SUMMARY
Adipocyte hypertrophy and hyperplasia are important processes in the development of obesity. To understand obesity and its associated diseases, it is important to elucidate the molecular mechanisms governing adipogenesis. MiR-375 has been demonstrated to inhibit differentiation of neurites and participate in the regulation of insulin secretion and blood homeostasis. However, it is unknown whether miR-375 plays a role in adipocyte differentiation.
To investigate the role of miR-375 in adipocyte differentiation, we compared miR-375 expression level between 3T3-L1 pre-adipocytes and adipocytes using miRNA microarray and quantitative real-time RT-PCR (qRT-PCR) analysis. Furthermore, we evaluated the effects of overexpression or inhibition of miR-375 on 3T3-L1 adipocyte differentiation.
In this study, we found that miR-375 expression was increased after induction of adipogenic differentiation. Overexpression of miR-375 enhanced 3T3-L1 adipocyte differentiation: as evidenced by its ability to increase mRNA levels of both CCAAT/enhancer binding proteinα (C/EBPα) and peroxisome proliferator-activated receptor gamma (PPARγ2) and induction of adipocyte fatty acid-binding protein (aP2) and triglyceride (TG) accumulation. Furthermore, we found overexpression of miR-375 suppressed phosphorylation levels of extracellular signal-regulated kinases 1/2 (ERK1/2). In contrast, Anti-miR-375 increased ERK1/2 phosphorylation levels and inhibited mRNA expression of C/EBPα, PPARγ2 and aP2 in 3T3-L1 adipocyte, accompanied by decreased adipocyte differentiation.
Taken together, these data suggest that miR-375 promotes 3T3-L1 adipocyte differentiation, possibly via modulating ERK - PPARγ2 - aP2 pathway.
Keywords: MiRNA-375, 3T3-L1 Adipocytes, Differentiation, Obesity, ERK1/2
INTRODUCTION
Epidemic obesity is now a major health challenge in both developed and developing countries.1 The excessive growth, differentiation and hypertrophy of adipocytes, the major cellular component in fat tissue, are fundamental processes in the development of obesity. 2 Thus adipocytes are emerging as major targets in the prevention and treatment of obesity, type 2 diabetes, and cardiovascular disease. 3 To this end, it is important to understand the molecular mechanisms governing adipogenesis. The mouse 3T3-L1 pre-adipocyte cell line is a widely-used adipocyte differentiation model. Upon treatment with a standard cocktail of hormones, the cells undergo growth arrest, clonal expansion and subsequent terminal differentiation into mature adipocytes. This process is characterized by the expression of adipocyte-specific proteins such as aP2 protein, which is under the control of two key transcription factors: C/EBPα and PPARγ. 4 PPARγ is one of the nuclear receptors and is phosphorylated at its Ser84 site by ERKs. The ERK-PPARγ pathway is thought to play a role in adipocyte differentiation. 5–6
MicroRNAs (miRNAs) are short non-coding RNAs that post-transcriptionally regulate gene expression. 7 MiRNAs have been involved in numerous physiological and pathological processes, including development,8–9 energy homeostasis,10 sugar and lipid metabolism,11–13 and tumorigenesis.8,14 Recent findings indicate some miRNAs play a role in regulating adipocyte differentiation. For instance, miR-143 was up-regulated after induction of differentiation in human pre-adipocytes and mouse 3T3-L1 cells, and antisense oligonucleotides against miR-143 blocked adipocyte differentiation.15–16 The miR-103 and miR-17/92 cluster accelerated adipocyte differentiation, 17–18 while let-7, miR-27 and miR-27b impaired adipogenesis.19–21 However, it is unknown whether miR-375 plays a role in adipocyte differentiation. Six years ago, miR-375 was identified as a pancreatic-specific miRNA implicated in the regulation of insulin secretion.22 Recent studies further demonstrated that miR-375 is also important in maintaining blood homeostasis and inhibiting neurite differentiation. 23–25 Based on these observations, we sought to test the hypothesis that miR-375 is important in the regulation of adipocyte differentiation using an in vitro 3T3-L1 pre-adipocyte model.
METHODS
Materials
3T3-L1 pre-adipocytes were purchased from the American Type Culture Collection (Manassas, VA); 3-isobutyl-1-methylxanthine (IBMX), insulin and dexamethasone from Sigma (St. Louis, MO); Lipofectamine 2000, Trizol reagent and SYBR Green I dye from Invitrogen (Carlsbad CA); pSilencer™3.1-H1 expression vectors from Ambion (Austin, TX); polyclonal goat anti-PPARγ2 and anti-aP2, polyclonal rabbit anti-ERK1/2 and anti-phospho-ERK1/2, and mouse monoclonal anti-β-actin from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture and induction of differentiation in 3T3-L1 pre-adipocytes
3T3-L1 pre-adipocytes were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere. To induce differentiation, post-confluent 3T3-L1 pre-adipocytes (Day 0) were switched from regular DMEM growth media to differentiation media (DM) containing 0.5 mmol/L IBMX, 10 mg/mL insulin and 1 μmol/L dexamethasone, for 48 hrs. Cells were then maintained in DMEM media containing 10% FBS plus 10 mg/mL insulin, and replenished every other day with fresh media and insulin until Day 9, when cells were fully differentiated into adipocytes.
MicroRNA microarray analysis
Microarray analysis was performed on 5 μg of total RNA isolated from either 3T3-L1 pre-adipocytes or fully-differentiated adipocytes, using the μParaflo microfluidic technology according to the manufacturer’s protocol (LC Sciences, TX, USA). Hybridization was performed overnight on a microfluidic chip. On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide-coding segment complementary to a target miRNA and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. The array covered all miRNA transcripts available in the Sanger miRBase database (release 10). Post-hybridization, detection was achieved using fluorescence labeling with tag-specific Cy3 and Cy5 dyes. Hybridization images were collected using a GenePix 4000B laser scanner (Molecular Device, CA, USA) and digitized using Array-Pro image analysis software (Media Cybernetics, MD, USA). The data were then analyzed after background subtraction and internal normalization. Fold changes in miRNA expression were calculated with the ratio of two sets of detected signals [log2(adipocyte/pre-adipocyte)]. Then, the differences in miRNA expression were assessed by Student t test. P < 0.05 was considered significant.
Measuring the expressing levels of miRNA-375 by quantitative real-time PCR (qRT-PCR)
The relative expression levels of miR-375 in various samples were determined using a TaqMan MicroRNA Assay kit (ABI, USA) according to the manufacturer’s instruction. Briefly, total RNAs from cells were extracted utilizing an RNeasy mini kit. Then 10 ng of total RNAs were processed for reverse transcription (RT) using the TaqMan MicroRNA Reverse Transcription kit (ABI, USA). The resulting cDNA products were diluted at 1:20, and 1.33 μl of the diluted cDNA were used for PCR reaction (in a total volume 20 μl) with 1 μl of TaqMan MicroRNA Assay Mix and 10 μl of 2× TaqMan Universal PCR Master Mix. PCR reactions were conducted at 95°C for 10 min and then followed by 40 cycles (each cycle was for 15 s at 95°C, 60 s at 60°C) using an ABI 7900HT fast real-time PCR system. The highly conserved snRNA U6 was used as an internal normalizing control. The relative expression ratio of MiR-375 was calculated using the 2−ΔΔCt method.26
Generating and selecting cell clones that stably express miR-375
Oligonucleotides corresponding to the murine precursor sequence of miR-375 were introduced into pSilencer 3.1-H1 vector to obtain pmiR-375 vector. The introduced sequences were: sense, 5′-GATCCTTTGTTCGTTCGGCTCGCGTGATTCAAGAG ATCACGCGAGCCGAACGAACAAATTTTTGGAAA-3′; and antisense, 5′-AGCT TTTCCAAAAATTTGTTCGTTCGGCTCGCGTGATCTCTTGAATCACGCGAGC CGAACGAACAAAG-3′. Empty pSilencer™ 3.1-H1 vector was used as a negative control (pNeg). PmiR-375 or pNeg vector was transfected into 3T3-L1 pre-adipocytes using the Lipofectamine 2000 according to the manufacturer’s instructions. The stably transfected cell lines were obtained by G418 selection (800 mg/ml for 15 days) in DMEM growth media.
Transfection of anti-miR375 oligonucleotides into 3T3-L1 cells
3T3-L1 cells were plated in a six-well plate for 1 day and reached confluence, then cells were transfected with 2′-O-me-375 oligo (anti-miR-375: 5′-CAGUACUUUU GUGUAGUACAA-3′), or a control oligo (anti-miR-Con: 5′-CAGUACUUUUGUG UAGUACAA-3′) (Ambion, Austin, TX, USA) using Lipofectamine 2000 (150 ng of oligonucleotides were used in each well).
Measuring the mRNA levels of C/EBPa, PPARγ2 and aP2 by qRT-PCR
The qRT-PCR was performed as described previously.27 Briefly, total RNA was extracted from cells with Trizol reagent according to the manufacturer’s instructions (Invitrogen). First-strand cDNA was synthesized from 1μg total RNA using an oligo(dT)20 primer. The mRNA levels of selected genes were quantified using SYBR green PCR master mix in a LightCycler Real Time PCR system (Bio-Rad, Hercules, CA, USA). The sequences of PCR primers were as follows: (i) CCAAT/enhancer binding protein α (C EBPα), forward 5′-CGCAAGAGCCGAGATAAAGC-3′ and reverse 5′-CGGTCATTGTCACTGGTCAACT-3′; (ii) peroxisome proliferators -activated receptor (PPARγ2) forward 5′-CGCTGATGCACTGCCTATGA-3′ and reverse 5′-AGAGGTCCACAGAGCTGATTC C-3′ ; (iii) adipocyte fatty acid binding protein 2 (aP2) forward 5′-CATGGCCAAGCCCAACAT-3′ and reverse 5′-CGCCC AGTTTGAAGGAAATC-3′; and (iv)β-actin forward 5′-TGTCCACCTTCCAGCAG ATGT-3′ and reverse 5′-AGCTCAGTAACAGTCCGCCTAGA-3′. The thermal cycling programme consisted of 45 cycles (each cycle was for 30 s at 95°C, 30 s at 58°C and 40 s at 72°C). With β-actin as an internal control each sample was measured twice using the comparative cycle threshold (CT) method.26
Oil Red O staining
Cells were washed twice in phosphate-buffered saline (PBS) and fixed in 10% formalin at room temperature for 10 min, then stained with Oil Red O (stock solution: 3 mg/mL dissolved in isopropanol; working solution: 60% Oil Red O stock solution and 40% distilled water) at 60°C for 10 min and then counterstained with haematoxylin for 1 min. Microscopic images were obtained from randomly selected fields using an Olympus microscope (Tokyo, Japan). For quantitative analysis, the stained lipid droplets were dissolved in 100% isopropanol; then absorbance at 510 nm was obtained.
Triglyceride assay
Cells were washed twice with PBS, collected in saline solution and then sonicated for homogenization. Total triglyceride levels in cell lysates were measured with high performance liquid chromatography (HPLC) (results were expressed as milligrams of triglyceride per milligram of cellular proteins).
Western blotting analysis
Cells were washed twice in ice-cold PBS, and lysed in a buffer containing 10 mmol/L HEPES (pH7.9), 5 mmol/L MgCl2, 10 mmol/L KCl and 0.5 % NP-40. Cell lysates were collected by centrifuging at 13000 g for 15 min at 4°C. Protein concentrations in the cell lysates were determined by BCA assay. Sample proteins (30–50 μg) were separated by 10 or 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes. The membranes were blocked for 60 min in a buffer containing 0.1% Tween-20 and 5% milk. Antibodies against ERK1/2, phosphorylated ERK1/2, PPARγ2 and aP2 were used to identify specific proteins or the specific status of protein, which were visualized by ECL method described previously.28 Intensity of each protein band of interest was quantified by densitometry.
Statistical analysis
Data are presented as the mean ± SD. Two groups were compared by unpaired Student’s t test and multiple groups were analyzed by one-way analysis of variance (ANOVA) followed by Dunnett’s test. P < 0.05 was considered significant. All statistical analyses were performed using SPSS Version11.0 software (SPSS, Chicago, IL).
RESULTS
MiR-375 was upregulated in 3T3-L1 adipocytes
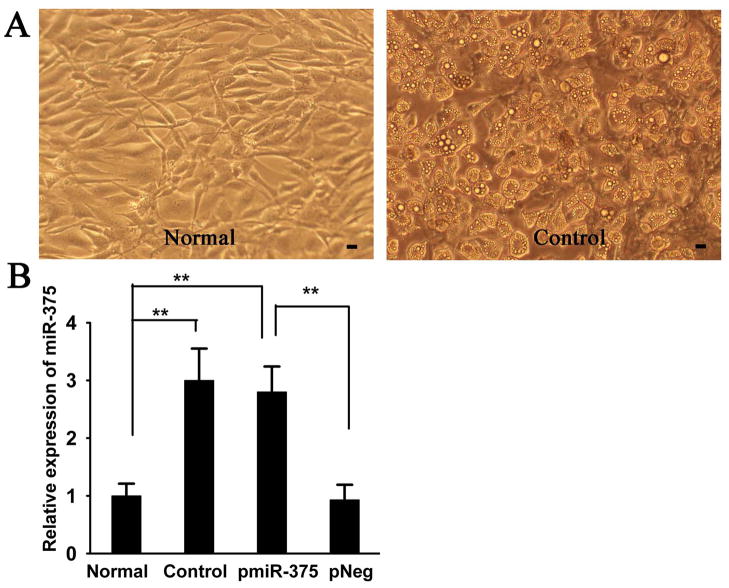
To study the potential role of miRNA in adipogenic differentiation, 3T3-L1 pre-adipocytes was induced to differentiate into mature adipocytes (Fig. 1A), then we performed miRNA microarray profiling in 3T3-L1 cells. A total of 28 miRNAs were detected to exhibit significantly differential expression in 3T3-L1 adipocytes (the Control group) compared to 3T3-L1 pre-adipocytes (the Normal group). Among them, miR-375 was shown to be significantly upregulated (about 2.65 folds) in 3T3-L1 adipocytes (Table 1). Consistent with the array results, qRT-PCR analysis further validated that miR-375 was significantly increased (about 3 folds) in Control group (differentiation group) compared to the Normal group (Fig 1. B). Together, these data demonstrate that expression level of miR-375 is elevated during adipogenic differentiation.
Fig. 1. MiR-375 expression is upregulated during 3T3-L1 adipocyte differentiation.
A:Representative microscopic images of 3T3-L1 pre-adipocytes (Normal) and 3T3-L1 adipocytes (Control); B: Relative expression ratio of miR-375 was measured by qRT-PCR as described in the Methods in Normal, Control, and 3T3-L1 pre-adipocytes that stably expressed pmiR-375 (pmiR-375) and pNeg (pNeg). The expression levels of miR-375 in the Normal cells were considered as 1, its relative expression levels in other groups were presented as a ratio with that of Normal cells. Data were expressed as the mean ± SD (n = 3). ** P< 0.01. Scale bar = 10 μm.
Table 1. Adipogenesis associated changes in miRNAs.
Relative levels of miRNAs were measured by miRNA microarray analysis in 3T3-L1 pre-adipocytes and 3T3-L1 adipocytes. Relative change was calculated as described in the Methods. P < 0.05 was considered as a significant difference. The value of positive logarithm indicates the gene was up-regulated, and negative logarithm indicates down-regulation.
| miRNA name | Folds of change | P value | miRNA name | Folds of change | P value |
|---|---|---|---|---|---|
| mmu- miR-143 | 2.67 | 0.005 | mmu-miR-23a | 1.87 | 0.031 |
| mmu-miR-375 | 2.65 | 0.008 | mmu-miR-23b | 2.08 | 0.013 |
| mmu-miR-21 | 1.67 | 0.017 | mmu-miR-296 | 1.26 | 0.009 |
| mmu-miR-27a | 1.37 | 0.025 | mmu-miR-290 | 2.17 | 0.015 |
| mmu-miR-27b | 2.10 | 0.032 | mmu-miR-24 | 1.08 | 0.012 |
| mmu-miR-22 | 1.07 | 0.031 | mmu-miR-29a | 1.21 | 0.011 |
| mmu-miR-185 | 1.93 | 0.019 | mmu-miR-298 | 1.45 | 0.007 |
| mmu-let-7a | 2.19 | 0.017 | mmu-miR-291a -5p | 1.28 | 0.023 |
| mmu-let-7b | 1.42 | 0.021 | mmu-miR-292-5p_MM2 | 2.56 | 0.036 |
| mmu-let-7c | 1.51 | 0.013 | mmu-miR-199a* | 2.37 | 0.031 |
| mmu-let-7e | 1.82 | 0.006 | mmu-miR-199b | 1.08 | 0.014 |
| mmu-let-7f | 1.56 | 0.028 | mmu-miR-292-3p | 1.58 | 0.022 |
| mmu-let-7g | 1.24 | 0.017 | spike_control-18 | −1.21 | 0.018 |
| mmu-miR-16 | 1.79 | 0.021 | mmu-miR-142-5p | −1.03 | 0.021 |
MiR-375 promoted 3T3-L1 adipocyte differentiation
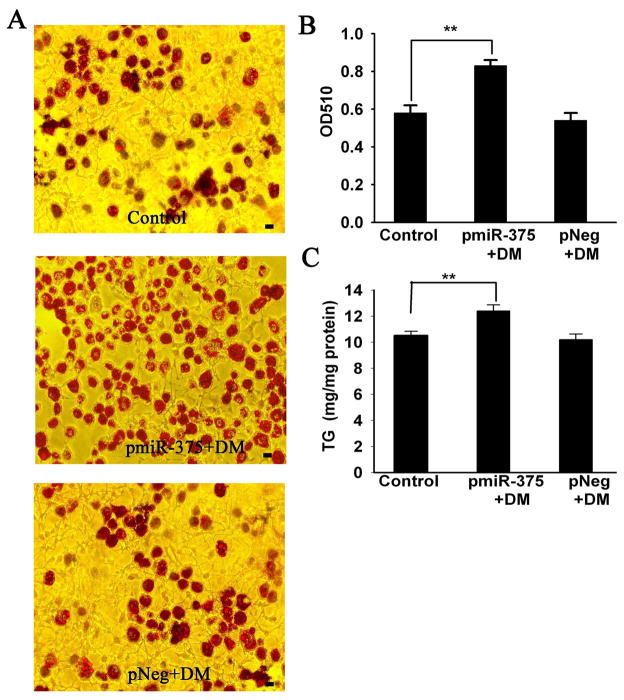
To investigate the role of miR-375 in 3T3-L1 adipocyte differentiation, we established 3T3-L1 cell lines with stable transfection of pmiR-375 and pNeg. QRT-PCR analysis showed that there were 3.0-fold increases of miR-375 in the pmiR-375 group as comparing to the pNeg group (vector control), and 2.8-fold increases compared with the Normal group (no vector and no differentiation). (Fig. 1B). These data verified success in overexpressing miR-375 in the selected cell line. Overexpression of the miR-375 significantly promoted adipocyte differentiation, as demonstrated by oil red O staining (Fig. 2A). As shown in Fig. 2A, total lipid accumulation significantly increased in the pmiR-375+DM group compared with the pNeg+DM group, which was further quantified by measuring optical density (OD) values at 510 nm (A510) in Oil Red O eluted solutions. There was a substantial (52.8%) increment in A510 readings in the pmiR-375+DM group as compared to the pNeg+DM group. Furthermore, triglyceride (TG) content measured by HPLC, was also increased (21.6%) in the pmiR-375+DM group in comparison to the pNeg+DM group. No difference in total lipid or TG was detected between the pNeg group+DM and Control group (Fig. 2).
Fig. 2. Overexpression of MiR-375 promoted adipocyte differentiation in 3T3-L1 cells.
3T3-L1 pre-adipocytes of Normal, pmiR-375 and pNeg were induced to differentiate into adipocytes with differentiation media (DM) as the Control group, pmiR-375+DM group and pNeg+DM group, respectively. The assays were performed on fully differentiated adipocytes (Day 9). (A) Oil Red O staining. (B) The amount of Oil Red O was quantified after extraction with isopropanol by reading optical density (OD) at 510 nm. (C) Triglyceride content was measured using the HPLC method. Data are expressed as the mean ± SD (n=3). ** P< 0.01. Scale bar = 10 μm.
MiR-375 up-regulated mRNA expressions of C/EBPα, PPARγ2 and aP2
Since adipogenesis is characterized by increased expression of key transcription factors (C/EBPα and PPARγ2) and adipocyte-specific genes (aP2), we then investigated the effects of miR-375 on the expression of these genes by qRT-PCR during the course of adipocyte differentiation. The elevated expression levels of C/EBPa, PPARγ2 and aP2 were initially observed 2 days after differentiation, and peaked in 5 days. MiR-375 significantly enhanced the mRNA levels of C/EBPα and PPARγ2 by 1.6- to 1.8-fold from day 2 to day 9 during adipocyte differentiation, as compared with the control group (Fig. 3A and 3B). Accordingly, MiR-375 also markedly increased aP2 expression (Fig. 3C). In contrast, there were no significant changes at mRNA expression levels of C/EBPα, PPARγ2 and aP2 between the control and pNeg+DM group. These results indicated that miR-375 stimulated the gene expression of adipogenic markers during 3T3-L1 adipocyte differentiation.
Fig. 3. MiR-375 up-regulated mRNA expressions of C/EBPα, PPARγ2 and aP2.
3T3-L1 cells were induced to differentiate as described in the methods. mRNA levels of C/EBPα, PPARγ2 and aP2 were examined by qRT-PCR at day 0, 2, 5, 9. Results were reported as change relative to the mRNA level of each gene on Day 0. Data are the mean ± SD (n = 3). * P < 0.05; ** P < 0.01.
Anti-miR-375 inhibited 3T3-L1 adipocyte differentiation
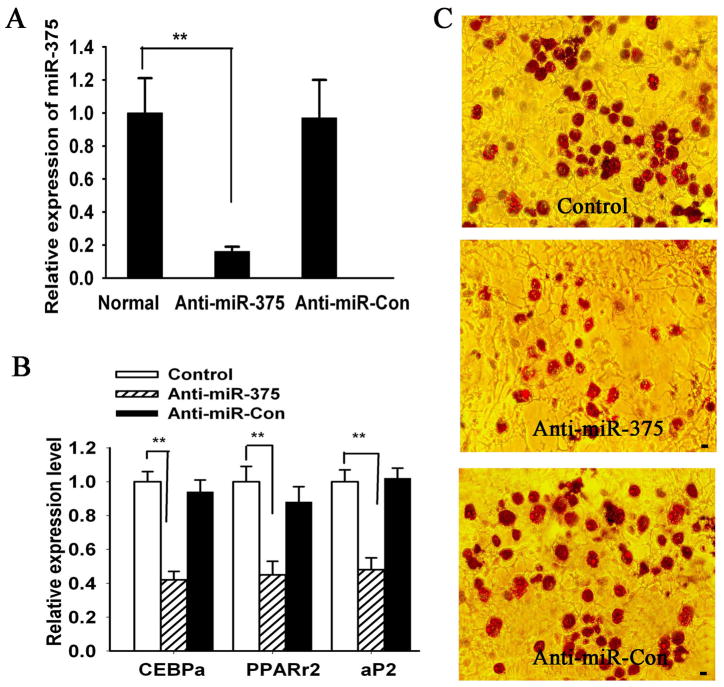
To further verify the effect of miR-375 on 3T3-L1 adipocyte differentiation, we transfected 3T3-L1 pre-adipocytes with a specific miR-375 oligos inhibitor (2′-O-me-375). QRT-PCR data validated that miR-375 was significantly decreased in cells treated with anti-miR-375 oligo as comparing to the control group. Oil red O staining and qRT-PCR analysis indicated that downregulation of miR-375 expression led to decreased adipogenic differentiation in 3T3-L1 cells (Fig. 4)
Fig 4. Inhibition of miR-375 decreased adipocyte differentiation in 3T3-L1 cells.
(A) MiR-375 levels were determined in control oligo (Anti-miR-Con) or 2′-O-me-375 (Anti-miR-375) transfected-3T3-L1 cells using qRT-PCR. Relative expression levels were obtained as described for Fig. 1B. (B) Total RNA was extracted from differentiated 3T3-L1 cells on Day 9. Gene expression of C/EBPα, PPARγ2 and aP2 was quantified by qRT-PCR for three independent experiments. Relative mRNA expression was calculated as relative change from the Control group level. (C) Oligonucleotide-transfected -3T3-L1 cells were grown to confluence, and adipogenic differentiation was induced for 9 days. Subsequently, the cells were stained with Oil Red O. Data are the mean ± SD (n = 3). ** P < 0.01. Scale bar = 10 μm.
Effect of miR-375 on the protein expression and activation of ERK1/2 in 3T3-L1 adipocytes
Some previous studies have suggested that ERK1/2 pathway was involved in adipocyte differentiation5–6. To investigate whether miR-375 regulates 3T3-L1 adipocyte differentiation via ERK1/2 pathway, we performed Western-blot analysis to examine the effect of miR-375 on the protein expression and activation status of ERK1/2 in 3T3-L1 adipocytes during induction period. No significant differences were observed in the total ERK levels among these groups. Nevertheless, the phosphorylation of ERK1/2 peaking at day 2 that was observed in the control group, miR-375 reduced or prevented the increase in ERK1/2 phosphorylation that was detected at day 2 compared to the control group and pNeg+DM group. However, anti-miR-375 significantly increased the phosphorylation of ERK1/2. In contrast, there were no significant differences in phosphorylation levels of ERK1/2 among the Control group, pNeg+DM group and anti-miR-Con group (Fig. 5).
Fig. 5. Effects of miR-375 on protein expression and activation of ERK1/2 in 3T3-L1 adipocytes.

Adipocyte differentiation was induced in 3T3-L1 cells in DM. Cell lysates were collected at indicated times of differentiation and subjected to Western blot analysis using antibodies specific for phosphorylated or total ERK1/2. Data are the mean ± SD (n = 3). * P < 0.05; ** P < 0.01.
Effect of miR-375 on PPARγ2 and aP2 protein expression in 3T3-L1 cells
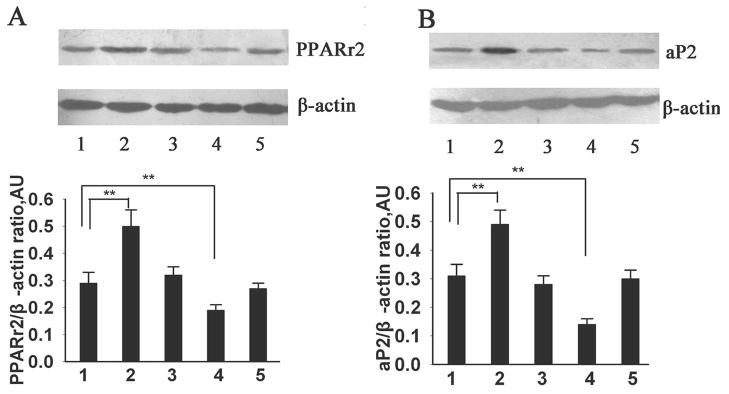
To determine whether miR-375 impacted on the downstream molecules of ERK signaling, we analyzed the protein expression of PPARγ2, a known key transcriptional factor in the adipogenic process. 4 In addition, we examined the protein expression of aP2, a hallmark of adipogenesis, and one of the targeted genes of PPARγ2 during adipocyte differentiation.4 As shown in Fig. 6, PPARγ2 and aP2 protein levels substantially increased on day 9 in the Control group, which further enhanced in the pmiR-375+DM group compared with the Control group and pNeg+DM group. However, the protein expression of PPARγ2 and aP2 was significantly reduced in the anti-miR-375 group compared with the Control group and anti-miR-Con group. There was no significant difference of PPARγ2 or aP2 protein expression among the control group, pNeg+DM group and anti-miR-Con group.
Fig. 6. Effects of miR-375 on PPARγ2 and aP2 protein expression in 3T3-L1 cells.
PPARγ2 and aP2 protein levels were measured by Western blot analysis in 3T3-L1 cells at differentiation day 9. Representative immunoblots and densitometric analysis on the specific protein levels normalized to internal β-actin level are reported: PPARγ2 in panel A and aP2 in panel B. Data are expressed as the mean ± SD (n=3). **P<0.01. 1: Control group; 2: pmiR-375+DM group; 3: pNeg+DM group; 4: Anti-miR-375 group; 5: Anti-miR-Con group
DISCUSSION
Adipocyte differentiation requires a highly orchestrated series of changes of gene expression in precursor cells. 4 Although PPARγ and C/EBPα have been identified as key regulators in adipogenesis, 4 the detailed molecular mechanisms remain to be fully elucidated. In the present study, we have identified miR-375 as a novel player in promoting adipocyte differentiation, as evidenced by its dramatic increase during adipogenic process, its ability to accelerate intracellular triglyceride accumulation (Fig. 1–2), as well as increasing mRNA and protein expression of several adipogenic marker genes (Fig. 3 & Fig. 6). Our results are in general agreement with recent publications in which various miRNAs were shown to participate in the regulation of adipocyte differentiation. 15–21 MiR-375 has been recently reported to negatively regulate glucose-stimulated insulin secretion by controlling the level of myotrophin.22 It also modulates β-cell functions by targeting PDK1 and maintains normal pancreatic α and β-cell mass.24, 29 MiR-375 has also been reported to mediate neurite differentiation, 25 However, our data reveal a new role for miR-375 in another physiological process, namely adipocyte differentiation.
What are the underlying mechanisms by which miR-375 promotes adipocyte differentiation? We found that overexpression of miR-375 decreased ERK1/2 phosphorylation, while anti-miR-375 significantly increased the phosphorylation of ERK1/2 in 3T3-L1 adipocytes (Fig. 5). Several laboratories have reported a role of ERK1/2 in regulating adipogenesis, but these conclusions have been controversial. Some studies reported that activation of ERK1/2 by various effectors blocked adipogenesis, 5, 30 whereas others indicated a requirement for rapid transient activation of ERK for either clonal expansion or adipocyte differentiation. 31–32 To explain these apparently contradictory results of ERK activation on adipogenesis, it has been speculated that activation of ERK might have opposing effects during adipogenesis. In early during adipocyte differentiation, ERK has to be activated for a proliferative step, but later in adipocyte differentiation, ERK needs to be shut off to prevent PPARγ phosphorylation. 33 Thus, it is likely that ERK activation is tightly and temporally controlled, since under certain conditions ERK activity may be required for adipocyte differentiation, while in other conditions ERK activity may impair adipocyte differentiation. Our data support the notion that the ERK1/2 pathway plays a negative role in 3T3-L1 adipocyte differentiation,5, 30 which we suggest is mediated by miR-375 (Fig. 5). Further, our data suggest that downstream targets of the ERK pathway are also regulated by miR-375. PPARγ2, a key adipogenic transcription factor, has been shown to be negatively regulated by ERK.34–35 Both the protein and mRNA levels of PPARγ2 were significantly increased in adipocytes overexpressing miR-375 as compared to those in adipocytes stably expressing pNeg (Fig. 2 & 6). In addition, the protein and mRNA levels of aP2, one of the target genes for PPARγ2,4 were also increased in the pmiR-375+DM group compared with the Control and pNeg+DM group (Fig. 2 & 6). By contrast, treatments with anti-miR-375 oligos significantly inhibited PPARγ2 and aP2 protein expression in these cells (Fig. 6). Taken together our data suggest that the increased expression of PPARγ2 and aP2 may be due to the release of inhibitory effects on PPARγ2 by ERK, whose activation is suppressed by miR-375 (Fig. 5). The exact molecular mechanisms by which ERK activation might suppress the expression of PPARγ2 remain to be delineated.
Recent reports indicate that myotrophin and PDK1 are targets of miR-375 in mediating insulin secretion and β-cell functions, respectively.22,24 Even though we demonstrated that miR-375 modulates the activation of ERK, but not its expression during adipocyte differentiation (Fig. 4), the exact target gene(s) of miR-375 remain to be identified during the process. At this time, it is difficult to determine whether the promotional effect of miR-375 on adipocyte differentiation is solely mediated through ERK pathway or through other unknown miR-375 targets. Further investigation is required.
In conclusion, the present study provides evidence that miR-375 promotes 3T3-L1 pre-adipocyte differentiation that maybe mediated through the ERK- PPARγ2 -aP2 pathway. Therefore, miR-375 is a potential target for designing therapeutic interventions in the treatment and prevention of obesity, type 2 diabetes, and metabolic syndrome.
Acknowledgments
The authors would like to thank Drs. Charles Henderson and Connie Mitchell of Palmer College of Chiropractic Florida for their kind proof reading on the manuscript. The work was supported by grants from the National Natural Science Foundation of China (81000328, 30971170), the National Major Basic Research Program of China (973 Program; 2006CB503808), the Natural Science Foundation of Hunan (07C0050), the Construct Program of the Key Discipline in Hunan Province and the Constructing Program of Key Disciplines in Hunan Province. LZ is supported by a National Institutes of Health K01AG21999 grant.
References
- 1.Haslam DW, James WP. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 2.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and Type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9 :367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nawrocki AR, Scherer PE. Keynote review: the adipocyte as a drug discovery target. Drug Discov Today. 2005;10:1219–1230. doi: 10.1016/S1359-6446(05)03569-5. [DOI] [PubMed] [Google Scholar]
- 4.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SW, Muise AM, Lyons PJ, et al. Regulation of adipogenesis by a transcriptional repressor that modulates MAPK activation. J Biol Chem. 2001;276:10199–10206. doi: 10.1074/jbc.M010640200. [DOI] [PubMed] [Google Scholar]
- 6.Kim KA, Kim JH, Wang YH, Sul HS. Pref-1 (Preadipocyte Factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol Cell Biol. 2007;27:2294–2308. doi: 10.1128/MCB.02207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Stefani G, Slack F. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 10.Teleman AA, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes & Dev. 2006;20:417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krützfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting Metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Gauthier BR, Wollheim CB. MicroRNAs: ‘ribo-regulators’ of glucose homeostasis. Nat Med. 2006;12:36–38. doi: 10.1038/nm0106-36. [DOI] [PubMed] [Google Scholar]
- 13.Poy MN, Spranger M, Stoffel M. MicroRNAs and the regulation of glucose and lipid metabolism. Diabetes Obes Metab. 2007;9:67–73. doi: 10.1111/j.1463-1326.2007.00775.x. [DOI] [PubMed] [Google Scholar]
- 14.Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14–3-3. Cancer Res. 2010;70:2339 – 2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 15.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 16.Kajimoto K, Naraba H, Iwai N. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA. 2006;12:1626–1632. doi: 10.1261/rna.7228806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58 :1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Li YC, Wang JH, et al. miR-17–92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. PNAS. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun T, Mingui F, Bookout AL, et al. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol. 2009;23:925 – 931. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Q, Gao Z, Alarcon RM, et al. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009;276 :2348–2358. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karbiener M, Fischer C, Nowitsch S, et al. MicroRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun. 2009;390 :247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 22.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 23.Lynn FC. Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab. 2009;20:452–459. doi: 10.1016/j.tem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 24.El OA, Baroukh N, Martens GA, et al. MiR-375 targets 3′-phosphoinositide-dependent protein kinase-1and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–2717. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelmohsen K, Hutchison ER, Lee EK, et al. MiR-375 inhibits differentiation of neurites by lowering HuD Levels. Molecular and cellular biology. 2010;30:4197–4210. doi: 10.1128/MCB.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Ling HY, Ou HS, Feng SD, et al. Changes in microRNA profile and effects of miR-320 in insulin-resistant 3T3-L1 adipocytes. Clin Exp Pharmacol Physiol. 2009;38:e32–39. doi: 10.1111/j.1440-1681.2009.05207.x. [DOI] [PubMed] [Google Scholar]
- 28.Ling HY, Hu B, Wang BX, et al. Effects of rosiglitazone on the proliferation of vascular smooth muscle cell induced by high glucose. Cardiovasc Drugs Ther. 2008;22:453–460. doi: 10.1007/s10557-008-6127-6. [DOI] [PubMed] [Google Scholar]
- 29.Poy MN, Hausser J, Trajkovski M, et al. miR-375 maintains normal pancreatic - and β-cell mass. PNAS. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanabe Y, Koga M, Saito M, et al. Inhibition of adipocyte differentiation by mechanical stretching through ERK-mediated downregulation of PPARr2. J Cell Sci. 2004;117:3605–3614. doi: 10.1242/jcs.01207. [DOI] [PubMed] [Google Scholar]
- 31.Boney CM, Smith RM, Gruppuso PA. Modulation of insulin-like growth factor I mitogenic signaling in 3T3-L1 preadipocyte differentiation. Endocrinology. 1998;139:1638–1644. doi: 10.1210/endo.139.4.5920. [DOI] [PubMed] [Google Scholar]
- 32.Prusty D, Park B, Davis KE, Farmer SR. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor r (PPARr) and C/EBPa gene expression during the differentiation of 3T3-L1 preadipocytes. J Biol Chem. 2002;277:46226 – 46232. doi: 10.1074/jbc.M207776200. [DOI] [PubMed] [Google Scholar]
- 33.Bost F, Aouadi M, Caron L, et al. The role of MAPKs in adipocyte differentiation and obesity. Biochimie. 2005;87:51–56. doi: 10.1016/j.biochi.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 34.Burgermeister E, Seger R. MAPK kinases as nucleo-cytoplasmic shuttles for PPARgamma. Cell Cycle. 2007;6:1539–48. doi: 10.4161/cc.6.13.4453. [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharya I, Ullrich A. Endothelin-1 inhibits adipogenesis: Role of phosphorylation of Akt and ERK1/2. FEBS Letters. 2006;580:5765–5771. doi: 10.1016/j.febslet.2006.09.032. [DOI] [PubMed] [Google Scholar]