(SICI)1097-0134(199606)25:2<143::AID-PROT1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 78.Spolar RS, Record MT. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 79.Savage HJ, Elliott CJ, Freeman CM, Finney JL. Lost hydrogen bonds and buried surface area: rationalising stability in globular proteins. J Chem Soc, Faraday Trans. 1993;89:2609–2617. [Google Scholar]
- 80.Fleming PJ, Rose GD. Do all backbone polar groups in proteins form hydrogen bonds? Protein Sci. 2005;14:1911–1917. doi: 10.1110/ps.051454805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karp DA, Gittis AG, Stahley MR, Fitch CA, Stites WE, Garcia-Moreno EB. High Apparent Dielectric Constant Inside a Protein Reflects Structural Reorganization Coupled to the Ionization of an Internal Asp. Biophysical Journal. 2007;92:2041–2053. doi: 10.1529/biophysj.106.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. Hydrogen Bonding Markedly Reduces the pK of Buried Carboxyl Groups in Proteins. J Mol Biol. 2006;362:594–604. doi: 10.1016/j.jmb.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 83.Pace CN, Shirley BA, McNutt M, Gajiwala K. Forces contributing to the conformation stability of proteins. FASEB J. 1996;76:75–83. doi: 10.1096/fasebj.10.1.8566551. [DOI] [PubMed] [Google Scholar]
- 84.Makhatadze GI, Privalov PL. On the entropy of protein folding. Protein Sci. 1996;5:507–10. doi: 10.1002/pro.5560050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boczko EM, Brooks CL., 3rd First-principles calculation of the folding free energy of a three-helix bundle protein. Science. 1995;269:393–6. doi: 10.1126/science.7618103. [DOI] [PubMed] [Google Scholar]
- 86.Brady GP, Sharp KA. Entropy in protein folding and in protein-protein interactions. Curr Opin Struct Biol. 1997;7:215–21. doi: 10.1016/s0959-440x(97)80028-0. [DOI] [PubMed] [Google Scholar]
- 87.Pace CN, Laurents DV, Thomson JA. pH dependence of the urea and guanidine hydrochloride denaturation of ribonuclease A and ribonuclease T1. Biochemistry. 1990;29:2564–2572. doi: 10.1021/bi00462a019. [DOI] [PubMed] [Google Scholar]
- 88.Pace CN. Single surface stabilizer. Nat Struct Biol. 2000;7:345–6. doi: 10.1038/75100. [DOI] [PubMed] [Google Scholar]
- 89.Reiner A, Henklein P, Kiefhaber T. An unlocking/relocking barrier in conformational fluctuations of villin headpiece subdomain. Proc Natl Acad Sci U S A. 2010;107:4955–60. doi: 10.1073/pnas.0910001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lei H, Wu C, Liu H, Duan Y. Folding free-energy landscape of villin headpiece subdomain from molecular dynamics simulations. Proc Natl Acad Sci U S A. 2007;104:4925–30. doi: 10.1073/pnas.0608432104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kubelka J, Henry ER, Cellmer T, Hofrichter J, Eaton WA. Chemical, physical, and theoretical kinetics of an ultrafast folding protein. Proc Natl Acad Sci U S A. 2008;105:18655–62. doi: 10.1073/pnas.0808600105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang JS, Wallin S, Shakhnovich EI. Universality and diversity of folding mechanics for three-helix bundle proteins. Proc Natl Acad Sci U S A. 2008;105:895–900. doi: 10.1073/pnas.0707284105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Frank BS, Vardar D, Buckley DA, McKnight CJ. The role of aromatic residues in the hydrophobic core of the villin headpiece subdomain. Protein Sci. 2002;11:680–687. doi: 10.1110/ps.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu H. PhD Dissertation. Texas A&M University; 2009. Understanding forces that contribute to protein stability: applications for increasing protein stability. [Google Scholar]
- 95.Hebert EJ, Grimsley GR, Hartley RW, Horn G, Schell D, Garcia S, Both V, Sevcik J, Pace CN. Purification of ribonucleases Sa, Sa2, and Sa3 after expression in Escherichia coli. Protein Expression and Purif. 1997;11:162–168. doi: 10.1006/prep.1997.0776. [DOI] [PubMed] [Google Scholar]
- 96.Shaw KL. PhD Dissertation. Texas A&M University; College Station, TX: 2000. Reversing the net charge of ribonuclease sa. [Google Scholar]
- 97.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 98.Grimsley GR, Huyghues-Despointes BMy, Pace CN, Scholtz JM. Measuring the conformational stability of a protein. In: Simpson RJ, editor. Purifying proteins for proteomics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2004. pp. 535–566. [Google Scholar]
- 99.Pace CN, Horn G, Hebert EJ, Bechert J, Shaw K, Urbanikova L, Scholtz JM, Sevcik J. Tyrosine hydrogen bonds make a large contribution to protein stability. J Mol Biol. 2001;312:393–404. doi: 10.1006/jmbi.2001.4956. [DOI] [PubMed] [Google Scholar]
- 100.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 101.Pace CN, Grimsley GR, Thomson JA, Barnett BJ. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. J Biol Chem. 1988;263:11820–11825. [PubMed] [Google Scholar]
- 102.McDonald IK, Thornton JM. Satisfying hydrogen bonding in globular proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 103.Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA a-self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.