Abstract
A cell's response to its environment is often determined by signaling through the actions of enzyme cascades. The ability to organize these enzymes into multiprotein complexes allows for a high degree of fidelity, efficiency and spatial precision in signaling responses.
Control of cell signaling events occurs at many levels. Classically, regulation of catalysis occurs via interactions with metabolites, cofactors or chemical messengers that allosterically modulate enzyme activity. Additionally, the post-translational modification of enzymes and effector proteins alters the binding properties and activity of these macromolecules. Together, these modifications act to adjust the flow of information through signal-transduction cascades.
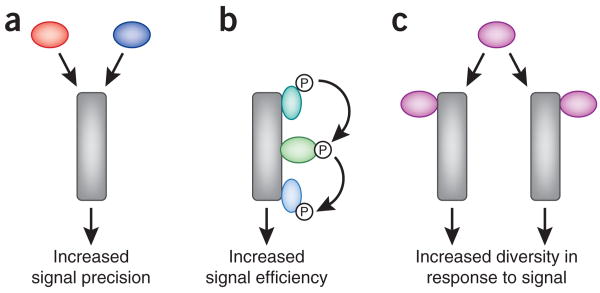
Protein-protein interactions also contribute to the control of cell signaling events. For example, the signal-dependent formation of multiprotein complexes creates local pockets of amplified enzyme activity1. The creation of these active signaling platforms allows spatially segregated changes in cellular behavior. Conversely, signaling components can also be released from these complexes upon activation, providing a mechanism for the relay of information from one cellular location to another. In the following sections, we will highlight some ways that cells process information through multiprotein complexes. For the sake of simplicity, we will focus on three ways that intracellular signals can be processed. These are (i) the integration of distinct chemical signals at the level of a signaling complex; (ii) the linear relay of signals through preassembled signaling scaffolds; and (iii) the spatial organization and segregation of parallel signaling units via the compartmentalization of broad-spectrum signaling enzymes (Fig. 1a–c).
Figure 1.
Schematic representation of mechanisms of signal transduction. (a) In signal integration, two independent inputs act on the same pathway to elicit a common outcome. (b) Scaffolding proteins allow the efficient relay of signals through successive enzymes in a pathway. (c) Specificity of signaling through broad-specificity enzymes is often assured by the spatial segregation of enzyme complexes.
Complexes in signal integration
The idea that two signals converge to elicit specific biological events is a common theme in cellular regulation. This is perhaps best exemplified by the synergistic actions of different second messengers such as cAMP and calcium in the regulation of cardiac contractility and insulin secretion from pancreatic β islets2,3. In both cases, the synchronization and integration of cAMP and calcium-responsive effectors enhances the speed and precision of these complex cellular events. However, the efficiency of signal integration is often increased when enzymes are anchored within the same signaling complex.
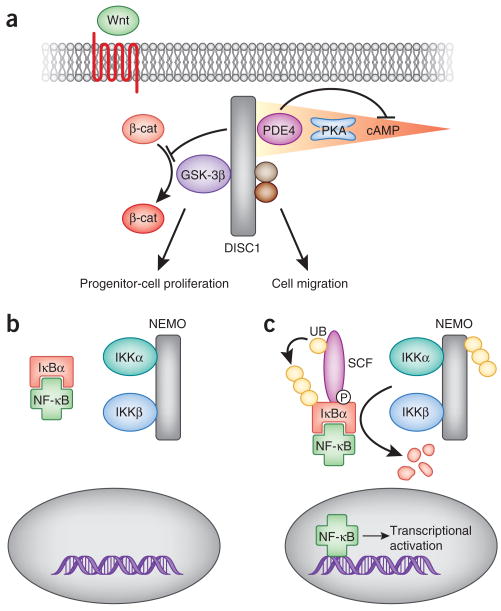
For example, Disrupted-in-Schizophrenia-1 (DISC1), a multifunctional scaffold protein, integrates signaling through a number of distinct pathways to regulate various aspects of neurodevelopment. Mutations in DISC1 are associated with the development of schizophrenia, a psychiatric disorder characterized by disturbances in cognition, perception and social interactions4,5. Although schizophrenia manifests in early adulthood, it is generally believed to be caused by defects in neurodevelopment6,7. Consistent with this, DISC1 is required for the parallel processing of signals from two pathways important for normal neuronal development and function—specifically, Wnt and cyclic AMP (cAMP) signaling. DISC1 has been shown to mediate signaling downstream of Wnts, as gene silencing of DISC1 leads to a reduction in Wnt responsiveness8,9 (Fig. 2a). Functionally, depletion of DISC1 results in decreased numbers of neural progenitor cells in the subventricular zone and dentate gyrus due to premature cell-cycle exit of these progenitors9. This phenotype is caused by disruptions in Wnt signaling, as it can be rescued by the expression of a degradation-resistant form of β-catenin or the pharmacological inhibition of glycogen synthase kinase 3β (GSK-3β)8,9. In addition, DISC1 has been implicated in the regulation of cAMP signaling through interactions with several members of the phosphodiesterase 4 (PDE4) family10 at centrosomes (Fig. 2a). In the presence of high levels of cAMP, the activity of PDE4s is increased following protein kinase A (PKA)-mediated phosphorylation. As a result, cAMP is metabolized by PDE4 and PKA activity is terminated11. Notably, missense mutations in DISC1, which alter the binding of PDE4 isoforms to the scaffold, result in schizophrenic and depressive behavioral phenotypes12. Although far from proven, there is reason to believe that DISC1 may provide an environment for the integration of cAMP and Wnt signals. This may occur at the level of GSK-3β, an enzyme that is inactivated upon PKA phosphorylation at Ser9 (ref. 13). Thus, DISC1 may prove to represent an example of a scaffold molecule that has the capacity to integrate distinct and independent upstream signaling pathways in a single cell. Whether all of its binding partners are present in a single complex at the same time is an avenue for further investigation, and the manner in which changes in the stability or composition of the DISC1 complex contribute to the onset of schizophrenia has yet to be fully understood. Nonetheless, the discovery that changes in this multienzyme complex can be a factor in the etiology of psychiatric disorder underscores the role of the cAMP and Wnt signaling cascades in the control of cognitive function, emotional responses and social behaviors.
Figure 2.
Signal integration: the DISC1 scaffold integrates Wnt and cAMP signaling to coordinate neurodevelopment. (a) DISC1 is required to regulate GSK-3β activity downstream of Wnt signaling, resulting in the stabilization and nuclear translocation of β-catenin (β-cat). This action of DISC1 is required for the regulation of progenitor-cell proliferation in the brain. Additionally, DISC1 coordinates PKA targets nuclear distribution protein nudE–like 1 (NDEL1) and LIS1 as well as PDE4 on the same molecular scaffold. This arrangement ensures tight control of cAMP-mediated signaling and likely has a role in cell migration. (b) Signaling through NF-κB requires the integration of phosphorylation and ubiquitination events. Under basal conditions, the nuclear localization signal (NLS) of NF-κB is masked by the binding of IκBs, holding NF-κB in the cytosol. (c) The nuclear translocation of NF-κB is initiated by the phosphorylation and ubiquitination of IκBs as a result of NEMO ubiquitination, IKK activation and recruitment of SCF E3 ligases to pIκB. UB, ubiquitin.
Another role of signal-integrating complexes is the incorporation of signal-termination enzymes. This introduces a temporal component to a signaling pathway, as the active state of the complex is transient. The transcription factor nuclear factor κB (NF-κB) is activated in response to extracellular stimuli and controls the transcription of genes involved in many cellular processes14. Under basal conditions, NF-κB exists in a cytosolic complex with two main inhibitory proteins, IκBα15,16 and IκBβ17 (Fig. 2b). Stimulation of the pathway results in the destabilization of the IκB proteins and the translocation of NF-κB into the nucleus. The stability of the cytosolic NF-κB–IκB complex depends on the activity level of the IκB kinase (IKK) complex18–21. This complex consists of two catalytic subunits, IKKα and IKKβ, and the master regulator, NF-κB essential modulator (NEMO)22. This complex is activated by NEMO ubiquitination23,24, which results in the phosphorylation of the NF-κB inhibitor IκB by the IKKs. This phosphorylation event recruits the Skp, Cullin, F-box–containing (SCF) family of ubiquitin ligases to IκB. The binding of the E3 ligase results in the polyubiquitination and subsequent degradation of IκB25–28 (Fig. 2c). The nuclear localization sequence of NF-κB is unmasked by this degradation, allowing its translocation to the nucleus. Similarly, signal termination depends on the integration of the ubiquitination and phosphorylation states of various pathway members. NEMO is deubiquitinated by the cylindromatosis tumor suppressor protein (CYLD), decreasing the activity of the IKK complex29. Meanwhile, IκB proteins are dephosphorylated by the phosphatase PP2A, preventing further recruitment of the SCF ubiquitin ligases. Together, these changes result in the stabilization of IκBs and the sequestration of NF-κB in the cytosol. Thus, activation and termination of NF-κB signaling require the integration of multiple signaling inputs such as phosphorylation and ubiquitination. Furthermore, the proximity of signal-termination elements such as PP2A and the E3 ubiqutin ligases within the NF-κB complex ensures that these events occur rapidly.
Scaffold proteins organize signal relay
The linear transfer, from one enzyme to the next, of signals that are organized into a protein scaffold is an efficient means of cellular communication. Scaffolding proteins often maintain such multienzyme complexes30. This process may be best exemplified by the molecular organization of eukaryotic mitogen-activated protein (MAP) kinase cascades31. Extracellular stimuli trigger the processive activation of these kinases when organized into three-tier enzyme cascades. Distinct signals trigger the first member of the cascade, a MAP kinase kinase kinase (MAP3K). This enzyme in turn phosphorylates and activates MAP kinase kinases (MAP2Ks). This intermediary enzyme phosphorylates the terminal MAP kinase (MAPK) that is then free to act on various downstream targets including other protein kinases, transcriptional factors and cytoskeletal components. A consensus view is that scaffolding proteins function to spatially organize MAP cascades in a manner that drives the flow of information from the initiator kinase to the terminal kinase in the complex. Prototypic examples of this configuration would include the kinase suppressor of Ras (KSR) scaffold, which organizes the Raf and MEK ERK kinase cascades32,33 and the Jun kinase interacting proteins (JIPs), which synchronize the activity of enzymes in the Jun N-terminal kinase cascade34. A common feature of these complexes is that the upstream kinases such as Raf and MKK1 have restricted substrate specificities and act exclusively on the next enzyme in the cascade. Thus, the binding of the initiator kinases to scaffolds places the enzymes in the vicinity of their respective targets. More importantly, the spatial grouping of successive signaling kinases by scaffolds places them in a context that facilitates the preferential relay of signals to the terminal enzyme in the chain.
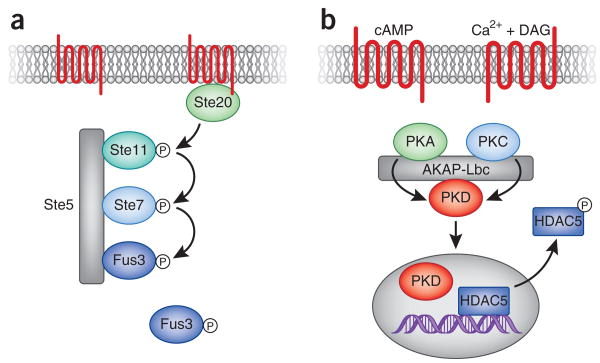
Another useful property of enzyme scaffolding is to segregate signals in a manner that prevents indiscriminate cross-talk between pathways. This concept is particularly important in unicellular organisms such as yeast, where a variety of cytoplasmic processes can be simultaneously modulated by different kinase scaffolds. In yeast, mating, invasive growth and responses to high osmolarity are all regulated by distinct MAP kinase pathways that share a common MAP3K called Sterile 11 (Ste11)35–37. Segregation of Ste11 activity involves binding to scaffolding proteins such as Pbs2 and Ste5. Recruitment of Ste11 into the osmosensing pathway requires interaction with Pbs2 (ref. 37). This protein not only scaffolds Ste11 but also acts as the MAP2K in this pathway. In contrast, Ste5 organizes Ste11 and the kinases Ste7 and Fus3 to direct signals through the yeast mating pathway (Fig. 3a). Recent evidence suggests that Ste5 also facilitates the activation of its kinase-binding partners38. Synthetic biology approaches have identified a regulatory domain in Ste5 that catalytically unlocks the Fus3 kinase for phosphorylation by Ste7 (ref. 39). This finding broadens the role of scaffolding proteins, as it suggests that Ste5 not only functions to organize successive components of a yeast MAPK cascade but also allosterically modifies the conformation of its bound kinases, making them more amenable to activation.
Figure 3.
Ste5 coordinates signal activation in the yeast mating pathway. (a) In response to the presence of pheromone, Ste20 is activated at the cell membrane and in turn activates its downstream target Ste11. This initiates a cascade of phosphorylation events leading to the activation and release of the terminal component Fus3 from the Ste5 scaffold. By spatially coordinating the kinases involved in the pheromone response, Ste5 ensures both the speed and accuracy of signaling. (b) AKAP-Lbc coordinates the activation and translocation of protein kinase D. The integration of calcium/phospholipid (diacylglycerol, DAG) and cAMP signals promotes the activation of PKA and PKC respectively. These enzyme activities act synergistically to promote the phosphorylation of PKD in the active-site loop and its release from the anchoring protein complex. The newly active enzyme is then free to translocate into the nucleus, where it can phosphorylate the DNA-modifying enzyme HDAC5. Phospho-HDAC5 then exits the nucleus, thereby promoting the relaxation of DNA. In cardiomyocytes, this leads to transcriptional activation of genes that propagate the hypertrophic responses.
In other contexts, scaffolding proteins can participate in the transfer of signals from one region of the cell to the next. The A kinase anchoring protein AKAP-Lbc forms a multiprotein complex that relays information from the plasma membrane to the nucleus in response to hypertrophic signals40,41. These signals, which include elevated adrenergic activity, lead to the reprogramming of cardiomyocyte gene expression by myocyte enhancer factor 2 (MEF2), known as the fetal gene response, and ultimately to increased cell size42,43. Initiation of the fetal gene response requires the phosphorylation and subsequent nuclear export of the MEF2 binding partner and transcriptional repressor HDAC5 (refs. 44,45). The lipid-responsive kinase protein kinase D1 (PKD1) is known to phosphorylate histone deacetylase 5 (HDAC5) in response to hypertrophic agents. This kinase is found in an AKAP-Lbc anchored multiprotein complex with two upstream activating kinases, PKA and protein kinase C (PKC) (Fig. 3b). In response to stimulation, PKC phosphorylates PKD1 on Ser744 and Ser748, leading to a small increase in PKD kinase activity46. Maximal activation of PKD1 occurs when PKC phosphorylation of PKD1 coincides with PKA-mediated phosphorylation of AKAP-Lbc46. The coincidence of these two signals leads to the release of active PKD1 from AKAP-Lbc and the translocation of the active kinase into the nucleus. Inside the nucleus, PKD1 phosphorylates HDAC5 at Ser498 and Ser660 (ref. 41). These phosphorylation events are critical for the recruitment of 14-3-3 proteins to HDAC5 and its subsequent nuclear export41 (Fig. 3b). Once released from HDAC5, MEF2 initiates the transcription of genes involved in the fetal gene response. Coanchoring of all three kinases is essential for the efficient relay of information, as markers of hypertrophy are not observed following stimulation in cells expressing AKAP-Lbc mutations, which disrupt binding to PKA, PKC or PKD.
Although kinase scaffolds come in all shapes and sizes, a unifying principle of the KSR, JIP, Ste5 and AKAP-Lbc scaffolds seems to be that the terminal kinase is released from the multiprotein complex once it has been activated. This final step is critical for the propagation of the signal. First, it insures that the activated kinase is free to diffuse through the cell to its site of action (frequently in the nucleus). Second, the persistent removal of the activated terminal kinase and its replacement by a dormant enzyme provides a mechanism to amplify the signal. Another implication of kinase scaffolding is that these enzyme complexes may represent important target sites for drug action. For example, the Raf/MEK/ERK trio modulates cell growth and proliferation. Research interest in this signaling pathway has been prompted by clinical evidence that activating mutations in Ras are found in 20–25% of all human tumors. Drugs that inhibit Raf, such as sorafenib, are prescribed to combat renal and hepatic carcinomas, whereas PLX4720 targets melanoma. Thus, these drugs must effectively target Raf in the context of the KSR-1 scaffold in order to have any therapeutic potential.
Spatial segregation of signals
As we have gained greater understanding of the molecules involved in cell signaling, it has become clear that sometimes the same enzymes are used in diverse pathways and different contexts. Spatial segregation of kinases, phosphatases and GTPases is necessary to preserve the specificity of these broad-spectrum enzymes. This is often achieved by their attachment to membranes, organelles and the cytoskeleton. This compartmentalization creates an environment in which a common chemical message can simultaneously evoke several independent but local cellular responses. Spatial segregation of receptors and enzymes is frequently used by signaling pathways that process extracellular queues such as hormonal input or respond to the generation of intracellular second messengers.
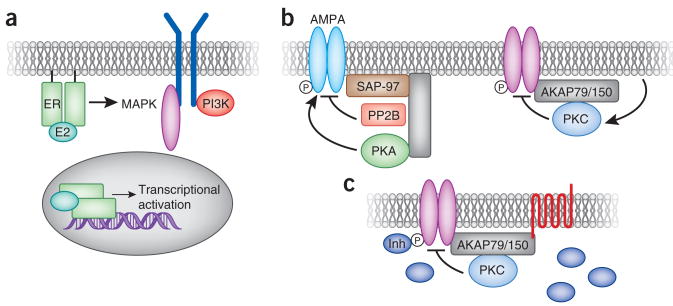
Hormones, such as estrogen and estradiol, readily diffuse across the cell membrane, where they encounter estrogen receptors (ERs). The cellular response to this estrogen pulse depends on the subcellular pool of the ER engaged. Much of the recent work in this field has focused on the different roles of plasma membrane47 and nuclear ERs (Fig. 4a). Although some debate remains regarding the contribution of low-affinity ERs such as GPR30, it is clear that the classical nuclear receptors ERα and ERβ are localized to and crucial for estrogen signaling at the plasma membrane48–50. Palmitoylation and subsequent interaction with caveolin-1 appear to promote the translocation of ERα and ERβ to the plasma membrane; however, the precise regulation of these events is currently unclear51,52. Nevertheless, the identification of the same subset of receptors at both the plasma membrane and the nucleus implies that cellular responses to estrogen are not solely determined by the receptor subtype but also by the subcellular location of the hormone–ER complex. For example, rapid activation of RAF/MEK/ERK and phosphoinositide 3 kinase (PI 3-kinase) is known to occur downstream of estrogen signaling53–57, leading to changes in signaling, cell motility and cell survival58,59. These effects require plasma-membrane rather than nuclear ERs50. Conversely, membrane-localized ERs are not sufficient to mediate the prolonged effects of estrogen, such as the upregulation of developmental programs and transcriptional control by nuclear receptors50. Thus, these two pools of receptors have distinct functions that are crucial for normal cellular behavior. Additionally, it has emerged that membrane ER signaling may synergize with nuclear receptors. In one example, crosstalk between membrane ERs and epidermal growth factor or insulin- like growth factor 1 leads to activation of downstream ERK signaling cascades60,61. In some circumstances, the upregulation of MAPK signaling leads to the phosphorylation of nuclear ERs on Ser305, triggering ligand-independent transcriptional activation62,63. Activation of PI 3-kinase signaling by membrane ERs upregulates transcriptional regulation via a different mechanism. Under resting conditions, the nuclear ERα is inactivated through a chronic, GSK-3β–mediated phosphorylation of Ser118 (ref. 64). However, the cross-activation of PI 3-kinase and AKT upon estrogen binding to membrane ERα leads to the phosphorylation of GSK-3β. This results in the inactivation of GSK-3β65 and the derepression of the nuclear ERα. These examples highlight how spatial segregation of similar effector proteins can be used to impose higher levels of cellular organization.
Figure 4.
Segregation of estrogen signaling events. (a) Estrogen binds receptors at the plasma membrane and in the nucleus, leading to distinct cellular outcomes. Interaction with plasma-membrane receptors leads to cross-activation of other signaling components, including growth factor receptors, MAPK and PI 3-kinase (PI3K). Interaction with nuclear receptors leads to transcriptional activation of estrogen-responsive genes. Segregation of AKAP79/150 signaling events. (b) AKAP79/150 is cross-linked to the AMPA-type glutamate receptors via interaction with the membrane associated guanylate kinase (MAGUK) scaffolding protein SAP-97. This configuration brings the kinase PKA and the phosphatase PP2B in proximity to the channel to control its phosphorylation state. A complex between the same anchoring protein and a different binding partner, PKC, facilitates the phosphorylation-dependent suppression of M channels. (c) An unusual property of the AKAP79/150–PKC interaction is that the anchored kinase is rendered insensitive to ATP-competitive inhibitors (Inh). This creates a local pool of the enzyme that is resistant to these pharmacological agents.
Signal segregation is evident in the action of the second messenger cAMP. This cyclic nucleotide freely diffuses from its site of synthesis at the plasma membrane to discrete compartments where it activates cAMP responsive kinases (PKA), ion channels and guanine nucleotide exchange factors (EPACs). Subcellular targeting of these ‘cAMP receptors’ is mediated by a family of AKAPs that tether PKA and EPACs in proximity to selected substrates66,67. However, another notable feature of AKAPs is the ability to organize groups of second messenger–regulated enzymes. For example, the neuronal anchoring protein AKAP79/150 anchors the cAMP-dependent kinase PKA, the Ca2+- and lipid-regulated PKC and the calmodulin-stimulated protein phosphatase PP2B/calcineurin68,69. A combination of biochemical and electrophysiological approaches has shown that distinct AKAP79/150 signaling complexes modulate the activity of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPA) and M/KCNQ ion channels70–73. Notably, the regulation of these channels requires a nonoverlapping subset of anchored enzymes.
The AMPA current propagates excitatory synaptic responses74,75. This ligand-gated ion channel requires the anchoring of PP2B and PKA for full activity76–78. Specifically, AKAP79/150–anchored PP2B is required for the attenuation of the AMPA current in response to tonic stimulation, whereas PKA is required for the maintenance of AMPA currents and channels at the cell surface71. Although the AKAP79/150–AMPA channel complex has the capacity to recruit PKC, this enzyme does not seem to be required for the control of this subset of excitatory synaptic responses. However, in the sympathetic nervous system, anchoring of PKC by AKAP79/150 is required for the attenuation of the hyperpolarizing K+ current (M current) through the M/KCNQ channel79. AKAP79/150 targets PKC where it can optimally respond to activating signals from the m1 muscarinic receptor and preferentially phosphorylate the KCNQ2 subunit to suppress the M current71,72. Whereas anchoring of PKA and PP2B is dispensable for the regulation of the M current, the PKC binding region of AKAP79/150 is both necessary and sufficient for M-current suppression71 (Fig. 4b). Another interesting feature of the M-current complex is that AKAP79/150 protects PKC from certain ATP-competitive inhibitors80. This implies that anchoring proteins such as AKAP79/150 not only segregate individual cell-signaling processes but also can change the pharmacological profile of certain protein kinases (Fig. 4c). Together, these data raise the possibility that AKAP79/150 segregates distinct second-messenger signals within a cell by tethering unique combinations of binding partners with different ion channels. An added level of regulation may be introduced if interaction with AKAPs or other interacting proteins can modify kinase susceptibility to pharmacological agents. This finding should have important consequences for drug discovery and research projects predicated on the selectivity of pharmacological protein kinase inhibitors.
Conclusion
As our appreciation of cellular architecture grows, it has become apparent that cells are not just bags of enzymes. Rather, eukaryotic cells require a high degree of molecular organization to integrate, relay or segregate the chemical signals that control all aspects of cellular behavior. Although the three modes of molecular organization discussed in this article offer unique advantages for efficient information transfer, it should be noted that these processes are not mutually exclusive. In fact, many signaling pathways use two or more of these mechanisms. For example, both DISC1 and NF-kB make use of the spatial segregation and the integration of multiple signaling inputs to fine tune cellular responses. Although DISC1 may integrate Wnt and cAMP signals, it is not yet clear that these messages influence the same biological event. In contrast, components of NF-kB signaling pathway use cytoplasmic phosphorylation and ubiquitination events to enact transcriptional activation in the nucleus. Certainly, the proximity of the phosphorylation and ubiquitination machinery ensures that changes in the stability of the NF-kB complex can be rapidly modulated. Although working in a different way, signaling from the membrane to the nucleus is a characteristic shared by AKAP-Lbc complexes. However, their mechanism of action shows hallmarks of all three modes of molecular organization. As a scaffold, AKAP-Lbc segregates protein kinase D from its nuclear targets by anchoring it in the cytosol. However, upon adrenergic stimulation, AKAP-Lbc integrates cAMP, calcium and phospholipid signals to mobilize successive enzymes in the PKD activation cascade. This results in the release and nuclear translocation of PKD, ultimately triggering transcriptional activation. In fact, translocation of active PKD from the cytoplasm to the nucleus is necessary to trigger the next phase in this transcriptional activation process. Thus, through the amalgamation of these simple regulatory steps, it is possible to coordinate and synchronize sophisticated cellular events in both space and time.
All in all, we have just begun to glimpse the complexity of the protein-protein interactions that underlie the cellular organization of cell-signaling cascades. Therefore, we would like to speculate on what the future might be for this field. Recent advances in technology hold promise for the future. Most signaling responses represent the coupling of individual chemical, physical or electrical events. Therefore, the simultaneous monitoring of more than one event will undoubtedly give more information about the order and dynamics of a given cellular response. For example, real-time visualization of AKAP79-anchored PKC phosphorylation with a fluorescent kinase activity reporter called CKAR can now be performed concurrently with measuring electrophysiological changes in muscarine-sensitive ion channels. The coincident detection of both steps suggests that AKAP79 not only directs PKC toward the ion channel but also synchronizes kinase activation to instantaneously reduce ion flow80,81. The advent, some time ago, of genetically encoded fluorescence reporters developed by Tsien and colleagues opens many more possibilities for this type of study82,83. In addition, the ability to monitor the impact of a signal input using systems biology approaches should provide deeper insight as to how a cell manages to simultaneously coordinate several distinct events. Mathamatical modeling of these signaling pathways may go hand in hand with these system-wide approaches by providing an in silico framework to test cell-based hypotheses. Together, these approaches may shed new light on the dynamics and flux of signaling cascades and point the way toward new drug targets.
Acknowledgments
We would like to thank members of the Scott lab for their critical review of this manuscript. This work was supported by the Foundation Leducq and by NIH grant DK54441 to J.D.S.
Footnotes
Competing Financial Interests: The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they're apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 3.Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 4.Millar JK, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 5.St Clair D, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 6.Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: update 2005. Mol Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 7.Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- 8.Brandon NJ, et al. Understanding the role of DISC1 in psychiatric disease and during normal development. J Neurosci. 2009;29:12768–12775. doi: 10.1523/JNEUROSCI.3355-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao Y, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3β/β-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millar JK, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 11.Murdoch H, et al. Isoform-selective susceptibility of DISC1/phosphodiesterase-4 complexes to dissociation by elevated intracellular cAMP levels. J Neurosci. 2007;27:9513–9524. doi: 10.1523/JNEUROSCI.1493-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clapcote SJ, et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Fang X, et al. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc Natl Acad Sci USA. 2000;97:11960–11965. doi: 10.1073/pnas.220413597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan F, Lenardo MJ. The nuclear signaling of NF-κB: current knowledge, new insights, and future perspectives. Cell Res. 2010;20:24–33. doi: 10.1038/cr.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beg AA, et al. IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 16.Baeuerle PA, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-κB transcription factor. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JE, Phillips RJ, Erdjument-Bromage H, Tempst P, Ghosh S. IκB-β regulates the persistent response in a biphasic activation of NF-κB. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 18.Regnier CH, et al. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 19.Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 20.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 21.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 22.Yamaoka S, et al. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 23.Tang ED, Wang CY, Xiong Y, Guan KL. A role for NF-κB essential modifier/ IκB kinase-γ(NEMO/IKKγ) ubiquitination in the activation of the IκB kinase complex by tumor necrosis factor-α. J Biol Chem. 2003;278:37297–37305. doi: 10.1074/jbc.M303389200. [DOI] [PubMed] [Google Scholar]
- 24.Zhou H, et al. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 25.Scherer DC, Brockman JA, Chen Z, Maniatis T, Ballard DW. Signal-induced degradation of IκB alpha requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiDonato J, et al. Mapping of the inducible IκB phosphorylation sites that signal its ubiquitination and degradation. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, Ghosh S. β-TrCP mediates the signal-induced ubiquitination of IκBβ. J Biol Chem. 1999;274:29591–29594. doi: 10.1074/jbc.274.42.29591. [DOI] [PubMed] [Google Scholar]
- 28.Yaron A, et al. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 29.Kovalenko A, et al. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 30.Zeke A, Lukacs M, Lim WA, Remenyi A. Scaffolds: interaction platforms for cellular signalling circuits. Trends Cell Biol. 2009;19:364–374. doi: 10.1016/j.tcb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 32.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 33.Brown MD, Sacks DB. Protein scaffolds in MAP kinase signalling. Cell Signal. 2009;21:462–469. doi: 10.1016/j.cellsig.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Elion EA. Pheromone response, mating and cell biology. Curr Opin Microbiol. 2000;3:573–581. doi: 10.1016/s1369-5274(00)00143-0. [DOI] [PubMed] [Google Scholar]
- 36.Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 37.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 38.Feng Y, Song LY, Kincaid E, Mahanty SK, Elion EA. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- 39.Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 40.Appert-Collin A, Cotecchia S, Nenniger-Tosato M, Pedrazzini T, Diviani D. The A-kinase anchoring protein (AKAP)-Lbc-signaling complex mediates α1 adrenergic receptor-induced cardiomyocyte hypertrophy. Proc Natl Acad Sci USA. 2007;104:10140–10145. doi: 10.1073/pnas.0701099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carnegie GK, et al. AKAP-Lbc mobilizes a cardiac hypertrophy signaling pathway. Mol Cell. 2008;32:169–179. doi: 10.1016/j.molcel.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molkentin JD, Dorn GW., II Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu Rev Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 43.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 44.Vega RB, et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sucharov CC, Langer S, Bristow M, Leinwand L. Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. Am J Physiol Cell Physiol. 2006;291:C1029–C1037. doi: 10.1152/ajpcell.00059.2006. [DOI] [PubMed] [Google Scholar]
- 46.Carnegie GK, Smith FD, McConnachie G, Langeberg LK, Scott JD. AKAP-Lbc nucleates a protein kinase D activation scaffold. Mol Cell. 2004;15:889–899. doi: 10.1016/j.molcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 47.Pietras RJ, Szego CM. Specific binding sites for oestrogen at the outer surfaces of isolated endometrial cells. Nature. 1977;265:69–72. doi: 10.1038/265069a0. [DOI] [PubMed] [Google Scholar]
- 48.Norfleet AM, Thomas ML, Gametchu B, Watson CS. Estrogen receptor-α detected on the plasma membrane of aldehyde-fixed GH3/B6/F10 rat pituitary tumor cells by enzyme-linked immunocytochemistry. Endocrinology. 1999;140:3805–3814. doi: 10.1210/endo.140.8.6936. [DOI] [PubMed] [Google Scholar]
- 49.Pedram A, Razandi M, Levin ER. Nature of functional estrogen receptors at the plasma membrane. Mol Endocrinol. 2006;20:1996–2009. doi: 10.1210/me.2005-0525. [DOI] [PubMed] [Google Scholar]
- 50.Pedram A, et al. Developmental phenotype of a membrane only estrogen receptor α (MOER) mouse. J Biol Chem. 2009;284:3488–3495. doi: 10.1074/jbc.M806249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Acconcia F, et al. Palmitoylation-dependent estrogen receptor alpha membrane localization: regulation by 17β-estradiol. Mol Biol Cell. 2005;16:231–237. doi: 10.1091/mbc.E04-07-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Razandi M, et al. Identification of a structural determinant necessary for the localization and function of estrogen receptor α at the plasma membrane. Mol Cell Biol. 2003;23:1633–1646. doi: 10.1128/MCB.23.5.1633-1646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watters JJ, Campbell JS, Cunningham MJ, Krebs EG, Dorsa DM. Rapid membrane effects of steroids in neuroblastoma cells: effects of estrogen on mitogen activated protein kinase signalling cascade and c-fos immediate early gene transcription. Endocrinology. 1997;138:4030–4033. doi: 10.1210/endo.138.9.5489. [DOI] [PubMed] [Google Scholar]
- 54.Migliaccio A, et al. Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol-receptor complex in MCF-7 cells. EMBO J. 1996;15:1292–1300. [PMC free article] [PubMed] [Google Scholar]
- 55.Migliaccio A, et al. Steroid-induced androgen receptor-oestradiol receptor β-Src complex triggers prostate cancer cell proliferation. EMBO J. 2000;19:5406–5417. doi: 10.1093/emboj/19.20.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simoncini T, et al. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature. 2000;407:538–541. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castoria G, et al. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J. 2001;20:6050–6059. doi: 10.1093/emboj/20.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartucci M, Morelli C, Mauro L, Ando S, Surmacz E. Differential insulin-like growth factor I receptor signaling and function in estrogen receptor (ER)-positive MCF-7 and ER-negative MDA-MB-231 breast cancer cells. Cancer Res. 2001;61:6747–6754. [PubMed] [Google Scholar]
- 59.Levin ER. Plasma membrane estrogen receptors. Trends Endocrinol Metab. 2009;20:477–482. doi: 10.1016/j.tem.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kahlert S, et al. Estrogen receptor α rapidly activates the IGF-1 receptor pathway. J Biol Chem. 2000;275:18447–18453. doi: 10.1074/jbc.M910345199. [DOI] [PubMed] [Google Scholar]
- 61.Filardo EJ, Quinn JA, Frackelton AR, Jr, Bland KI. Estrogen action via the G protein-coupled receptor, GPR30: stimulation of adenylyl cyclase and cAMP-mediated attenuation of the epidermal growth factor receptor-to-MAPK signaling axis. Mol Endocrinol. 2002;16:70–84. doi: 10.1210/mend.16.1.0758. [DOI] [PubMed] [Google Scholar]
- 62.Balasenthil S, Barnes CJ, Rayala SK, Kumar R. Estrogen receptor activation at serine 305 is sufficient to upregulate cyclin D1 in breast cancer cells. FEBS Lett. 2004;567:243–247. doi: 10.1016/j.febslet.2004.04.071. [DOI] [PubMed] [Google Scholar]
- 63.Balasenthil S, et al. p21-activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells. J Biol Chem. 2004;279:1422–1428. doi: 10.1074/jbc.M309937200. [DOI] [PubMed] [Google Scholar]
- 64.Medunjanin S, et al. Glycogen synthase kinase-3 interacts with and phosphorylates estrogen receptor α and is involved in the regulation of receptor activity. J Biol Chem. 2005;280:33006–33014. doi: 10.1074/jbc.M506758200. [DOI] [PubMed] [Google Scholar]
- 65.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 66.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 67.Dodge-Kafka KL, et al. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coghlan VM, et al. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 69.Klauck TM, et al. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 70.Bhattacharyya S, Biou V, Xu W, Schluter O, Malenka RC. A critical role for PSD-95/AKAP interactions in endocytosis of synaptic AMPA receptors. Nat Neurosci. 2009;12:172–181. doi: 10.1038/nn.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoshi N, Langeberg LK, Scott JD. Distinct enzyme combinations in AKAP signalling complexes permit functional diversity. Nat Cell Biol. 2005;7:1066–1073. doi: 10.1038/ncb1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bal M, Zhang J, Hernandez CC, Zaika O, Shapiro MS. Ca2+/calmodulin disrupts AKAP79/150 interactions with KCNQ (M-Type) K+ channels. J Neurosci. 2010;30:2311–2323. doi: 10.1523/JNEUROSCI.5175-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu Y, et al. Age-dependent requirement of AKAP150-anchored PKA and GluR2-lacking AMPA receptors in LTP. EMBO J. 2007;26:4879–4890. doi: 10.1038/sj.emboj.7601884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scannevin RH, Huganir RL. Postsynaptic organization and regulation of excitatory synapses. Nat Rev Neurosci. 2000;1:133–141. doi: 10.1038/35039075. [DOI] [PubMed] [Google Scholar]
- 75.Bredt DS, Nicoll RA. AMPA receptor trafficking at excitatory synapses. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 76.Tavalin SJ, et al. Regulation of GluR1 by the A-kinase anchoring protein 79 (AKAP79) signaling complex shares properties with long-term depression. J Neurosci. 2002;22:3044–3051. doi: 10.1523/JNEUROSCI.22-08-03044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banke TG, et al. Control of GluR1 AMPA receptor function by cAMP-dependent protein kinase. J Neurosci. 2000;20:89–102. doi: 10.1523/JNEUROSCI.20-01-00089.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Esteban JA, et al. PKA phosphorylation of AMPA receptor subunits controls synaptic trafficking underlying plasticity. Nat Neurosci. 2003;6:136–143. doi: 10.1038/nn997. [DOI] [PubMed] [Google Scholar]
- 79.Hoshi N, et al. AKAP150 signaling complex promotes suppression of the M-current by muscarinic agonists. Nat Neurosci. 2003;6:564–571. doi: 10.1038/nn1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoshi N, Langeberg LK, Gould CM, Newton AC, Scott JD. Interaction with AKAP79 modifies the cellular pharmacology of PKC. Mol Cell. 2010;37:541–550. doi: 10.1016/j.molcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prince J, Ahn N. The case of the disapprearing drug target. Mol Cell. 2010;37:455–456. doi: 10.1016/j.molcel.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 82.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 83.Tsien RY. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture) Angew Chem Int Edn Engl. 2009;48:5612–5626. doi: 10.1002/anie.200901916. [DOI] [PubMed] [Google Scholar]