Summary
HLA-A*0201-restricted virus-specific CD8+ cytotoxic T lymphocytes (CTLs) do not appear to control HIV effectively in vivo. To enhance the immunogenicity of a highly conserved subdominant epitope, TV9 (TLNAWVKVV, p24 Gag19–27), mimotopes were designed by screening a large combinatorial nonapeptide library with TV9-specific CTLs primed in vitro from healthy donors. A mimic peptide with a low binding affinity to HLA-A*0201, TV9p6 (KINAWIKVV), was studied further. Parallel cultures of in vitro-primed CTLs showed that TV9p6 consistently activated crossreactive and equally functional CTLs as measured by cytotoxicity, cytokine production and suppression of HIV replication in vitro. Comparison of TCRB gene usage between CTLs primed from the same donors with TV9 or TV9p6 revealed a degree of clonal overlap in some cases and an example of a conserved TCRB sequence encoded distinctly at the nucleotide level between individuals (a “public” TCR); however, in the main, distinct clonotypes were recruited by each peptide antigen. These findings indicate that mimotopes can mobilize functional crossreactive clonotypes that are less readily recruited from the naïve T cell pool by the corresponding wildtype epitope. Mimotope-induced repertoire diversification could potentially override subdominance under certain circumstances and enhance vaccine-induced responses to conserved but poorly immunogenic determinants within the HIV proteome.
Keywords: HIV vaccine, CD8+ cytotoxic T lymphocyte, agonist peptide, in vitro immunization
Introduction
Virus-specific CD8+ cytotoxic T lymphocytes (CTLs) play a central role in the control of HIV infection [1, 2]. However, the ability of such CTLs to impose immune pressure on HIV varies according to specificity, with some responses seemingly exerting no influence on the virus [3]. Strong linkages have been reported between a few HLA–B class I alleles and control of infection [4]. The impact of these molecules on disease outcome is mediated largely by potent CTL activities that target highly conserved determinants [5], the majority of which reside within the p24 Gag protein [2]. Aside from their character, these CTL responses may be effective because the selected escape mutations exact a significant cost to viral fitness, thereby favoring the host even after immune evasion [2]. Thus, the issue of whether similar epitope targets restricted by common HLA alleles remain within the HIV proteome to allow for meaningful population coverage is fundamental to the prospects for a broadly applicable T cell-based vaccine.
Virus-specific CTL responses restricted by the most prevalent allele, HLA-A*0201, do not appear to suppress HIV effectively in vivo [6], consistent with the notion that “inactive” epitopic forms may have been fixed in the circulating viral strains [7, 8]. Paradoxically, however, there is one well-defined epitope in p24 Gag (TV9, TLNAWVKVV, residues 19–27) that shares many features in common with known protective determinants. Thus, TV9 is highly conserved across HIV clades, with only one common variant in which valine at position 9 is “conservatively” replaced by isoleucine [9]. This epitope resides in the first α-helix at the N-terminus of p24 and overlaps by five residues with the protective HLA–B*57-restricted ISPRTLNAW (IW9) determinant recognized early by slow progressors [10]. Despite the small number of reports of TV9 reactivity in patients, there are indications that CTL responses to this epitope can control virus [11]. Above all, an elevated and sustained TV9-specific response was noted in one person who remained uninfected despite parenteral exposure to a highly replicating HIV strain [12]. The functional sensitivity of the in vivo TV9 reactivity, a factor considered important for the inhibition of HIV in vivo [13], was also relatively high [11]. Thus, the potential of TV9 as a vaccine target deserves further exploration, particularly since there may be few alternatives within the HIV proteome for the most prevalent HLA class I allele.
Previously, we studied the pre-infection TCR repertoire for TV9 by priming naive CD8+ T cells from healthy seronegative donors [11]. Stable, homogeneous and immunologically reactive TV9-specific CTL (TV9-CTL) cultures were generated from most donors, thereby suggesting that the cognate TCR repertoire is not limiting in HLA-A*0201 carriers. TV9-CTLs were polyfunctional and suppressed HIV replication in vitro. Thus, while in vitro immunization suggests that TV9 is potentially highly immunogenic, data from patients indicate that it would be necessary to devise immunization strategies that selectively elicit clonotypes with high functional sensitivities to overcome the typical subdominance of this epitope in the context of viral infection. Here, we identified mimic peptides (“mimotopes”) by probing a large positional scanning synthetic combinatorial nonapeptide library (PS-SCL) with homogeneous, well characterized in vitro-primed TV9-CTLs [14]. We selected a peptide, TV9p6 (KINAWIKVV), with the lowest binding affinity to HLA-A*0201 for detailed analysis, reasoning that an unstable peptide-HLA-A*0201 complex and the attendant low epitope density would preferentially select clonotypes with high functional sensitivity. Parallel CD8+ T cell cultures specific for TV9 and TV9p6 were established to compare specificity and cross-recognition based on functional assays that included cytotoxicity, cytokine production and suppression of HIV replication in vitro. Concomitant repertoire analyses were conducted using a quantitative molecular approach to evaluate antigen-driven clonotype selection in relation to these functional attributes.
Results
Prediction of TV9 mimotopes by screening a positional scanning synthetic nonapeptide combinatorial library
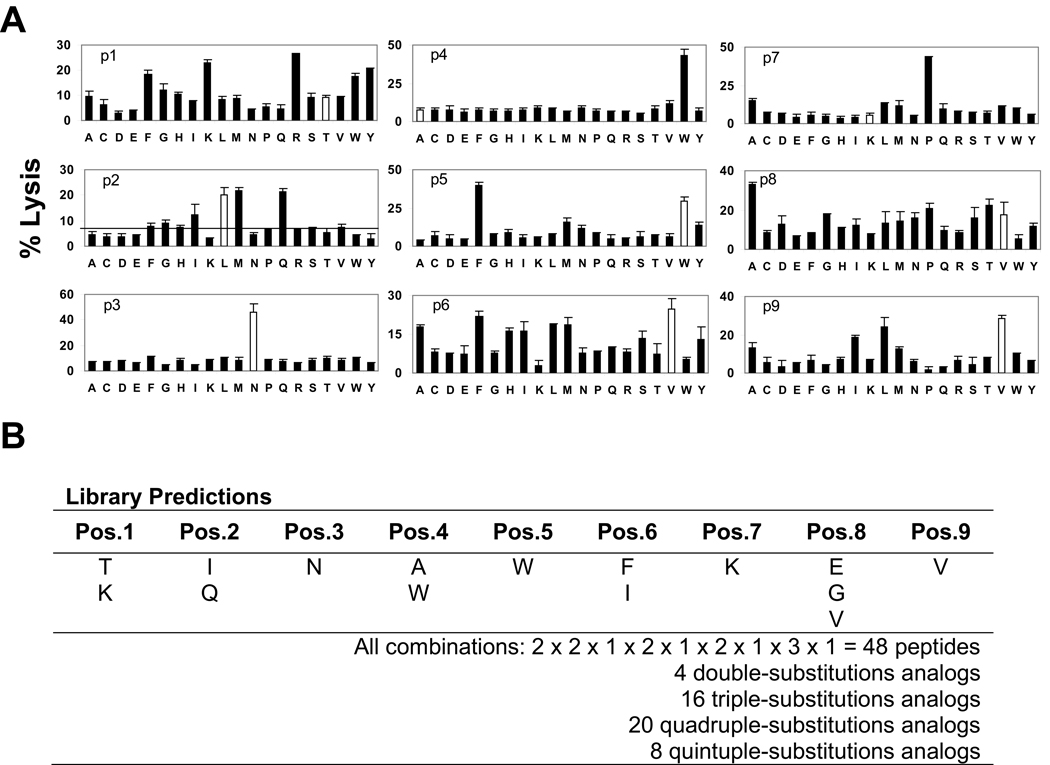
A polyclonal TV9-CTL culture (TV9-1, index culture) was screened for cytolytic activity with 180 nonapeptide library mixtures as described previously [14]. Each well contained 104 CTLs and 2×103 51Cr-labeled TAP-deficient, HLA-A*0201-expressing T2 cells with 100 µg/mL of a peptide mixture. Significant and reproducible differences in lysis were observed between mixtures. Figure 1A is representative of three scans performed. Remarkably, the percent lysis with some mixtures, each containing 1.7 × 1010 peptides at femtomolar concentrations, was equivalent to that observed with the TV9 peptide (30 to 50% versus 60% at 1 µg/mL). However, T cell activation may be achieved via degenerate stimulation by many peptides within each mixture [15].
Figure 1.
Scanning a nonapeptide PS-SCL to identify TV9 agonist peptides. (A) Index TV9-1 cytotoxicity elicited by the 180 mixtures of a nonapeptide PS-SCL. Each graph, designated p1–p9, represents a set of 20 mixtures having the defined amino acid listed on the x-axis at a given position. The y-axis denotes the % lysis of T2 cells in the presence of each mixture. The hollow bars represent the native TV9 amino acid sequence. Each mixture was assayed in triplicate and scanning was repeated three times. (B) Design of 48 candidate peptides. The rationale for the selection of a particular active mixture in each position is discussed in the text.
Six mixtures defined with residues corresponding to the TV9 sequence (hollow bars) were among the most stimulatory mixtures for sublibraries within that position. They were L in position 2 (P2), N in P3, W in P5, V in P6, and V in P8 and P9. Results with the P2 and P9 anchor positions confirmed the power and accuracy of this assay. L and M are known preferred residues in P2 of HLA-A*0201; the scans showed that these mixtures were equally stimulatory. Likewise, mixtures that contained the preferred anchor residues in P9 (I, L, and V) were almost equally active. N was the only stimulatory mixture in P3, suggesting that it may be a primary TCR contact site. P1 and P6 appeared degenerate, with at least five mixtures that were more or equally stimulatory than matching TV9 residues. Curiously, the mixtures corresponding to the native A and K in P4 and P7 were less stimulatory than those for W and P, respectively, suggesting that the natural residues are not optimal for recognition by TV9-1 CTLs.
Based on the library scans, candidate variant peptides were designed for subsequent testing. To limit their number, substitutions were not made in positions thought to be important for TCR recognition (P3, P5 and P7, [16]) or in P9. Substitutions made in P1, P2, P4, P6 and P8 were mostly conservation or semi-conservative, except for the non-conservative replacement of A by W in P4 (Figure 1B). A panel of 48 peptides with two to five replacements in the TV9 sequence was synthesized.
Recognition of candidate peptides by TV9-CTL cultures
The candidate peptides were tested for recognition by TV9-CTLs from five donors. Cytotoxicity was measured at four concentrations (1, 10, 100 and 1000 ng/mL) to estimate relative antigenicity. A peptide was considered cross-recognized if it elicited ≥50% of the lysis observed at the two highest concentrations of TV9.
Thirteen of the 48 peptides tested (27%) were cross-recognized by TV9-1 CTLs and by at least one other culture (Group I, Table I). Four peptides (TV9p6, TV9p29, TV9p30 and TV9p18) were recognized by all cultures. The ability of non-index TV9-CTLs to recognize 6 to 28 peptides encoding two to five substitutions suggests that recognition of the TV9/HLA-A*0201 complex is quite degenerate. Curiously, TV9-2 and TV9-4 CTLs were sensitized by 6 and 18 sequences not recognized by index CTLs (Group II, Table I), thereby suggesting differences in the degree of repertoire overlap between donors.
Table I.
Recognition of predicted candidate peptides, presented by T2 target cells, by the index and four allogeneic HLA-A*0201+ TV9-CTL cultures as determined by cytotoxicity. Positive recognition for TV9 and reactive artificial sequences is boldfaced and shaded. Nd, not determined.
| Candidate Peptide Name and Sequence |
Residues Substituted |
TV9-1 (index) | TV9-2 | TV9-3 | TV9-4 | TV9-5 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Lysis at Peptide Concentrations (ng/mL) |
|||||||||||||||||||||||||||||||
| 1 | 10 | 100 | 1000 | 1 | 10 | 100 | 1000 | 1 | 10 | 100 | 1000 | 1 | 10 | 100 | 1000 | 1 | 10 | 100 | 1000 | ||||||||||||
| Native | TV9 | T | L | N | A | W | V | K | V | V | 0 | 3 | 18 | 42 | 50 | 19 | 73 | 83 | 81 | 7 | 34 | 64 | 76 | 50 | 78 | 85 | 78 | 7 | 35 | 59 | 44 |
| Group I | p30 | – | I | – | – | – | I | – | – | – | 2 | 0 | 6 | 29 | 45 | 2 | 36 | 80 | 75 | 2 | 37 | 56 | 60 | 0 | 5 | 58 | 53 | 0 | 36 | 67 | 76 |
| p6 | K | I | – | – | – | I | – | – | – | 3 | 0 | 8 | 31 | 41 | 34 | 73 | 77 | 78 | 10 | 34 | 51 | 51 | 0 | 0 | 30 | 45 | 2 | 8 | 52 | 76 | |
| p29 | – | I | – | – | – | I | – | G | – | 3 | 15 | 37 | 47 | 64 | 29 | 63 | 77 | 74 | 9 | 40 | 48 | 59 | 6 | 57 | 68 | 55 | 5 | 48 | 56 | 86 | |
| p5 | K | I | – | – | – | I | – | G | – | 4 | 0 | 20 | 44 | 51 | 64 | 84 | 84 | 85 | 14 | 30 | 30 | 34 | 1 | 0 | 25 | 53 | 8 | 4 | 13 | 58 | |
| p42 | – | Q | – | – | – | I | – | – | – | 2 | 2 | 7 | 27 | 41 | 9 | 63 | 75 | 74 | 0 | 24 | 46 | 48 | 0 | 9 | 59 | 83 | 2 | 2 | 29 | 28 | |
| p18 | K | Q | – | – | – | I | – | – | – | 3 | 0 | 11 | 29 | 44 | 29 | 66 | 75 | 73 | 7 | 28 | 41 | 46 | 0 | 0 | 43 | 81 | 0 | 5 | 35 | 49 | |
| p41 | – | Q | – | – | – | I | – | G | – | 3 | 13 | 32 | 42 | 52 | 47 | 79 | 84 | 77 | 8 | 31 | 40 | 53 | 0 | 37 | 65 | 80 | 0 | 3 | 28 | 28 | |
| p17 | K | Q | – | – | – | I | – | G | – | 4 | 4 | 33 | 44 | 61 | 66 | 83 | 86 | 82 | 11 | 24 | 29 | 28 | 0 | 15 | 69 | 87 | 0 | 2 | 3 | 14 | |
| p40 | – | Q | – | – | – | I | – | E | – | 3 | 0 | 17 | 39 | 46 | 33 | 73 | 78 | 70 | 17 | 26 | 32 | 34 | 0 | 0 | 0 | 1 | 0 | 5 | 5 | 14 | |
| p16 | K | Q | – | – | – | I | – | E | – | 4 | 0 | 21 | 36 | 50 | 6 | 52 | 76 | 80 | 23 | 23 | 28 | 30 | 0 | 0 | 0 | 0 | 2 | 1 | 6 | 13 | |
| p35 | – | I | – | W | – | I | – | G | – | 4 | 1 | 3 | 24 | 47 | 0 | 1 | 6 | 35 | 0 | 0 | 0 | 0 | 79 | 79 | 81 | 77 | 0 | 2 | 36 | 41 | |
| p47 | – | Q | – | W | – | I | – | G | – | 4 | 4 | 17 | 30 | 47 | 2 | 6 | 30 | 54 | 0 | 0 | 0 | 7 | 56 | 83 | 85 | 83 | 0 | 2 | 5 | 18 | |
| p23 | K | Q | – | W | – | I | – | G | – | 5 | 0 | 2 | 24 | 43 | 1 | 0 | 4 | 11 | 0 | 0 | 0 | 0 | 76 | 92 | 90 | 93 | 0 | 2 | 5 | 18 | |
| Group II | p28 | – | I | – | – | – | I | – | E | – | 3 | 0 | 1 | 14 | 46 | 19 | 64 | 77 | 72 | 31 | 37 | 34 | 37 | 0 | 0 | 0 | 0 | 0 | 14 | 25 | nd |
| p36 | – | I | – | W | – | I | – | – | – | 3 | 3 | 0 | 0 | 4 | 3 | 8 | 15 | 43 | 0 | 0 | 0 | nd | 34 | 66 | 78 | 73 | 0 | 4 | 8 | 29 | |
| p34 | – | I | – | W | – | I | – | E | – | 4 | 0 | 0 | 0 | 0 | 2 | 3 | 3 | 12 | 0 | 0 | 0 | 4 | 1 | 13 | 56 | 67 | 0 | 0 | nd | 7 | |
| p4 | K | I | – | – | – | I | – | E | – | 4 | 0 | 0 | 19 | 47 | 43 | 72 | 79 | 74 | 25 | 32 | 32 | 34 | 0 | 0 | 0 | 1 | 8 | 0 | 9 | 36 | |
| p12 | K | I | – | W | – | I | – | – | – | 4 | 0 | 0 | 0 | 3 | 1 | 9 | 15 | 51 | 0 | 0 | 2 | 11 | 33 | 83 | 87 | 89 | 0 | 3 | 16 | 31 | |
| p10 | K | I | – | W | – | I | – | E | – | 5 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 11 | 37 | 1 | 0 | 10 | nd | |
| p11 | K | I | – | W | – | I | – | G | – | 5 | 0 | 0 | 0 | 30 | 0 | 4 | 13 | 16 | 0 | 0 | 0 | 0 | 57 | 84 | 84 | 82 | 3 | 2 | 14 | nd | |
| p39 | – | Q | – | – | – | F | – | – | – | 2 | 0 | 0 | 0 | 11 | 2 | 2 | 30 | 52 | 0 | 0 | 1 | 20 | 0 | 0 | 1 | 19 | 0 | 7 | 23 | 52 | |
| p37 | – | Q | – | – | – | F | – | E | – | 3 | 0 | 0 | 0 | 16 | 0 | 0 | 16 | 37 | 0 | 16 | 26 | 32 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 7 | |
| p38 | – | Q | – | – | – | F | – | G | – | 3 | 0 | 6 | 26 | 38 | 0 | 0 | 12 | 32 | 0 | 0 | 1 | 17 | 0 | 3 | 38 | 68 | 0 | 6 | 9 | 47 | |
| p15 | K | Q | – | – | – | F | – | – | – | 3 | 0 | 0 | 17 | 40 | 15 | 66 | 69 | 69 | 0 | 0 | 7 | 35 | 0 | 0 | 5 | 27 | 1 | 8 | 15 | 38 | |
| p13 | K | Q | – | – | – | F | – | E | – | 4 | 0 | 0 | 0 | 29 | 0 | 12 | 48 | 57 | 1 | 19 | 28 | 29 | 0 | 0 | 0 | 0 | 6 | 6 | 9 | 14 | |
| p14 | K | Q | – | – | – | F | – | G | – | 4 | 0 | 0 | 8 | 36 | 19 | 8 | 38 | 72 | 0 | 0 | 0 | 17 | 0 | 0 | 9 | 42 | 5 | 0 | 4 | 14 | |
| p45 | – | Q | – | W | – | F | – | – | – | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 54 | 0 | 14 | 22 | 27 | 31 | 77 | 84 | 79 | 0 | 0 | 0 | 1 | |
| p43 | – | Q | – | W | – | F | – | E | – | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 7 | 12 | 65 | 74 | 82 | 0 | 4 | 0 | 2 | |
| p44 | – | Q | – | W | – | F | – | G | – | 4 | 0 | 0 | 0 | 32 | 0 | 1 | 0 | 4 | 0 | 0 | 0 | 3 | 46 | 71 | 76 | 83 | 0 | 0 | 0 | 4 | |
| p21 | K | Q | – | W | – | F | – | – | – | 4 | 0 | 0 | 0 | 0 | 3 | 0 | 10 | 48 | 0 | 20 | 29 | 39 | 43 | 89 | 89 | 95 | 1 | 4 | 9 | 16 | |
| p19 | K | Q | – | W | – | F | – | E | – | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 47 | 87 | 83 | 0 | 3 | 11 | 23 | |
| p20 | K | Q | – | W | – | F | – | G | – | 5 | 0 | 0 | 0 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 54 | 96 | 85 | 91 | 0 | 6 | 9 | 19 | |
| p48 | – | Q | – | W | – | I | – | – | – | 3 | 0 | 0 | 0 | 3 | 2 | 7 | 20 | 51 | 0 | 0 | 0 | 10 | 32 | 68 | 84 | 83 | 0 | 3 | 20 | 5 | |
| p46 | – | Q | – | W | – | I | – | E | – | 4 | 0 | 0 | 0 | 1 | 3 | 2 | 16 | 31 | nd | 2 | 27 | 34 | 40 | 78 | 86 | 87 | 0 | 0 | 5 | 6 | |
| p24 | K | Q | – | W | – | I | – | – | – | 4 | 0 | 0 | 0 | 3 | 0 | 1 | 15 | 31 | 0 | 0 | 4 | 17 | 34 | 66 | 92 | 90 | 0 | 2 | 4 | 9 | |
| p22 | K | Q | – | W | – | I | – | E | – | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 12 | 52 | 2 | 6 | 6 | 29 | |
| p27 | – | I | – | – | – | F | – | – | – | 2 | 0 | 0 | 0 | 14 | 1 | 4 | 33 | 61 | 0 | 0 | 3 | 21 | 0 | 0 | 0 | 5 | 1 | 5 | 37 | nd | |
| p25 | – | I | – | – | – | F | – | E | – | 3 | 0 | 1 | 0 | 7 | 1 | 1 | 7 | 14 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | nd | |
| p26 | – | I | – | – | – | F | – | G | – | 3 | 0 | 0 | 0 | 23 | 0 | 6 | 12 | 27 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 36 | 0 | 1 | 7 | nd | |
| p3 | K | I | – | – | – | F | – | – | – | 3 | 0 | 0 | 0 | 34 | 12 | 63 | 68 | 58 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 1 | 0 | 31 | 68 | |
| p1 | K | I | – | – | – | F | – | E | – | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 37 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 10 | |
| p2 | K | I | – | – | – | F | – | G | – | 4 | 0 | 0 | 0 | 13 | 0 | 6 | 52 | 64 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 10 | 19 | |
| p33 | – | I | – | W | – | F | – | – | – | 3 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 13 | 0 | 4 | 29 | 36 | 9 | 47 | 73 | 77 | 0 | 0 | 13 | 6 | |
| p31 | – | I | – | W | – | F | – | E | – | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 64 | 64 | 0 | 7 | 7 | 13 | |
| p32 | – | I | – | W | – | F | – | G | – | 4 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 8 | 0 | 0 | 0 | 1 | 19 | 59 | 84 | 78 | 0 | 4 | 5 | 20 | |
| p9 | K | I | – | W | – | F | – | – | – | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 6 | 29 | 38 | 0 | 23 | 79 | 86 | 8 | 9 | 13 | nd | |
| p7 | K | I | – | W | – | F | – | E | – | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 14 | 7 | 4 | 3 | 40 | |
| p8 | K | I | – | W | – | F | – | G | – | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 18 | 81 | 87 | 0 | 5 | 9 | 43 | |
| No. of the 48 artificial sequences recognized | 14 | 16 | 6 | 28 | 6 | ||||||||||||||||||||||||||
Immunogenicity of four broadly cross-recognized peptides
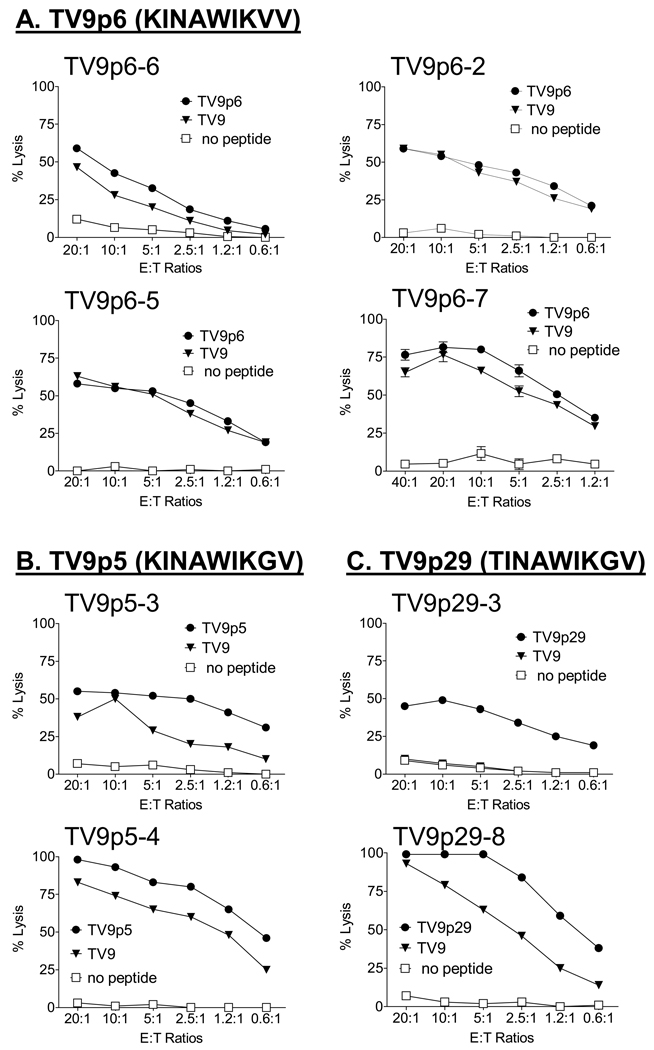
The ability of the first four peptides listed in Table I to prime TV9-crossreactive CTLs was determined (Figure 2). TV9p30 was not immunogenic in three different donors (data not shown). TV9p6 immunized CTLs (TV9p6-2, TV9p6-5, TV9p6-6 and TV9p6–7) that cross-recognized TV9 consistently (Figure 2A). TV9p5 was also immunogenic and elicited TV9-crossreactive CTLs from two donors (Figure 2B). TV9p29 was immunogenic but induced crossreactive CTLs from only one of two cultures (p29-8, Figure 2C). That double to quadruple substituted mimotopes stimulated crossreactive CTLs is consistent with our contention that the available naive T cell pool for TV9 may be broad [11].
Figure 2.
Evaluation of the ability of three agonist peptides (TV9p6, TV9p5 and TV9p29) listed in Table I to induce antigen-specific, TV9-crossreactive CTLs by in vitro immunization of naïve CD8+ T cells from two to four healthy seronegative donors. Cultures are labeled as TV9pX-Y, where TV9pX identifies the inducing peptide and the numeral Y denotes a distinct donor. (A) TV9p6-6, TV9p6-2, TV9p6-5 and TV9p6–7; (B) TV9p5-3 and TV9p5-4; and, (C) TV9p29-3 and TV9p29-8. CD8+T cells were assayed for specificity and cross-recognition of TV9 using chromium release assays at effector:target (E:T) ratios ranging from 20:1 to 0.6:1. T2 cells without peptide were included as negative controls in each assay. Data are shown as mean ± SEM of triplicate assays and are representative of two independent experiments; in most cases, the error bars are smaller than the plot symbols.
Binding affinities of TV9 and its analogs to HLA-A2 class I antigens
Table II shows the binding affinities of TV9 and its mimotopes to members of the HLA-A2 supertype. Since HIV-specific CTL epitopes restricted by protective HLA–B alleles [17] show lower average peptide binding affinities than those of nonprotective HLA-A-restricted epitopes [18], we selected TV9p6 with the lowest affinity to determine whether it is possible to consolidate the TV9 response around higher avidity clonotypes.
Table II.
Binding affinities of TV9 and its mimic peptides to HLA-A2 supertype molecules. Dash indicates IC50 >10,000 nM.
| Peptide | Sequence | # Substitutions | IC50 (nM) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A*0201 | A*0202 | A*0203 | A*0206 | A*6802 | |||||||||||
| TV9 | T | L | N | A | W | V | K | V | V | Native | 701 | 179 | 108 | 1,965 | 23,937 |
| TV9p5 | K | I | N | A | W | I | K | G | V | 4 | 536 | 433 | 10 | 224 | 36,760 |
| TV9p6 | K | I | N | A | W | I | K | V | V | 3 | 5,120 | 2,950 | 56 | 13,449 | -- |
| TV9p29 | T | I | N | A | W | I | K | G | V | 3 | 1,062 | 706 | 15 | 2,248 | 269 |
| TV9p30 | T | I | N | A | W | I | K | V | V | 2 | 4,228 | 1,835 | 112 | 12,581 | 6,218 |
| SL9 | S | L | Y | N | T | V | A | T | L | Native | 367 | 79 | 19 | -- | -- |
CD8+ cultures to TV9p6 and to TV9
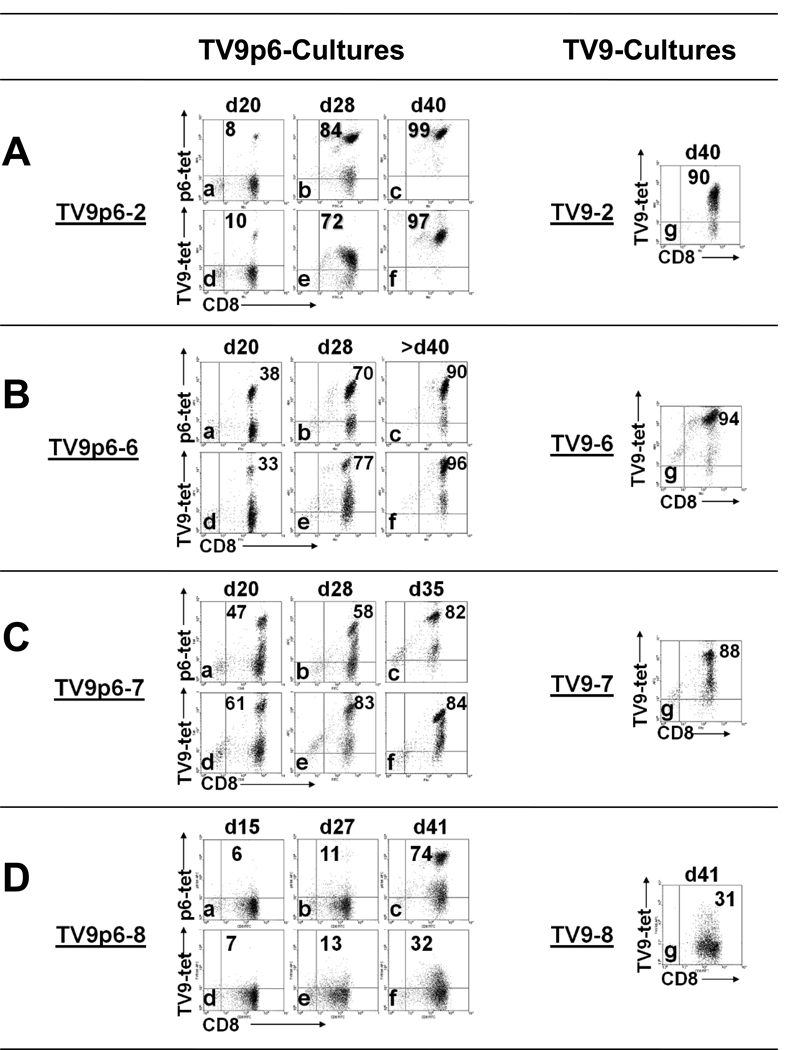
Parallel CD8+ cultures to TV9p6 or TV9 were generated from four donors and induction of peptide-specific T cells was monitored with tetramers (Figure 3). For donors 2, 6 and 7, the results were similar (Figure 3A–C). TV9p6-specific T cells were detected as early as day 20. By day 35, the TV9p6-cultures were essentially homogeneous by tetramer staining. Moreover, the staining patterns of TV9p6-T cells with both TV9p6 and TV9 tetramers were essentially identical, indicating that TV9p6-T cells recognized TV9. Parallel TV9-cultures showed ~90% TV9-T cells by day 40.
Figure 3.
Specificity and crossreactivity of TV9p6 CTLs monitored over time by tetramer staining. TV9p6-tetramer binding by the CD8+ cultures TV9p6-2, TV9p6-6, TV9p6–7 and TV9p6–8 at three time points is shown in Panels A-a to A–c, B-a to B–c, C-a to C-c and D-a to D-c. Crossreactivity of the TV9p6-cultures was assessed concomitantly by staining with TV9-tetramer (A–d to A–f, B–d to B–f, C–d to C–f and D–d to D–f). For comparison, TV9-tetramer staining of the CD8+ parallel cultures TV9-2, TV9-6, TV9-7 and TV9-8 is shown in Panels A–g, B–g, C–g and D–g, respectively. Numbers in the upper right quadrant represent the percentage of CD8+ tetramer+ stained cells. An irrelevant tetramer was used to gate for specific tetramer binding. Data are representative of at least three independent experiments.
Differential tetramer staining was observed in the TV9p6–8 culture (Figure 3D). The majority (74%) of the cells stained intensely and exclusively with the TV9p6-tetramer on day 41; only weak and poorly distinguished staining was observed with the TV9-tetramer. A similar pattern of low intensity staining was detected in the parallel TV9-8 culture, thereby indicating that the naïve repertoire for TV9 from this individual might consist primarily of low avidity clonotypes. Thus, TV9p6 primed both TV9-specific clonotypes and a distinct non-crossreactive population in this donor.
CD8 coreceptor dependence of cytotoxicity mediated by TV9- and TV9p6-CTLs
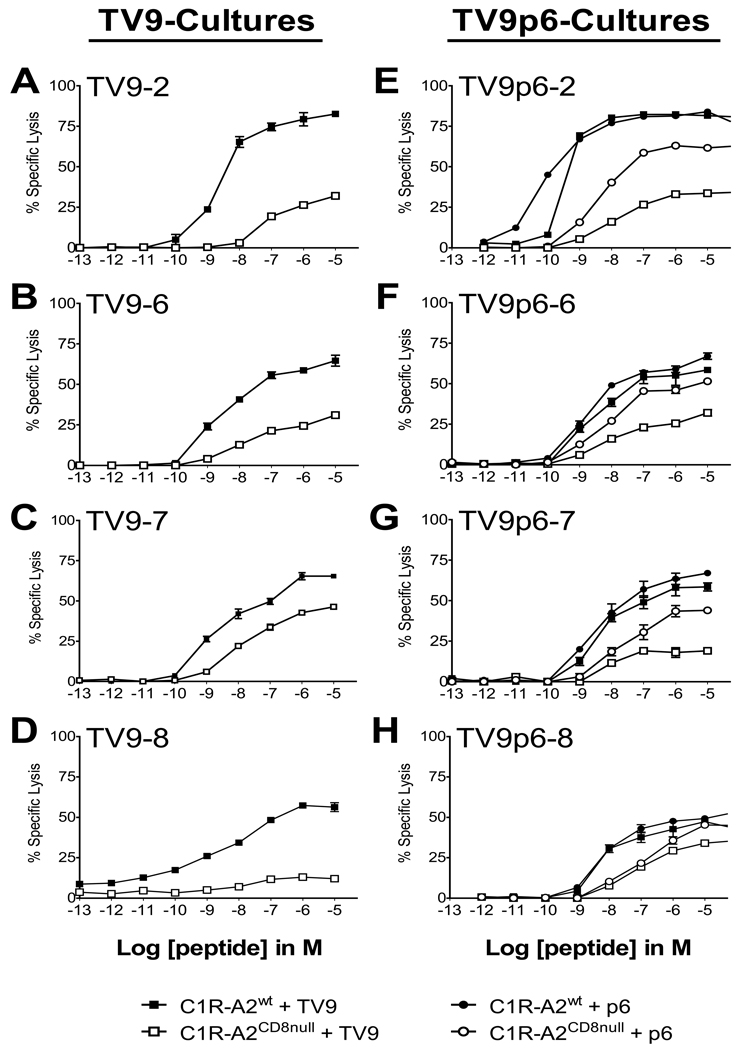
Homogeneous TV9- and TV9p6-CTL cultures were assessed for their dependence on CD8 compensation using peptide-pulsed CIR target cells expressing wildtype HLA-A*0201 (C1Rwt) or point-mutated HLA-A*0201 that cannot bind CD8 (C1RCD8null) [19] in chromium release cytotoxicity assays. Lysis of TV9-pulsed C1RCD8null cells by TV9-CTLs was substantially reduced compared to lysis of TV9-pulsed C1Rwt cells (Figures 4A–D), thereby indicating that abrogation of CD8 binding impaired but did not obviate downstream functional activation. Figures 4E–H compare lysis mediated by parallel TV9p6-cultures over an effectual range of TV9p6 or TV9 peptide concentrations. TV9p6-T cell cultures were equally cytotoxic to C1Rwt cells pulsed with either peptide (filled symbols). Cytotoxicity was reduced without CD8 participation (open symbols), although this loss was markedly less pronounced for the cognate peptide in most cases. The use of paired target cells revealed that TV9p6-T cells exhibited greater intrinsic avidity than TV9-T cells for their cognate pMHCI complexes (p=0.037; comparison of percent change in cytotoxicity at 10−7 M with C1R-A2wt versus C1R-A2CD8null cells for TV9-CTLs and TV9p6-CTLs using an unpaired two-tailed Student T test) [20]. However, there was no significant difference between the CD8 dependencies of TV9-CTLs and TV9p6-CTLs with respect to lysis of target cells pulsed with the natural peptide. Importantly, these data show that CD8 binding to pMHCI can fully compensate for any “sensitivity” disadvantages associated with TV9 recognition in terms of the ability of TV9p6-T cells to kill in vitro.
Figure 4.
CD8 dependency of cytotoxic activity mediated by TV9- and TV9p6-CTLs. Parallel cultures to TV9 or TV9p6, established by in vitro immunization using peptide-pulsed DCs, were identified as TV9-Y or TV9p6-Y, where the numeral Y denotes a distinct donor. (A) TV9-2, (B) TV9-6, (C) TV9-7, (D) TV9-8 and parallel (E) TV9p6-2, (F) TV9p6-6, (G) TV9p6–7 and (H) TV9p6–8 CD8+ cultures were assayed for functional sensitivity in a chromium release assay using C1R-A2wt and C1R-A2CD8null targets loaded with a range of concentrations of TV9p6 or TV9 peptide (10−5 to 10−13M) at an E:T ratio of 10:1 (–■–■–,–●–●–: C1R-A2wt pulsed with TV9 and TV9p6, respectively; –□–□–,–○–○–: C1R-A2CD8null pulsed with TV9 and TV9p6, respectively). Lysis of C1R cells alone was subtracted to give percent specific lysis. Data are shown as mean ± SEM of triplicate assays and are representative of two independent experiments; in most cases, the error bars are smaller than the plot symbols.
Functional profile of TV9- and TV9p6-CTLs
As polyfunctionality of HIV-specific CD8+ T cells appears to correlate with viral control in vivo [21, 22], the ability of cultured T cells to degranulate and secrete IL-2, TNF-α and IFN-γ was assessed after stimulation with peptide-pulsed C1R-A2wt cells (Table III). Five of the six cultures were polyclonal, with tetramer+ cells staining with more than one TCR Vβ mAb; only TV9p6-1 was potentially monoclonal. The percentage of tetramer+ TV9- or TV9p6-T cells corresponded to that expressing surface CD107a/b upon antigen encounter. IFN-γ was produced by most CD107a/b-expressing cells, a proportion of which also produced IL-2 and TNF-α. Thus, both TV9p6- and TV9-CTLs exhibited a degree of polyfunctionality.
Table III.
CD107a/b mobilization and production of cytokines after stimulation with TV9p6 or TV9 loaded onto C1R-A2wt cells in three sets of parallel CTL cultures. No staining was detected using an irrelevant peptide (YV9).
| CTL cultures (% Tetramer+ Cells) |
Vβ Staining (% Tetramer+ Cells) |
Stimulating Peptide |
% T cells in culture staining for | |||
|---|---|---|---|---|---|---|
| CD107a/b | CD107a/b IL-2 |
CD107a/b TNF-α |
CD107a/b IFN-γ |
|||
| TV9p6-1 (100) | Vβ2 (100) | TV9p6 | 81 | 33 | 33 | 81 |
| TV9 | 71 | 22 | 23 | 69 | ||
| TV9-1 (98) | Vβ8 (32), Vβ17 (19), Vβ21.3 (4), Vβ22.3 (4) | TV9 | 81 | 8 | 9 | 72 |
| TV9p6-4 (52) | Vβ2 (40), Vβ8 (2), Vβ14 (1), Vβ21.3 (2) | TV9p6 | 68 | 20 | 10 | 48 |
| TV9 | 61 | 15 | 9 | 46 | ||
| TV9-4 (23) | Vβ8 (2), Vβ14 (1), Vβ22 (2), Vβ9 (14) | TV9 | 81 | 1 | 11 | 41 |
| TV9p6-5 (40) | Vβ8 (3), Vβ12 (1),Vβ22 (4) | TV9p6 | 44 | 25 | 21 | 20 |
| TV9 | 25 | 2 | 8 | 9 | ||
| TV9-5 (57) | Vβ22 (50) | TV9 | 61 | 34 | 48 | 61 |
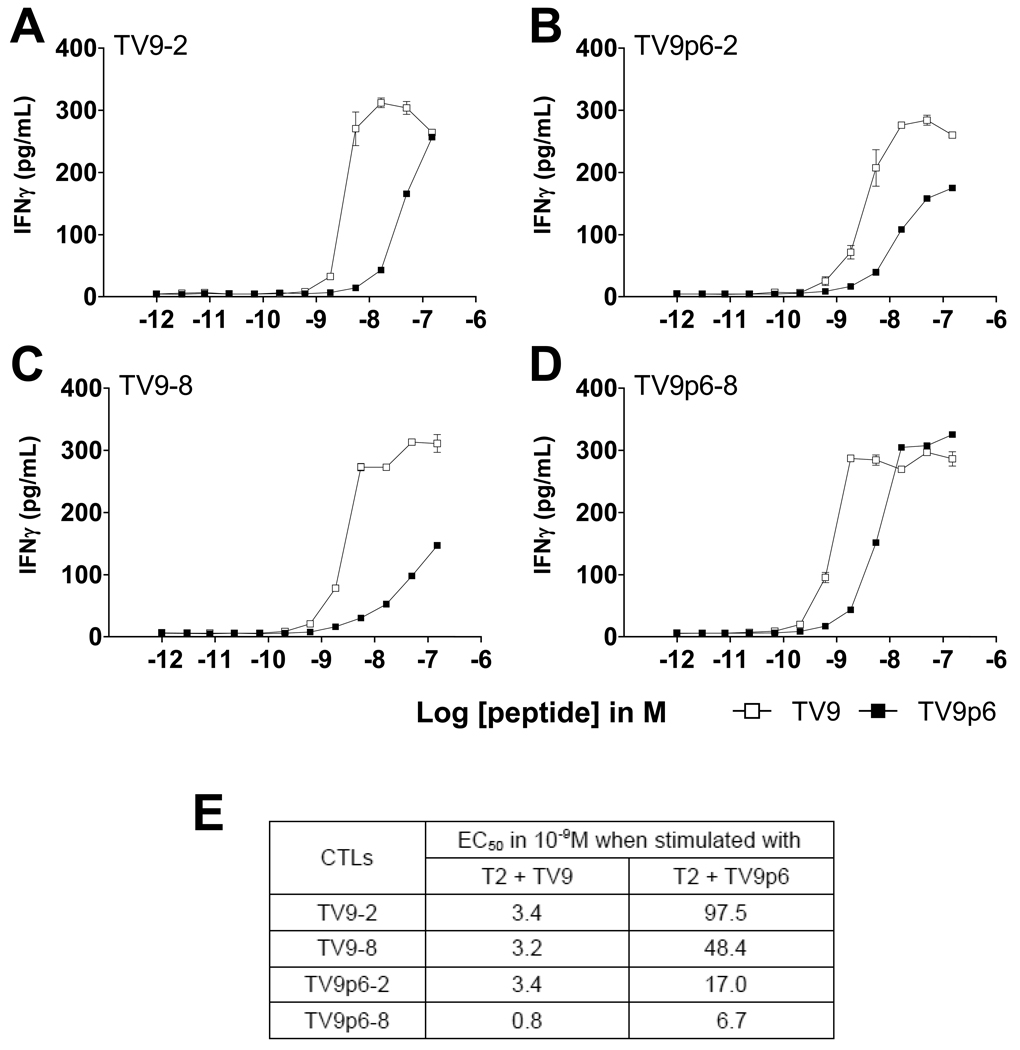
We next quantified the amount of IFN-γ secreted by parallel TV9- and TV9p6-CTLs after antigenic stimulation over a range of concentrations (Figure 5). Interestingly, both TV9- and TV9p6-CTL cultures showed greater sensitivity to stimulation by TV9 compared to TV9p6 as shown by the lower EC50 values for TV9. Although this may reflect in part the different binding affinities of TV9 and TV9p6 peptides to HLA-A*0201 (Table II), these data suggest that CTLs elicited by the artificial TV9p6 sequence would release IFN-γ as readily as those sensitized by the natural epitope when engaging HIV-infected cells.
Figure 5.
Functional sensitivity for two sets of parallel cultures determined by IFN-γ secretion. (A) TV9-2, (B) TV9p6-2, (C) TV9-8 and (D) TV9p6–8 cultures were stimulated at an E:T ratio of 1:10 with T2 cells pulsed with either TV9 or TV9p6. IFN-γ secretion into the supernatant was quantified by ELISA after 48 hours. (E) EC50 values were calculated for the TV9 and TV9p6 peptides in each case with Graphpad Prism V software. Data are shown as mean ± SEM of triplicate assays and are representative of two independent experiments; in most cases, the error bars are smaller than the plot symbols.
TV9p6-CTLs suppress HIV replication in vitro
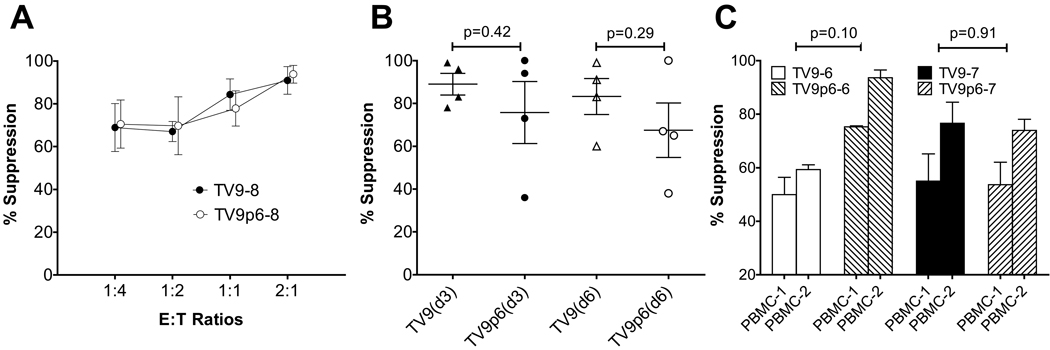
The ability of TV9- and TV9p6-CTLs to suppress replication of NL4-3.1 and JR-CSF in vitro was assessed (Figure 6). Both viruses encode for TV9. Increasing suppression of NL4-3.1 replication in T1 cells on day 3 was observed with increasing E:T ratios of TV9-8 and TV9p6–8 CTLs (Figure 6A). Moreover, no significant difference in suppression was observed over the range of E:T ratios. Similar antiviral efficacy between four sets of parallel TV9- and TV9p6-CTL cultures was observed at two time points (Figure 6B). Lastly, all CTL cultures inhibited the replication of JR-CSF in non-CD8+ PBMCs (Figure 6C). Thus, mimotope-primed CTLs inhibited HIV replication in vitro at least as efficiently as those activated by the wildtype peptide.
Figure 6.
TV9p6-CTLs suppress HIV replication at least as efficiently as TV9-CTLs. (A) Percent suppression of NL4-3.1 replication in T1 cells at day 3 post-infection by tetramer+ T cells at different E:T ratios as indicated. Data are shown as mean ± SEM of triplicate assays and are representative of two independent experiments. (B) Suppression of NL4-3.1 virus in T1 cells by TV9- and TV9p6-CTL cultures. The average percent suppression by TV9-2, -6, -7 and -8 T cells at day 3 and 6 are indicated by ▲ and Δ, respectively. Likewise, suppression mediated by TV9p6-2, -6, -7 and -8 T cells at the two time points are denoted by the symbols ● and ○, respectively. The E:T ratio for this assay was 2:1. Data are representative of two independent experiments. The concentrations of p24 in infected T1 cell cultures in the absence of CTLs were 0.3 ± 0.03 ng/mL and 0.8 ± 0.1 ng/mL on day 3 and 93 ± 14 ng/mL and 684 ± 48 ng/mL on day 6. (C) Suppression of HIV JR-CSF replication in two acutely infected, allogeneic HLA-A*0201-matched CD8-depleted PBMC cultures by two parallel CTL cultures (TV9-6 and TV9p6-6; TV9-7 and TV9p6–7) at day 9 post-infection. The concentrations of p24 in infected PBMC-1 and PBMC-2 cultures in the absence of CTLs were 311 ng/mL and 210 ng/mL, respectively. The p values comparing suppression by TV9- and TV9p6-CTL were determined using an unpaired, two-tailed Student T test with Graphpad Prism V software.
TCRBV gene usage by parallel cultures of TV9- and TV9p6-CTLs
To characterize further the nature of TV9- and TV9p6-CTLs, we examined clonotype usage within TV9 tetramer-binding CD8+ T cell populations in parallel cultures from several donors using a template-switch anchored RT-PCR to amplify all expressed TCRB gene products without bias as described previously [23] (Table IV). All TV9-CD8+ T cell populations were oligoclonal, comprising 6 or fewer TCR clonotypes; distinct hierarchical frequencies were also apparent. Furthermore, despite starting naïve T cell pools derived from the same source, parallel TV9- and TV9p6-CTL cultures were largely distinct in terms of TV9-specific clonal composition. However, a degree of intra-individual clonal overlap was observed between TV9-6 and TV9p6-6 cultures and between TV9-7 and TV9p6–7 cultures (light shading, italicized sequences). Importantly, this demonstrates that at least some identical clonotypes were present within the initial naïve T cell pools and provides evidence that differential priming was not solely a result of stochastic "sampling" bias. Of note, a public clonotype, differentially encoded at the nucleotide level, was shared between two individuals (TV9p6-6 and TV9-9; darker shading, bold sequences).
Table IV.
TCRBV and TCRJ usage, CDR3 amino acid sequence and percent frequency of TV9-specific CD8+ T cell clonotypes in parallel CTL cultures.
| CTLs | HLA Class I Alleles | Vβ (Arden) | IMGT TRB | CDR3 | TRBJ | Frequency |
|---|---|---|---|---|---|---|
| TV9-7 | A*0201,A11 B18,B27 | 13 | 6-1 | CASSEEGAWNTGELF | 2-2 | 90 |
| 9 | 3-1 | CASSHTSGGLKDTQY | 2–3 | 10 | ||
| TV9p6-7 | 7 | 4-3 | CASSPRGDWEEKAGELF | 2-2 | 77 | |
| 13 | 6-1 | CASSEEGAWNTGELF | 2-2 | 15 | ||
| 4 | 29-1 | CSVEDPDRGYEQY | 2–7 | 8 | ||
| TV9-6 | A*0201,A29 B40,B44 | 14 | 27 | CASSRRGDGFQPQH | 1–5 | 36 |
| 13 | 6-6 | CASRDFYNEQF | 2-1 | 29 | ||
| 2 | 20-1 | CSAFSSGTGGVTGELF | 2-2 | 14 | ||
| 7 | 4-3 | CASSQDPSPSTDTQY | 2–3 | 7 | ||
| 6 | 7–8 | CASSLWGLAPDNEQF | 2-1 | 7 | ||
| 14 | 27 | CASSLWGGPMETQY | 2–5 | 7 | ||
| TV9p6-6 | 2 | 20-1 | CSAFSSGTGGVTGELF | 2-2 | 82 | |
| 6 | 7-2 | CASSLVPQGEEQY | 2–7 | 6 | ||
| 2 | 20-1 | CSAFSSGTGGVTGEPF | 2-2 | 6 | ||
| 2 | 20-1 | CSAFSSGTEGVTGELF | 2-2 | 6 | ||
| TV9-9 | A*0201,A24 B15,B40 | 6 | 7-2 | CASSLVPQGEEQY | 2–7 | 88 |
| 6 | 7-2 | CASSLVPQGEEPY | 2–7 | 12 | ||
| TV9-3 | A2 | 3 | 28-0 | CASSYRGQSYEQY | 2–7 | 94 |
| 4 | 29-1 | CSGRGDSYTEAF | 1-1 | 4 | ||
| 5, 31 | 5–8 | CASSEYRGPSSYEQY | 2–7 | 1 | ||
| 3 | 28-0 | CASSCRGQSYEQY | 2–7 | 1 | ||
| TV9-2 | A*0201 B15,B40 | 2 | 20-1 | CSARDAGGDYGYT | 1–2 | 69 |
| 8 | 12-4 | CASSDTGELF | 2-2 | 26 | ||
| 9 | 3-1 | CASSQVKIGQGNEKLF | 1–4 | 6 | ||
| TV9p6-2 | 2 | 20-1 | CSASRDSYEQY | 2–7 | 100 | |
Discussion
The value of using conserved regions of the HIV proteome for the purposes of vaccination has been recognized for some time [24–26]. However, although these domains are populated with CD8+ T cell epitopes, most have not been correlated with protection in carriers of the majority of HLA class I alleles. We propose that the use of conserved regions within HIV as vaccines for non-protective class I alleles will depend on whether it is feasible to improve the intrinsic immunogenicity of these natural sequences. This may be achieved by various means, such as modulating the usual patterns of immunodominance [27], improving processing and presentation [28, 29], or stimulating with mimotopes [14]. Indeed, there is suggestive evidence that protection can be achieved by vaccine-induced responses to a subdominant HLA-A*02-restricted epitope [30]. TV9 is an example of a conserved subdominant epitope with characteristics of a useful vaccine target. Our data with CTLs primed in vitro from healthy donors revealed that it may be necessary to stimulate high avidity clonotypes selectively within the structurally diverse TV9-T cell precursor pools [11]. In this study, we identified and tested mimotopes of TV9 for the purpose of recruiting more appropriate HIV-specific CD8+ T cells. This approach is based on three principles: (i) a single TCR can respond to many different peptides [31]; (ii) the pool of precursor T cell clonotypes specific for a particular epitope can be very diverse [32, 33]; and, (iii) the requirements for T cell priming and the triggering of downstream effector functions are different [34].
We screened a PS-SCL with a TV9-CTL culture to enable de novo design of mimotopes that not only stimulate TV9-CTLs but also immunize crossreactive CTLs in vitro. The peptide library that we used consists of complex nonapeptide mixtures with one defined amino acid in one of the sequence positions and mixtures of all proteogenic amino acids excepting cysteine (which can be oxidized to crosslink peptides) in the remaining positions [35]. Up to 323 × 109 different peptides are represented in this library format. This strategy has been applied most extensively in autoimmune diseases and with promising clinical observations in cancer [36]. We have previously applied this approach to the “chronic immunodominant” HLA-A*0201-restricted SL9 (p17 Gag77–85) epitope and identified a mimotope that primed qualitatively superior CTLs from the naïve repertoires of healthy donors in vitro and triggered more extensive expansion of antigen-experienced CD8+ T cells from patients [14].
Very large numbers of mimotopes with different sequences and T cell activation potencies can be identified for each epitope, with each mimotope addressing a different repertoire that intersects around the natural HIV epitope. Here, we specifically explored whether a variant peptide (TV9p6) with significantly lower binding affinity to HLA-A*0201 than the native epitope, presumably leading to a reduced pMHC density on the surface of APCs, would focus the subdominant TV9-CTL response. In fact, a reciprocal relationship between binding affinity and functional sensitivity was recently reported for a cancer epitope and its agonist [37]. CTL cultures specific for TV9 and TV9p6 were almost always crossreactive as determined by a shared capacity to lyse target cells loaded over a broad range of peptide concentrations, thereby suggesting considerable overlap between their recognition properties despite differences in three of nine positions between the two peptides. Moreover, CTLs primed with both specificities consistently suppressed HIV replication in vitro, thereby indicating that they cross-recognized naturally processed and presented TV9. Although altered peptide ligands for other CTL epitopes can trigger differential responses [14], modification of the responding TCR repertoire by TV9p6 did not significantly alter any of the CTL functions or properties assessed, including antigen sensitivity measured by cytotoxicity or avidity measured by the intensity of tetramer staining. Moreover, the fact that highly functional CTLs were generated by TV9p6 indicates that either CTL activation does not necessarily depend on peptide binding affinity to class I antigens or, alternatively, that the low binding affinity of TV9p6 to HLA-A*0201 exceeded a minimal threshold. Our finding is in line with recent reports showing that neither immunodominance nor the efficacy of CTL responses correlates with the binding affinity of the epitopic peptides to class I antigens [18, 38]. Indeed, substantially greater selection pressure on HIV is imposed by HLA–B compared to HLA-A alleles [17] despite the lower average peptide binding affinity of the former [18].
The quality of a virus-specific CD8+ T cell response ultimately rests on the availability of “appropriate” clonotypes in the host T cell repertoire [39]. In HIV infections, most studies have relied on the analysis of patient responses to infections in vivo. Although informative, they provide only partial insight into the diversity of the immune repertoire among individuals. Analysis of the pre-infection virus-specific T cell pools will help complete the picture and may be particularly cogent for the design of prophylactic vaccines. For example, we noted a non-crossreactive, TV9p6-tetramer binding population in one of the four donors shown in Figure 3, indicating inter-individual differences in the intersection between the TCR repertoires specific for TV9 and TV9p6. This finding underscores the difficulty in selecting appropriate epitope variants that will raise vigorous crossreactive CD8+ T cells without inadvertently activating responder cells that are not specific for the virus. These non-HIV responses can be potentially deleterious, perhaps working against the emergence of new responses to viral variants [40, 41]. On the other hand, subsequent HIV infection may selectively activate only the virus-specific memory pool generated by pre-emptive vaccination with mimotopes, thus promoting immunological control of HIV. The use of artificial sequences may be particularly important for “obligate” subdominant epitopes, which are recognized by intrinsically small T cell precursor populations or suppressed by emerging T cells that target other epitopes. This study examines one strategy to heighten the intrinsic immunogenicity of known conserved subdominant determinants.
Molecular analysis of TCRB gene expression showed that CTL cultures specific for TV9 and TV9p6 were oligoclonal and generally dominated by one or two clonotypes (Table IV). Although no TCR motifs were apparent, biased usage of certain sequences was found in two of three parallel cultures (TV9-6/TV9p6-6 and TV9-7/TV9p6–7). This suggests considerable clonal overlap and a high precursor frequency of certain crossreactive clonotypes. Conservation within the CDR3 loop encoded distinctly at the nucleotide level was also detected between individuals, indicating a common antigen-specific “public TCR”. A recent study showed that usage of public clonotypes within a particular protective Gag-specific CD8+ T cell response in acute SIV infection can act as a molecular signature of biological outcome [42]. These findings emphasize the need to develop immunization strategies that selectively recruit efficacious epitope-specific CD8+ T cell clonotypes [43].
In summary, despite its low binding affinity to HLA-A*0201, TV9p6 did not selectively mobilize high avidity clonotypes, although it did effectively activate fully functional HIV-specific CTLs. Comparison of TCRB gene usage between CD8+ T cell populations specific for TV9 and TV9p6 showed that distinct clonotypes were recruited in each case from the same initial naïve T cell pool. Thus, mimotope-based vaccination represents a potentially viable approach to recruit additional cognate CD8+ T cell clonotypes that could overcome the intrinsic limitations of repertoire mobilization and thereby help to override natural patterns of epitope subdominance.
Materials and methods
Healthy seronegative donors
Heparinized blood was collected from healthy seronegative HLA-A*0201+ volunteers. High resolution HLA genotyping was performed at the Department of Transfusion Medicine, National Institutes of Health. This study was approved by the Human Investigation Committee at Wayne State University School of Medicine and the University of Texas at El Paso. All subjects provided written informed consent.
Generation of ex vivo primed peptide-specific CD8+ T cell cultures from healthy seronegative donors
The procedures for the generation of peptide-specific CTLs by in vitro immunization of naïve CD8+ T cells have been reported [44]. In brief, DCs were derived from adherent peripheral blood monocytes after culturing for 7 days in complete medium (RPMI 1640 medium containing 10% autologous serum, 100 U/mL penicillin, 100 µg/mL streptomycin and 2 mM L-glutamine) supplemented with GM-CSF (1000 U/mL; Leukine Sargramostim, Bayer HealthCare Pharmaceuticals, Wayne, NJ) and IL-4 (500 U/mL; Peprotech, Rocky Hill, NJ). Positively-selected autologous CD8+ T cells (Dynabeads; Dynal) were primed with irradiated (4,000 cGy) DCs pulsed for 2 hours with 10 µg/mL peptide at a T cell-DC ratio of 5:1 in 48-well cluster plates. T cells were subsequently re-stimulated every 7 to 10 days with autologous monocytes pulsed with peptides [14, 44]. Ten ng/mL IL-7 (Genzyme) was added on the day of priming and re-stimulation; 20 U/mL IL-2 was added one and four days later, respectively.
Target cell lines
Cytotoxicity or intracellular cytokine secretion was assessed against peptide-pulsed T2 cells (TAP-deficient T-B lymphoblast hybrids [45]) or C1R cells expressing either full-length wildtype HLA-A*0201 (C1R-A2wt) or a point-mutated variant (C1R-A2CD8null) that does not bind the CD8 coreceptor [46]. The HIV-1 replication-permissive T1 cell line expressing HLA-A*0201 was used for viral suppression assays. All cell lines were maintained in RPMI with 10% heat-inactivated fetal calf serum.
Flow cytometric analysis
Directly conjugated mAbs to CD8 (FITC- and PE-RPA-T8), CD107a (FITC-H4A3), CD107b (FITC-H4B4), HLA-A2 (FITC-BB7.2), IFN-γ (PE-B27), IL-2 (PE-MQ1-17H12), and TNF-α (PE-MAb11) were purchased from BD Pharmingen (San Diego, CA). Vβ family-specific mAbs were obtained from Beckman Coulter (Miami, FL). Tetramer stains were performed on ice for 30 min; cells were then washed and stained for CD8 at 4°C for an additional 20 min. Staining with an irrelevant tetramer was used as a control. Stained cells were acquired with a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) and data analyzed with WinMDI software. Intracellular cytokine production and degranulation were determined after stimulation for 4 hours with peptide-pulsed (10 µg/mL) T2 or C1R cells at 37°C as described previously [47]. Gating was performed on CD8+ T cells and greater than 10,000 events were collected for each sample. TV9- and TV9p6-tetramers were obtained from the NIH Tetramer Core Facility, Atlanta, GA.
Nonapeptide library and assay for cytotoxicity with library mixtures
A TV9-CTL culture [11] was used to scan an L-amino acid nonapeptide PS-SCL to identify candidate mimotope sequences [14]. All nonapeptides present in the library contained a free N-terminus and an amidated C-terminus. The PS-SCL comprised 180 mixtures in an OX8 format, where O represents one of the 20 L-amino acids in a defined position and X represents all natural L-amino acids except L-cysteine at each of the remaining positions. Each mixture was composed of 1.7 × 1010 peptides and the total X9 library consisted of some 323 × 109 different nonapeptides in approximately equimolar concentration. For stimulation of the index TV9-1, CTLs were washed and resuspended at 1 × 105 cells/mL in complete medium 7 days after specific re-stimulation. Then, 100 µL of this cell suspension was added to triplicate wells of 96-well U-bottom plates containing 2 × 103 51Cr-labeled T2 cells and the various peptide library mixtures (100 µg/mL). Cells were cultured for 4 hours at 37°C prior to harvesting and calculation of percent lysis determined with respect to control wells. Each mixture was assayed in triplicate and scanning was repeated three times. The library was prepared at Multiple Peptide Systems, San Diego, CA.
Peptides
TV9 (TLNAWVKVV) and agonist peptides p30 (TINAWIKVV), TV9p6 (KINAWIKVV), p29 (TINAWIKGV), and p5 (KINAWIKGV) peptides were purchased at >90% purity from Genemed Synthesis (San Francisco, CA).
MHC-peptide binding assays
Quantitative assays to measure the binding of peptides to HLA A*0201, A*0202, A*0203, A*0206, and A*6802 molecules were based on the inhibition of binding of a radiolabeled standard peptide as described previously [48]. Peptides were tested at six different concentrations in three or more independent assays and the concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide (IC50) was calculated. Under the conditions used, where [radiolabeled probe] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values.
IFN-γ detection by ELISA
IFN-γ secretion was determined by ELISA with an OptEIA Set (BD Pharmingen) in supernatants of T cells re-stimulated with peptide-pulsed T2 cells for 48 hours. The range of sensitivity for IFN-γ was 15 to 500 pg/mL. EC50 values (concentrations of peptide required to elicit 50% of the maximal reactivity) were calculated with GraphPad Prism V software.
In vitro viral suppression assays
The capacity of TV9-CTLs to inhibit HIV infection was determined in vitro. HIV JR-CSF and NL4-3.1 stocks were generated and titers were determined as previously described [49]. Infected PBMCs or T1 cells (infected by NL4-3.1 virus at 500 pg p24 per 106 cells) were cultured at 50,000 cells per flat-bottomed well in 96-well plates with CTLs at the indicated ratios in complete medium containing IL-2 (25 U/mL). PBMCs from HLA-A*0201+, HIV-uninfected donors were depleted of CD8+ cells (Miltenyi Biotec, Auburn, CA), activated with phytohemagglutinin A in complete medium containing IL-2 (25 U/mL) for 2 days and then infected by overnight incubation with HIV JR-CSF at 80 TCID50/mL (GenBank Accession Number AAB05598). Supernatants were tested for p24 antigen concentration by ELISA (HIV-1 p24CA antigen capture assay kit, AIDS and Cancer Virus Program, NCI Frederick Cancer Research) every 3 days up to day 9. The range of sensitivity for p24 was 7.5 to 400 pg/mL.
Molecular analysis of expressed TCRB gene products by quantitative clonotypic PCR
Tetramer-binding CD8+ T cells were sorted viably to >98% purity for RNA-based analysis of TCRB gene expression using an unbiased template-switch anchored RT-PCR as described previously [23]. The IMGT nomenclature is used for all molecular analyses conducted in this study.
Acknowledgments
This work was supported by the Michigan Life Sciences Corridor Program 1659 (J.K-M.), NIH R01-AI064069 and R01-AI77413 (J.K-M.), and NCRR 5G12RR008124 (J.K-M). We acknowledge support to S.E.B. and D.B.W. from the AIDS and Infectious Disease Science Center, NIH R43DA021175 (S.E.B.) and NIH R44DA015212 (D.B.W.). D.A.P. is a Medical Research Council (UK) Senior Clinical Fellow.
Abbreviations used in this paper
- APCs
antigen-presenting cells
- DCs
dendritic cells
- TCR
T cell receptor
- tetramer
tetrameric HLA-A*0201-peptide complex
- TV9
HIV p24 Gag19–27 epitope (TLNAWVKVV)
- TV9-
TV9-specific
Footnotes
Conflict of interest: The authors have declared no financial or commercial conflict of interest.
References
- 1.Walker BD, Burton DR. Toward an AIDS vaccine. Science. 2008;320:760–764. doi: 10.1126/science.1152622. [DOI] [PubMed] [Google Scholar]
- 2.Allen TM, Altfeld M. Crippling HIV one mutation at a time. J Exp Med. 2008;205:1003–1007. doi: 10.1084/jem.20080569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins DI. The hope for an HIV vaccine based on induction of CD8+ T lymphocytes--a review. Mem Inst Oswaldo Cruz. 2008;103:119–129. doi: 10.1590/s0074-02762008000200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 5.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brander C, Walker BD. Gradual adaptation of HIV to human host populations: good or bad news? Nat Med. 2003;9:1359–1362. doi: 10.1038/nm941. [DOI] [PubMed] [Google Scholar]
- 7.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 8.Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, Pillay T, Hilton L, Thobakgale C, Ramduth D, Draenert R, Le Gall S, Luzzi G, Edwards A, Brander C, Sewell AK, Moore S, Mullins J, Moore C, Mallal S, Bhardwaj N, Yusim K, Phillips R, Klenerman P, Korber B, Kiepiela P, Walker B, Goulder P. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201:891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korber BTM, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI. HIV Molecular Immunology. Los Alamos, New Mexico: Publisher: Los Alamos National Laboratory, Theoretical Biology and Biophysics; 2005. 2005. [Google Scholar]
- 10.Goulder PJ, Bunce M, Krausa P, McIntyre K, Crowley S, Morgan B, Edwards A, Giangrande P, Phillips RE, McMichael AJ. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res Hum Retroviruses. 1996;12:1691–1698. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- 11.Schaubert KL, Price DA, Frahm N, Li J, Ng HL, Joseph A, Paul E, Majumder B, Ayyavoo V, Gostick E, Adams S, Marincola FM, Sewell AK, Altfeld M, Brenchley JM, Douek DC, Yang OO, Brander C, Goldstein H, Kan-Mitchell J. Availability of a diversely avid CD8+ T cell repertoire specific for the subdominant HLA-A2-restricted HIV-1 Gag p2419-27 epitope. J Immunol. 2007;178:7756–7766. doi: 10.4049/jimmunol.178.12.7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Missale G, Papagno L, Penna A, Pilli M, Zerbini A, Vitali P, Pieroni G, Urbani S, Uggeri J, Pinheiro S, Rowland-Jones S, Ferrari C. Parenteral exposure to high HIV viremia leads to virus-specific T cell priming without evidence of infection. Eur J Immunol. 2004;34:3208–3215. doi: 10.1002/eji.200424889. [DOI] [PubMed] [Google Scholar]
- 13.Bennett MS, Ng HL, Dagarag M, Ali A, Yang OO. Epitope-dependent avidity thresholds for cytotoxic T-lymphocyte clearance of virus-infected cells. J Virol. 2007;81:4973–4980. doi: 10.1128/JVI.02362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kan-Mitchell J, Bajcz M, Schaubert KL, Price DA, Brenchley JM, Asher TE, Douek DC, Ng HL, Yang OO, Rinaldo CR, Jr, Benito JM, Bisikirska B, Hegde R, Marincola FM, Boggiano C, Wilson D, Abrams J, Blondelle SE, Wilson DB. Degeneracy and repertoire of the human HIV-1 Gag p17(77–85) CTL response. J Immunol. 2006;176:6690–6701. doi: 10.4049/jimmunol.176.11.6690. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Gran B, Pinilla C, Markovic-Plese S, Hemmer B, Tzou A, Whitney LW, Biddison WE, Martin R, Simon R. Combinatorial peptide libraries and biometric score matrices permit the quantitative analysis of specific and degenerate interactions between clonotypic TCR and MHC peptide ligands. J Immunol. 2001;167:2130–2141. doi: 10.4049/jimmunol.167.4.2130. [DOI] [PubMed] [Google Scholar]
- 16.Garcia KC, Teyton L, Wilson IA. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 17.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA–B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 18.Rao X, Costa AI, van Baarle D, Kesmir C. A comparative study of HLA binding affinity and ligand diversity: implications for generating immunodominant CD8+ T cell responses. J Immunol. 2009;182:1526–1532. doi: 10.4049/jimmunol.182.3.1526. [DOI] [PubMed] [Google Scholar]
- 19.Hutchinson SL, Wooldridge L, Tafuro S, Laugel B, Glick M, Boulter JM, Jakobsen BK, Price DA, Sewell AK. The CD8 T cell coreceptor exhibits disproportionate biological activity at extremely low binding affinities. J Biol Chem. 2003;278:24285–24293. doi: 10.1074/jbc.M300633200. [DOI] [PubMed] [Google Scholar]
- 20.Wooldridge L, Lissina A, Vernazza J, Gostick E, Laugel B, Hutchinson SL, Mirza F, Dunbar PR, Boulter JM, Glick M, Cerundolo V, van den Berg HA, Price DA, Sewell AK. Enhanced immunogenicity of CTL antigens through mutation of the CD8 binding MHC class I invariant region. Eur J Immunol. 2007;37:1323–1333. doi: 10.1002/eji.200636765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai KL, Karandikar NJ, Casazza JP, Koup RA. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- 24.Wilson CC, McKinney D, Anders M, MaWhinney S, Forster J, Crimi C, Southwood S, Sette A, Chesnut R, Newman MJ, Livingston BD. Development of a DNA vaccine designed to induce cytotoxic T lymphocyte responses to multiple conserved epitopes in HIV-1. J Immunol. 2003;171:5611–5623. doi: 10.4049/jimmunol.171.10.5611. [DOI] [PubMed] [Google Scholar]
- 25.Wang YE, Li B, Carlson JM, Streeck H, Gladden AD, Goodman R, Schneidewind A, Power KA, Toth I, Frahm N, Alter G, Brander C, Carrington M, Walker BD, Altfeld M, Heckerman D, Allen TM. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J Virol. 2009;83:1845–1855. doi: 10.1128/JVI.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letourneau S, Im EJ, Mashishi T, Brereton C, Bridgeman A, Yang H, Dorrell L, Dong T, Korber B, McMichael AJ, Hanke T. Design and preclinical evaluation of a universal HIV-1 vaccine. PLoS ONE. 2007;2:e984. doi: 10.1371/journal.pone.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenzer S, Wee E, Burgevin A, Stewart-Jones G, Friis L, Lamberth K, Chang CH, Harndahl M, Weimershaus M, Gerstoft J, Akkad N, Klenerman P, Fugger L, Jones EY, McMichael AJ, Buus S, Schild H, van Endert P, Iversen AK. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat Immunol. 2009;10:636–646. doi: 10.1038/ni.1728. [DOI] [PubMed] [Google Scholar]
- 28.Draenert R, Le Gall S, Pfafferott KJ, Leslie AJ, Chetty P, Brander C, Holmes EC, Chang SC, Feeney ME, Addo MM, Ruiz L, Ramduth D, Jeena P, Altfeld M, Thomas S, Tang Y, Verrill CL, Dixon C, Prado JG, Kiepiela P, Martinez-Picado J, Walker BD, Goulder PJ. Immune Selection for Altered Antigen Processing Leads to Cytotoxic T Lymphocyte Escape in Chronic HIV-1 Infection. J Exp Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milicic A, Price DA, Zimbwa P, Booth BL, Brown HL, Easterbrook PJ, Olsen K, Robinson N, Gileadi U, Sewell AK, Cerundolo V, Phillips RE. CD8+ T cell epitope-flanking mutations disrupt proteasomal processing of HIV-1 Nef. J Immunol. 2005;175:4618–4626. doi: 10.4049/jimmunol.175.7.4618. [DOI] [PubMed] [Google Scholar]
- 30.Heckerman D, Frahm N, Pereyra F, Dubey S, Geraghty D, Carlson J, Robertson M, McElrath J, Casimiro D, Walker BD. Vaccine-induced targeting of epitopes associated with spontaneous control of HIV viral replication is associated with lower set-point viral loads in HIV-infected participants from the STEP trial. 16th Conference on Retroviruses and Opportunistic Infections.2009. [Google Scholar]
- 31.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today. 1998;19:395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 32.Kedzierska K, La Gruta NL, Turner SJ, Doherty PC. Establishment and recall of CD8+ T-cell memory in a model of localized transient infection. Immunol Rev. 2006;211:133–145. doi: 10.1111/j.0105-2896.2006.00386.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhong W, Dixit SB, Mallis RJ, Arthanari H, Lugovskoy AA, Beveridge DL, Wagner G, Reinherz EL. CTL recognition of a protective immunodominant influenza A virus nucleoprotein epitope utilizes a highly restricted Vbeta but diverse Valpha repertoire: functional and structural implications. J Mol Biol. 2007;372:535–548. doi: 10.1016/j.jmb.2007.06.057. [DOI] [PubMed] [Google Scholar]
- 34.Udaka K, Wiesmuller KH, Kienle S, Jung G, Walden P. Self-MHC-restricted peptides recognized by an alloreactive T lymphocyte clone. J Immunol. 1996;157:670–678. [PubMed] [Google Scholar]
- 35.Pinilla C, Rubio-Godoy V, Dutoit V, Guillaume P, Simon R, Zhao Y, Houghten RA, Cerottini JC, Romero P, Valmori D. Combinatorial peptide libraries as an alternative approach to the identification of ligands for tumor-reactive cytolytic T lymphocytes. Cancer Res. 2001;61:5153–5160. [PubMed] [Google Scholar]
- 36.Sharav T, Wiesmuller KH, Walden P. Mimotope vaccines for cancer immunotherapy. Vaccine. 2007;25:3032–3037. doi: 10.1016/j.vaccine.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 37.Hou Y, Kavanagh B, Fong L. Distinct CD8+ T cell repertoires primed with agonist and native peptides derived from a tumor-associated antigen. J Immunol. 2008;180:1526–1534. doi: 10.4049/jimmunol.180.3.1526. [DOI] [PubMed] [Google Scholar]
- 38.Bihl F, Frahm N, Di Giammarino L, Sidney J, John M, Yusim K, Woodberry T, Sango K, Hewitt HS, Henry L, Linde CH, Chisholm JV, 3rd, Zaman TM, Pae E, Mallal S, Walker BD, Sette A, Korber BT, Heckerman D, Brander C. Impact of HLA–B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol. 2006;176:4094–4101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- 39.Davenport MP, Price DA, McMichael AJ. The T cell repertoire in infection and vaccination: implications for control of persistent viruses. Curr Opin Immunol. 2007;19:294–300. doi: 10.1016/j.coi.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Klenerman P, Zinkernagel RM. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. 1998;394:482–485. doi: 10.1038/28860. [DOI] [PubMed] [Google Scholar]
- 41.Larke N, Im EJ, Wagner R, Williamson C, Williamson AL, McMichael AJ, Hanke T. Combined single-clade candidate HIV-1 vaccines induce T cell responses limited by multiple forms of in vivo immune interference. Eur J Immunol. 2007;37:566–577. doi: 10.1002/eji.200636711. [DOI] [PubMed] [Google Scholar]
- 42.Price DA, Asher TE, Wilson NA, Nason MC, Brenchley JM, Metzler IS, Venturi V, Gostick E, Chattopadhyay PK, Roederer M, Davenport MP, Watkins DI, Douek DC. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med. 2009;206:923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, Letvin NL, Franchini G, Wolinsky SM, Koup RA, Douek DC. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Kan-Mitchell J, Bisikirska B, Wong-Staal F, Schaubert KL, Bajcz M, Bereta M. The HIV-1 HLA-A2-SLYNTVATL is a help-independent CTL epitope. J Immunol. 2004;172:5249–5261. doi: 10.4049/jimmunol.172.9.5249. [DOI] [PubMed] [Google Scholar]
- 45.Salter RD, Cresswell P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. Embo J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purbhoo MA, Boulter JM, Price DA, Vuidepot AL, Hourigan CS, Dunbar PR, Olson K, Dawson SJ, Phillips RE, Jakobsen BK, Bell JI, Sewell AK. The human CD8 coreceptor effects cytotoxic T cell activation and antigen sensitivity primarily by mediating complete phosphorylation of the T cell receptor zeta chain. J Biol Chem. 2001;276:32786–32792. doi: 10.1074/jbc.M102498200. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell MS, Lund TA, Sewell AK, Marincola FM, Paul E, Schroder K, Wilson DB, Kan-Mitchell J. The cytotoxic T cell response to peptide analogs of the HLA-A*0201-restricted MUC1 signal sequence epitope, M1.2. Cancer Immunol Immunother. 2007;56:287–301. doi: 10.1007/s00262-006-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidney J, Southwood S, Oseroff C, Del Guercio MF, Sette A, Grey H. Measurement of MHC/peptide interactions by gel filtration Current protocols in immunology. New York: Wiley; 1998. pp. 18.13.11–18.13.19. [DOI] [PubMed] [Google Scholar]
- 49.Yang OO, Kalams SA, Rosenzweig M, Trocha A, Jones N, Koziel M, Walker BD, Johnson RP. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J Virol. 1996;70:5799–5806. doi: 10.1128/jvi.70.9.5799-5806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]