Abstract
Influenza A virus causes seasonal epidemics, sporadic pandemics and is a significant global heath burden. Influenza virus is an enveloped virus that contains a segmented negative strand RNA genome. Assembly and budding of progeny influenza virions is a complex, multistep process that occurs in lipid raft domains on the apical membrane of infected cells. The viral proteins hemagglutinin (HA) and neuraminidase (NA) are targeted to lipid rafts, causing the coalescence and enlargement of the raft domains. This clustering of HA and NA may cause a deformation of the membrane and the initiation of the virus budding event. M1 is then thought to bind to the cytoplasmic tails of HA and NA where it can then polymerize and form the interior structure of the emerging virion. M1, bound to the cytoplasmic tails of HA and NA, additionally serves as a docking site for the recruitment of the viral RNPs and may mediate the recruitment of M2 to the site of virus budding. M2 initially stabilizes the site of budding, possibly enabling the polymerization of the matrix protein and the formation of filamentous virions. Subsequently, M2 is able to alter membrane curvature at the neck of the budding virus, causing membrane scission and the release of the progeny virion. This review investigates the latest research on influenza virus budding in an attempt to provide a step-by-step analysis of the assembly and budding processes for influenza viruses.
Keywords: Influenza Virus, Viral Like Particle, Assembly, Budding, Filamentous Virion, Membrane Curvature
Introduction
Influenza is a major cause of morbidity and mortality around the world. It is estimated that infection with seasonal strains of influenza virus results in the death of over 50,000 people per year (Thompson et al., 2003). In addition to the seasonal strains, pandemic strains of influenza virus are capable of circling the globe rapidly, causing severe disease, such as that caused by the 1918 pandemic strain that resulted in over 50 million deaths during a two year period. Pandemic strains, such as the currently circulating 2009 H1N1 strain that has infected over 22 million people (C.D.C., 2009), arise from the reassortment of the eight different genome RNA segments of two strains of influenza virus within a single host cell.
The eight, negative-sense, RNA segments of the influenza virus genome encode 11 different proteins, of which 8 are packaged into the infectious, enveloped, virion (Palese and Shaw, 2007). On the viral surface are the two main antigenic determinants of the virus, the spike glycoproteins; hemagglutinin (HA) and neuraminidase (NA). HA mediates viral entry into cells and has receptor binding and membrane fusion activity. NA mediates enzymatic cleavage of the viral receptor at late stages of infection, allowing for the release of progeny virions. A third integral membrane protein, M2, is a multi-functional, proton-selective, ion channel which has roles both in virus entry as well as in assembly and budding. Inside the viral envelope, the matrix protein (M1) provides structure to the virion and bridges interactions between the viral lipid membrane and the ribonucleoprotein (RNP) core. The RNP core is composed of the RNA polymerase complex proteins, PB1, PB2 and PA, and the nucleocapsid protein (NP) which mediates binding and packaging of the viral genome. During virus replication three other proteins are expressed that are not incorporated into the mature virion. Non-structural protein 1 (NS1) is a multi-functional protein with a major role in evasion of the host immune system. NS2 (NEP) plays a crucial role in mediating the export of viral RNPs from the cell nucleus during replication. Additionally, many strains of influenza virus, but not all, express a protein designated PB1-F2 which is transcribed from a second reading frame in PB1. The PB1-F2 protein is involved in the induction of host-cell apoptosis.
The influenza virion is pleiomorphic, forming spherical virions that are ~100 nm in diameter as well as filamentous virions that are ~100 nm in diameter but reaching over 20 μm in length. Whereas several laboratory strains produce solely spherical virions it is thought that in vivo human infection produces predominantly filamentous virions (Chu et al., 1949; Kilbourne and Murphy, 1960). Indeed, samples isolated directly from the human upper respiratory tract show mainly filamentous forms of influenza virus (Chu et al., 1949; Kilbourne and Murphy, 1960). Additionally, analysis of lung sections from fatal cases of the 2009 H1N1 pandemic strain show the presence of filamentous virions (Nakajima et al., 2010) and passage of the 2009 H1N1 strain in tissue culture shows that the virus retains the ability to form filamentous virions during repeated passage (Itoh et al., 2009; Neumann et al., 2009).
Filamentous forms of influenza virus have been noted in the literature for many years (Ada et al., 1958; Burnet and Lind, 1957; Choppin, 1963; Choppin et al., 1961; Choppin et al., 1960; Chu et al., 1949; Kilbourne and Murphy, 1960; Morgan et al., 1956), although the functional significance of these filamentous virions has never been ascertained. Recent work has shown that both the filamentous and the spherical forms of influenza virus contain only one copy of the viral genome (Calder et al., 2010; Noda et al., 2006; Rossman et al., 2010a). The virions appear to contain similar ratios of most viral proteins and appear to be comparably infectious (Roberts et al., 1998). Interestingly, serial passage of filamentous isolates of influenza virus in eggs causes a loss of the filamentous morphology (Choppin et al., 1960). Indeed, the two most widely used laboratory spherical strains, A/WSN/33 and A/PR/8, have been passaged extensively in eggs. Thus, the loss of filament-forming ability may be an adaptation to growth in eggs and not specifically applicable to human infection, although it should be noted that even infections with filamentous strains produce both filamentous as well as spherical virions. It is not clear if certain infection conditions will select for one form of the virus over the other, thus research into both the filamentous as well as the spherical forms of influenza virus is necessary.
For filamentous virus, in addition to unknown methods of transmission and pathogenesis, the mechanism by which the filaments enter cells is unknown. Research into spherical virions has demonstrated that influenza virus enters cells by receptor-mediated endocytosis. HA, on the surface of the virus, binds to sialic acid moieties on surface exposed glycoproteins and glycolipids, triggering endocytosis of the virus. The mechanism by which influenza virus triggers endocytosis is unknown; however, recent data suggest that spherical virus particles binding to cells may trigger the activation of receptor tyrosine kinases, such as the epidermal growth factor receptor (Eierhoff et al., 2010), causing cellular signaling that results in the de novo formation of clathrin coated pits (Rust et al., 2004) and the subsequent uptake of influenza virions. For filamentous virions, however, entry may require a different mechanism as the viral filaments are too large to fit in a standard clathrin-coated pit. Studies on the entry mechanism of the filamentous Ebola virus have shown that the virus enters cells via macropinocytosis (Nanbo et al., 2010; Saeed et al., 2010). Thus, we speculate that filamentous influenza virions may also utilize macropinocytosis. This would provide an explanation for previous data that showed that influenza viruses can utilize, an as-yet-undefined, non-clathrin, non-caveolin entry pathway (Rust et al., 2004; Sieczkarski and Whittaker, 2002).
Nonetheless, it has been shown that the entry of both forms of influenza virus requires endosomal acidification (Sieczkarski and Whittaker, 2005). The low pH environment of the endosome is necessary, both for triggering HA-mediated fusion of the viral-endosomal membranes and for activating the M2 ion channel. The M2 proton-selective ion channel, on endosomal acidification, mediates proton conduction into the virion core, causing dissociation of the RNP core from the M1 protein, which allows for its subsequent import into the nucleus and the start of viral replication (reviewed in Helenius, 1992; Lamb and Pinto, 2005; Pinto and Lamb, 2006; Skehel, 1992). Upon replication of the viral genome and expression of the viral proteins, assembly begins at lipid raft domains of the apical plasma membrane, leading to the production of the next generation of influenza virions.
HA, Lipid Rafts and the Initiation of Virus Budding
Influenza virus, both spherical as well as filamentous forms, utilize lipid raft domains in the plasma membrane of infected cells as sites of virus assembly and budding (Chen et al., 2007; Chen et al., 2005; Leser and Lamb, 2005; Takeda et al., 2003). Lipid rafts are variably-sized, cholesterol and sphingolipid-enriched, regions of the plasma membrane (Brown and Rose, 1992). Lipid rafts concentrate proteins within defined regions of the plasma membrane thus serving as functional domains (reviewed in Lingwood and Simons, 2010). Alternatively, expression of raft-associated proteins, such as the influenza virus HA protein, can cause a coalescence of lipid raft domains, forming a ‘barge of rafts’ or the viral ‘budozone’ (Schmitt and Lamb, 2005). Lipid rafts facilitate the budding of several different viruses, including HIV-1, Ebola virus and influenza virus (Brown and Rose, 1992; Cary and Cooper, 2000; Simons and Ikonen, 1997; Simons and Toomre, 2000; Suomalainen, 2002). For influenza virus, HA and NA are intrinsically associated with lipid raft domains, whereas the M2 protein is excluded from these domains (Chen et al., 2007; Leser and Lamb, 2005; Rossman et al., 2010a; Takeda et al., 2003). HA is a homotrimeric glycoprotein containing a 529 residue ectodomain that mediates viral attachment and entry. The HA 27 residue transmembrane (TM) domain, in conjunction with three palmitoylated cysteine residues (one located in the TM domain and two in the 10 amino acid cytoplasmic tail), mediates lipid raft association (Chen et al., 2005; Scheiffele et al., 1997; Takeda et al., 2003; Zhang et al., 2000b). The lipid raft association of HA is essential for efficient viral replication, as mutations within the HA TM domain that eliminate raft association significantly attenuate viral replication (Chen et al., 2005). Additionally, recent data has shown that influenza virus infection induces the expression of the, interferon-inducible, anti-viral protein, viperin (Wang et al., 2007). Expression of viperin causes a destabilization and reduction in lipid raft domains, leading to attenuation of influenza virus replication (Wang et al., 2007). Thus, it is possible that in virus-infected cells targeting HA to lipid raft domains serves to concentrate HA, allowing for the initiation of virus budding and this is antagonized by viperin-mediated disruption of lipid raft.
The exact mechanism of virus bud initiation is not currently known. However, in a virus-like particle (VLP) system, wild-type (wt), raft-associated, HA protein buds from cells in vesicles that resemble virions, without requiring the expression of any other viral proteins (Chen et al., 2007). Thus, HA appears to possess the ability to alter membrane curvature, which in conjunction with lipid raft-mediated concentration effects may enable the initiation of virus budding. In the VLP system, plasmid-based expression of HA in 293T cells, leads to the alteration of membrane curvature, the completion of budding and the release of a virus-like particle. Similarly, single-protein expression of NA or M2 lead to the release of VLPs, albeit at lower levels than HA (Chen et al., 2007; Lai et al., 2010). Combining the expression of HA, NA, M2 and M1 significantly enhanced VLP release.
Interestingly, budding in virus-infected cells does not appear to directly mirror the effects of VLP budding (Nayak et al., 2009). The reasons for the discrepancy between VLP and virus budding is not clear; however, VLPs generated from transfected cells lack normal virus-regulation of protein synthesis, resulting in aberrant protein levels that may affect assembly and budding. Even when expressing all viral proteins, transfected 293T cells produce only spherical VLPs, while expression of the same viral proteins during virus-infection leads to predominantly filamentous virions (Chen et al., 2007). This alteration in virion morphology may be attributed to differential modification of virus proteins during infection as compared to transfection, differential interactions between viral and host proteins, or alterations in viral protein expression or localization. Additionally, whereas in the VLP system HA is able to mediate single-protein budding, during virus infection budding requires additional viral proteins, such as M2. Deletion of the M2 protein or mutation of the M2 cytoplasmic tail inhibits virus budding (Chen et al., 2008; Cheung et al., 2005; Iwatsuki-Horimoto et al., 2006; McCown and Pekosz, 2005; McCown and Pekosz, 2006; Rossman et al., 2010a). It is possible that during virus budding HA is able to modify membrane curvature and initiate budding, however, viral protein assembly, such as M1 binding to the HA cytoplasmic tail or the recruitment of the RNPs, may prevent HA from completing the budding event.
This disconnect, between the ability of HA to mediate single-protein budding and its dependence on additional proteins to mediate budding during virus infection, is similar to previous results seen in the budding of endosomes into the multi-vesicular body (MVB), as mediated by the host endosomal sorting complex required for transport (ESCRT) system (Wollert et al., 2009). In an artificial system mimicking MVB budding, the ESCRT-I and -II complexes are able to modify membrane curvature and provide a scaffold for the recruitment of cargo proteins; however, addition of the ESCRT-III complex is required to mediate membrane scission and the release of the budding vesicle (Wollert and Hurley, 2010; Wollert et al., 2009). Similarly, HA may initiate influenza virus budding, however, viral protein assembly may block the ability of HA to complete budding, necessitating the recruitment of other viral components.
The role of HA in budding initiation, but not completion, is supported by deletion and mutation studies. Previous work has shown that deletion or mutation of HA does not appreciably alter the number of virions that bud from the virus-infected cell, although the resulting particles are defective, as HA is necessary for viral entry (Chen et al., 2005; Pattnaik et al., 1986). This suggests that in the absence of HA, other viral proteins, such as NA, are able to initiate budding, compensating for the lack of HA-initiated budding in these viruses. Interestingly mutation of other virus proteins, such as M2, impairs the release of viral particles, although it does not prevent the formation of virus buds on the surface of the infected cell (Chen et al., 2008; Cheung et al., 2005; Iwatsuki-Horimoto et al., 2006; McCown and Pekosz, 2005; McCown and Pekosz, 2006; Rossman et al., 2010a; Rossman et al., 2010b). Thus, M2 may be necessary for the completion of virus budding, while not required for its initiation.
M1 and Virus Assembly
If HA is able to initiate, but not complete, virus budding then it is possible that M1 mediates both the restriction on HA budding as well as the recruitment of viral proteins necessary to complete budding. M1 is a 252 amino acid protein that contains three domains of bundled α-helices separated by short linker sequences, with the middle domain mediating oligomerization and association with the RNP (Arzt et al., 2001; Arzt et al., 2004; Elster et al., 1994; Harris et al., 2001; Noton et al., 2007; Sha and Luo, 1997; Shishkov et al., 1999; Wakefield and Brownlee, 1989). M1 has been postulated to crosslink the cytoplasmic tails of HA and NA, possibly mediating incorporation into the budding virion; however, the domains required for this interaction have not been determined and may span the entire protein (Noton et al., 2007; reviewed in Schmitt and Lamb, 2005). Mutational analysis of the cytoplasmic tails of HA and NA has suggested that binding to the cytoplasmic tails mediates the recruitment of M1 into the forming virion, although M1 appears capable of being recruited by either NA and HA, suggesting that there is some redundancy in the roles of HA and NA during virus budding (Ali et al., 2000; Barman et al., 2004; Jin et al., 1997; Zhang et al., 2000a). Interestingly, the virions that result from cytoplasmic tail mutations in both HA and NA display greatly altered morphology along with a significant reduction in M1 and RNP incorporation (Jin et al., 1997; Zhang et al., 2000a).
In the virion, M1 interacts with both the plasma membrane and with NP in the form of the RNP (Bui et al., 1996; Noton et al., 2007; Zhang and Lamb, 1996). This is supported by recent cryo-electron tomography data that indicates M1 provides structure and support for the viral membrane. M1 was seen to form a helical net under the viral membrane, with regularly spaced holes that are possible places for the insertion of, and interaction with, the cytoplasmic tails of HA and NA (Calder et al., 2010). Thus, reduction in M1 incorporation may impair RNP recruitment and alter virion structure.
Further evidence for the role of M1 in virion structure comes from work investigating the genetic requirements of virus filament formation, where it has been shown that specific sequence variants of the M1 protein are able to confer the ability to form filamentous virions (Bourmakina and Garcia-Sastre, 2003; Burleigh et al., 2005; Elleman and Barclay, 2004; Roberts et al., 1998). Recent work has also shown that the pitch of the helical turn of the M1 protein is different between spherical and filamentous strains (Calder et al., 2010), further suggesting that the structure of the M1 protein determines the ability of the virus to form filamentous or spherical virions. It is of note that M1 within the cell is soluble and monomeric (Ruigrok et al., 2001). It has been suggested that binding to the cytoplasmic tails of HA and NA allows for M1 to associate with lipid raft membranes, triggering a conformational change that enables M1 polymerization at the site of virus budding (Gomez-Puertas et al., 2000; Ruigrok et al., 2001). Thus, M1 polymerization at sites of budding may provide the mechanism for the elongation of filamentous virions.
Interestingly, it has been reported previously that, in a vaccinia virus (Gomez-Puertas et al., 2000), or baculovirus (Latham and Galarza, 2001) based VLP system, M1 is capable of mediating single-protein VLP budding of filamentous VLPs. This ability of M1 to mediate VLP budding was shown to be dependent on the virus expression system and cannot be replicated via transfection based expression (Chen et al., 2007). This implies that the ability of M1 to mediate VLP budding is dependent on the expression of an unknown viral protein. It has also been suggested that M1 can mediate VLP budding if the membrane association of M1 is induced (Wang et al., 2010), although as seen with HA VLP budding (Chen et al., 2007), the relevance of M1 VLP budding to actual virus budding has not been determined. Additionally, it is not clear if RNP binding to M1 has any effect on the ability of M1 to cause budding. Two studies using cryo-electron microscopy have shown that the viral RNPs localize to one end of filamentous virions (Calder et al., 2010; Noda et al., 2006). We speculate that RNP binding to M1, which in turn is associated with HA and NA in lipid raft domains, may provide a trigger that causes the initiation of budding or perhaps causes M1 polymerization, leading to the elongation of the budding virion. However, no mechanistic role for NP in budding has been determined.
Due to its helical structure and ability to bind to membranes, M1 may possess an intrinsic ability to alter membrane curvature and cause membrane budding. It is possible that during virus budding, association of M1 with the cytoplasmic tails of HA and NA recruits M1 to the membrane, where the ability of M1 to alter membrane curvature combines with the HA-mediated initiation of virus budding to increase the efficiency of virus budding. The combination of HA and M1 in a plasmid-based VLP system enhanced the ability of HA to mediate VLP release; however, the greatest VLP release occurred with the further addition of M2 (Chen et al., 2007). Thus, the assembly of M1 at sites of budding, along with other viral proteins such as NP, may block M1 mediation of budding in a manner that is analogous to that of HA. As discussed below completion of virus budding requires an additional protein, M2, to mediate membrane scission and virion release.
M2 and Membrane Scission
In conjunction with interactions with HA, NA, NP and the plasma membrane, M1 also binds to the M2 ion channel protein. M2 is a 97 amino acid protein that associates in the membrane as a homotetramer (Holsinger and Lamb, 1991; Sugrue and Hay, 1991). The 19 amino acid TM domain forms the pore of the ion channel (Lamb et al., 1985; Pinto et al., 1997; Pinto et al., 1992; Zebedee et al., 1985), while the first 17 residues of the cytoplasmic tail form a, membrane-parallel, amphipathic helix (Holsinger and Lamb, 1991; Lamb et al., 1985; Nguyen et al., 2008; Pinto et al., 1992; Rossman et al., 2010a; Schnell and Chou, 2008; Tian et al., 2003; Zebedee et al., 1985). Recent work has suggested that the M2 cytoplasmic tail, and specifically its amphipathic helix residues 46–62, may play an important role in virus assembly and budding (Chen et al., 2008; Iwatsuki-Horimoto et al., 2006; McCown and Pekosz, 2005; McCown and Pekosz, 2006; Rossman et al., 2010a; Rossman et al., 2010b).
Whereas earlier studies mapped the genetic requirement for filamentous virion formation to the M1 protein (Bourmakina and Garcia-Sastre, 2003; Burleigh et al., 2005; Elleman and Barclay, 2004; Roberts et al., 1998), other work has shown that M2 may modify the ability of M1 to permit filament formation (Hughey et al., 1992; Hughey et al., 1995; Iwatsuki-Horimoto et al., 2006; McCown and Pekosz, 2006; Roberts et al., 1998; Rossman et al., 2010a; Zebedee and Lamb, 1988; Zebedee and Lamb, 1989). Truncation of the A/Udorn/72 M2 protein at residue 70, or mutation of the amphipathic helix, leads to a loss of filaments on the surface of influenza virus-infected cells (McCown and Pekosz, 2006; Rossman et al., 2010a). When M2 protein was reconstituted into large unilamellar vesicles containing high levels of cholesterol, which resemble lipidic virus budding domains, the M2 amphipathic helix induced negative membrane curvature (Rossman et al., 2010b). At sites of virus budding, this induction of negative curvature may stabilize the site of budding long enough to allow for filament polymerization (Rossman et al., 2010a; Rossman et al., 2010b). It has been speculated that the cytoplasmic tail of M2, between resides 70–77, plays an important role in binding to M1 (Chen et al., 2008; Grantham et al., 2010; McCown and Pekosz, 2006). Thus, mutation of M2, such that it can no longer bind to M1, may impair filament formation by preventing the M2-mediated stabilization of the virus budding site.
The M2 cytoplasmic tail was also shown to be important for the efficient production of infectious viral particles, as mutations between residues 70–77 impaired vRNP incorporation into budding virions (Grantham et al., 2010; McCown and Pekosz, 2005; McCown and Pekosz, 2006). However, it is not known if the cytoplasmic tail of M2 binds NP directly or if alterations in the M2-M1 interaction are responsible for the reduction in vRNP packaging. Additional evidence for the interaction between M2 and M1 comes from the observation that treatment of influenza virus-infected cells with the M2 ectodomain-specific monoclonal antibody (mAb) 14C2 causes a loss of filament formation (Hughey et al., 1995; Roberts et al., 1998) and causes a growth restriction (Zebedee and Lamb, 1988). Virus mutants that escape antibody growth restriction contain mutations in M1 or in the M2 cytoplasmic tail (Hughey et al., 1992; Zebedee and Lamb, 1988). Interestingly, it has been reported recently that M2 may also associate directly with HA in an M1-independent manner (Thaa et al., 2010). Thus, the recruitment of M2 to sites of virus budding may involve associations with both M1 and HA.
In addition to filament formation and vRNP recruitment, the interaction between M1 and M2, or M2 and HA, may play another important role during virus budding, the mediation of membrane scission. While HA, and possibly M1, are capable of altering membrane curvature and initiating the budding event, membrane scission and the release of the budding virion requires the M2 protein (Rossman et al., 2010b). Viruses such as HIV-1, Ebola virus and the paramyxovirus PIV-5 utilize the host ESCRT complex to facilitate membrane scission and to complete the budding process (reviewed in Carlton and Martin-Serrano, 2009; Chen and Lamb, 2008; Welsch et al., 2007). Late domain sequences, frequently found in viral matrix proteins, mediate recruitment of ESCRT-I or ESCRT-II complex proteins to the budding virus. It was reported that the influenza virus M1 protein sequence YRKL (residues 100–103) fulfilled the criteria of a late domain, including position independence of the sequence and the ability to replace the sequence with a know late domain sequence, PTAP (Hui et al., 2003). It was also reported that this influenza virus-specific late domain interacted with the ESCRT-I complex protein, vacuolar protein sorting-associated protein (VPS) 28 (Hui et al., 2006a). Disruption of the late domain sequence in M1 or siRNA knock down of VPS28 significantly attenuated virus budding (Hui et al., 2006a); however, these results were unable to be replicated and have since been retracted (Hui et al., 2006b). Further research has shown that whereas M1 does appear to interact with VPS28, influenza virus budding is not dependent on the host ESCRT complex, thus necessitating an alternate means of membrane scission (Bruce et al., 2009; Chen and Lamb, 2008; Watanabe and Lamb, 2010).
The host ESCRT-III complex mediates membrane scission by assembling a spiral of the protein Snf7 around the neck of a budding vesicle, progressively constricting the budding neck and forcing membrane scission (Wollert et al., 2009). Recent work from our laboratory indicates that influenza virus utilizes the amphipathic helix of the M2 protein to alter membrane curvature at the budding neck of the virus (Rossman et al., 2010b). Modification of membrane curvature by proteins that contain amphipathic helixes is an established phenomenon (Donaldson, 2008; Epand et al., 1995; Gouttenoire et al., 2009; Horvath et al., 2007; Min et al., 2008; Saarikangas et al., 2009; Schwartz et al., 2004). Interestingly, the M2 protein appears to modify membrane curvature in a cholesterol-dependant manner (Rossman et al., 2010b).
The M2 cytoplasmic tail contains two partial, overlapping, cholesterol recognition amino acid consensus (CRAC) motifs and the ability of M2 to bind cholesterol has been demonstrated (Rossman et al., 2010a; Schroeder et al., 2005). However, when expressed in the absence of other viral proteins, M2 is localized to non-raft areas of the apical plasma membrane (Leser and Lamb, 2005; Zhang et al., 2000b). It is speculated that binding to M1 may recruit M2 to the, lipid raft-enriched, sites of virus budding where M2 would then be able to bind cholesterol (Chen et al., 2008; Rossman et al., 2010a). This cholesterol binding may stabilize the association of M2 at the site of virus budding, allowing for the incorporation of M2 into the budding virion. Additionally, M2-cholesterol association may allow for M2 to alter membrane curvature at the site of virus budding (Rossman et al., 2010b). However, ablation of the CRAC domain only causes minor decreases in pathogenicity of the mutant influenza viruses in mice. Thus, the significance of cholesterol binding is not known (Stewart et al., 2010). The M2 cytoplasmic tail also contains a caveolin-1 binding domain (CBD) and binding to caveolin-1 has been demonstrated both in vitro and in virus-infected cells (Sun et al., 2010; Zou et al., 2009); thus, caveolin-1 binding may further support M2 localization at the, lipid raft-enriched, sites of virus budding.
Experiments utilizing large and giant unilamellar vesicles have shown that in a low cholesterol environment, equivalent to the bulk phase of the plasma membrane, reconstituted M2 appears to cause positive membrane curvature, while in the presence of high levels of cholesterol, such as those found in the lipid raft-enriched sites of virus budding, M2 mediates negative membrane curvature (Rossman et al., 2010b). Thus, the M1-mediated recruitment of M2 to sites of virus budding may trigger the induction of negative membrane curvature which may stabilize the HA and M1 induction of positive membrane curvature caused during budding initiation. This stabilization may allow for virus filament formation (Rossman et al., 2010a) and may prolong the budding event, allowing for the proper recruitment of the remaining viral proteins and the virus genome. Interestingly, mutations of M2 that reduce the interaction with M1 also reduce the incorporation of NP into the budding virion (Chen et al., 2008), possibly because M2 is not being recruited to lipid raft domains and is no longer stabilizing the site of virus budding.
In a virus-infected cell, M2 localizes to the boundary between raft and bulk plasma membrane domains surrounding the sites of virus budding (Rossman et al., 2010b). As the virus buds from ‘barges of lipid rafts’ (the budozone) on the surface of infected cells, the remaining patch of raft membrane will eventually be reduced as they are incorporated into the budding virion. The non-raft localized M2 is under represented in virions as compared to its high level of expression in cells (Lamb et al., 1985; Leser and Lamb, 2005; Zebedee and Lamb, 1988). During budding, perhaps due to its concentration at the lipid phase boundary, M2 will eventually be localized to the neck of the budding virion, at the boundary between the lipid raft-rich virion and the remaining bulk plasma membrane phase (Rossman et al., 2010b; Schroeder et al., 2005). At this point, M2 in the lower-cholesterol environment of the plasma membrane, would cause positive membrane curvature (Rossman et al., 2010b). Positive membrane curvature at the neck of the budding virion would be sufficient to cause membrane scission, possibly through modification of the line tension between the two lipid phases, resulting in the release of the budding virus. Although it is not possible to directly determine the number of M2 tetramers required to mediate membrane scission or the number of tetramers found at the neck of the budding virus, it has been estimated that by 6 hours post-infection there are 3.33 × 107 molecules of M2 found on the cell surface (Lamb et al., 1985), which is similar to the 5 × 107 molecules of HA found on the cell surface (Gething and Sambrook, 1981). As there is extensive colocalization between M2 and HA at sites of virus budding (Rossman et al., 2010a), the majority of the 3.33 × 107 molecules of M2 will be at, or near, the neck of the budding virus; thus likely providing a concentration of M2 tetramers that is sufficient to mediate membrane scission.
An alternative model of membrane scission and virus release has been proposed recently. The release of budding influenza virions was observed to be attenuated when the expression of the small GTP-binding protein Rab11 was reduced (Bruce et al., 2010). Rab11, and its interacting proteins, were also shown recently to be involved in the release of budding respiratory syncytial virus, and thus was considered to reflect an alternative means for membrane scission and virion release, though the mechanism for scission has not been determined (Brock et al., 2003; Utley et al., 2008). However, Rab11 is also known to be involved in the trafficking of proteins to the apical membrane in polarized cells (reviewed in Jing and Prekeris, 2009), and siRNA knockdown of Rab11 expression reduced the levels of M2 found on the apical membrane during influenza virus infection (Rossman et al., 2010b). Further studies are necessary to determine if Rab11 plays an active role in membrane scission during influenza virus infection or if Rab11 is essential for the proper transport of M2 which then mediates membrane scission and virion release.
Recent studies further support the idea of M2-mediated membrane scission. Mutation or deletion of M2 significantly attenuates the production of infectious virions as well as the release of virus particles (Chen et al., 2008; Cheung et al., 2005; Iwatsuki-Horimoto et al., 2006; McCown and Pekosz, 2005; McCown and Pekosz, 2006; Rossman et al., 2010a). Examination of cells infected with mutant viruses that are deleted for the M2 protein, or contain truncations of the M2 cytoplasmic tail, show that although virus budding begins virus release is blocked. The resulting incompletely budded virions posses a ‘beads on a string’ morphology that is also found in ESCRT-dependent viruses that are unable to undergo membrane scission due to inhibition of the ESCRT system (Yuan et al., 2000). It appears that both spherical as well as filamentous strains of influenza virus require the M2 protein for membrane scission, as mutation of M2 in a spherical strain also results in a scission-defective morphology (Iwatsuki-Horimoto et al., 2006; McCown and Pekosz, 2006). Interestingly, it was shown recently that cholesterol extraction of virus-infected cells by treatment with methyl-β-cyclodextrin (MβCD) caused a rapid release of viral particles and a reduction in their infectivity (Barman and Nayak, 2007). MβCD treatment would eliminate the M2-cholesterol association, allowing for M2 to induce positive membrane curvature, causing a release of budding virions (Rossman et al., 2010b). This premature triggering of membrane scission could cause virion release prior to the recruitment and organization of all required viral proteins and the RNPs, possibly explaining the reduced infectivity of the released virions (Barman and Nayak, 2007). Given the high degree of conservation of the M2 protein and the observation that amphipathic helices from several different strains of influenza virus can alter membrane curvature, it appears that mediation of membrane scission is a highly conserved function of the M2 protein (Rossman et al., 2010b), a function that is essential for the final stage of virus budding, virion release.
Model of Influenza Virus Budding
The mechanism of influenza virus budding and assembly has been investigated for many years. However, it has been difficult to generate a model of the mechanistic progressing of virus budding. Perhaps one of the difficulties is the disconnect between VLP budding and virus budding. In many other model systems, VLP budding is thought to portray accurately the basic mechanism of virus budding. However, for influenza virus, there appear to be many inconsistencies between virus and VLP budding, such as the observation that influenza virus forms filamentous virions, while the VLPs are spherical (Chen et al., 2007). Single protein expression experiments show that HA (Chen et al., 2007), NA (Lai et al., 2010), M2 (Chen et al., 2007; Rossman et al., 2010b) and membrane-targeted M1 (Gomez-Puertas et al., 2000; Wang et al., 2010) are all capable of causing VLP budding. This raises the question as to the nature of the driving force for influenza virus budding? The answer may not be as simple as with other viruses, rather multiple influenza virus proteins may all provide a driving force for budding. It is possible that the ability of multiple proteins to mediate budding may provide a level of redundancy, allowing the virus to survive loss of function in individual proteins to mediate budding. Alternatively, the spatial-temporal organization of viral protein assembly may allow each protein to provide its additive effect on budding in a sequential manner driving budding in a defined, organized fashion.
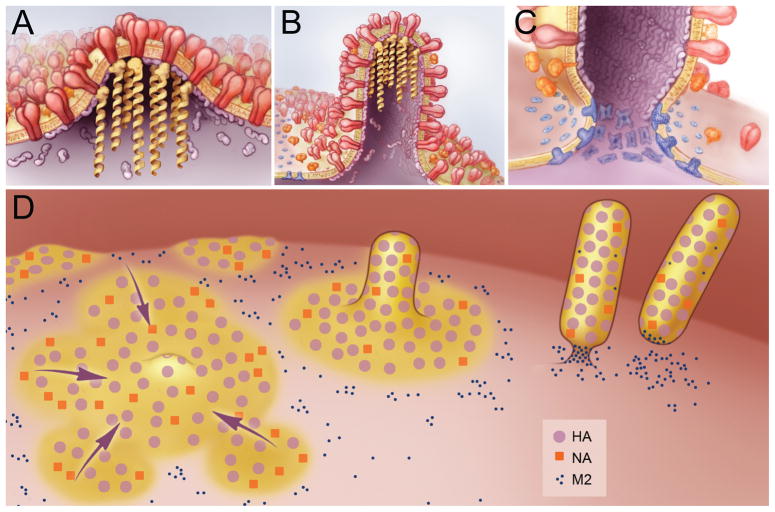
Between the different viral proteins capable of mediating budding, only HA and NA appear capable of initiating the budding event. M1 is unable to cause budding without membrane targeting provided by HA, NA or M2 (Gomez-Puertas et al., 2000; Wang et al., 2010) and M2 would be unable to initiate budding as it is delayed in expression in the infectious cycle as compared to HA, NA and M1 (Hughey et al., 1992; Valcarcel et al., 1991). This leaves HA and NA to initiate the budding event. As HA and NA are both targeted to lipid raft domains and both proteins appear to be able to alter membrane curvature causing budding, the concentration of HA and NA in lipid rafts may serve to provide a local alteration in membrane curvature that starts the budding process (Fig. 1a, d). The cytoplasmic tails of HA and NA may then serve as docking sites for M1 (Fig. 1a, b). Membrane recruitment of M1, through HA/NA cytoplasmic tail binding, may enable its polymerization, facilitating the formation of filamentous virions (Fig. 1b, d). Additionally, membrane-bound M1 may then mediate alterations in membrane curvature, further progressing the budding event. Alternatively, the binding of M1 to HA/NA cytoplasmic tails may block their ability to alter membrane curvature, necessitating the addition of M1-mediated alternations in membrane curvature to continue the budding process. Recruitment of NP and the RNPs to M1, at the site of budding, may mask the ability of M1 to alter membrane curvature, preventing the conclusion of budding as would occur during VLP budding (Fig. 1a). This block in budding may necessitate M2-mediated membrane scission for completion.
Figure 1.
Model of Influenza Virus Budding. A) The initiation of virus budding caused by clustering of HA (shown in red) and NA (shown in orange) in lipid raft domains. M1 (shown in purple) is seen binding to the cytoplasmic tails of HA and NA and serves as a docking site for the vRNPs (shown in yellow). B) Elongation of the budding virion caused by polymerization of the M1 protein, resulting in a polarized localization of the vRNPs. M2 (shown in blue) is recruited to the periphery of the budding virus though interactions with M1. C) Membrane scission caused by the insertion of the M2 amphipathic helix at the lipid phase boundary, altering membrane curvature at the neck of the budding virus and leading to release of the budding virus. D) Overview of the budding of influenza viruses, showing the coalescence of HA and NA containing lipid rafts (shown in yellow), the formation of a filamentous virion and membrane scission caused by M2 clustered at the neck of the budding virus.
The delayed expression of M2 (Zebedee et al., 1985) may allow for the initiation of budding, and for proper virion assembly, before the mechanism of scission and virion release is provided. Furthermore, the recruitment of M2 to sites of budding, by M1, puts M2 in a cholesterol-rich environment, where M2 stabilizes the site of budding instead of causing membrane scission (Fig. 1b). This would allow for proper assembly of the budding virion before M2 is localized to the neck of the budding virion, placing it at the boundary between the lipid raft-enriched virion and the bulk plasma membrane phases (Fig. 1c, d). At the budding neck, M2 may exert its own positive membrane curvature by inserting its amphipathic helix into the membrane and modifying the line tension between the two lipid phases. This further alteration of membrane curvature may provide the final force needed to mediate membrane scission and the release of the budding virion (Fig. 1c, d).
Following the completion of membrane scission, the virion may still be tethered to the cell membrane due to the interaction of virion-associated HA and cell-surface sialic acid moieties. NA is then able to play the final role in virus budding, cleaving sialic acid off the cell surface, preventing the HA-receptor interaction and freeing the budded virion. Interestingly, recent cryo-electron tomography experiments have shown that NA is concentrated at one location on budded virions, which may reflect the role of NA in mediating the final release of virions from the cell surface (Calder et al., 2010).
Influenza virus budding requires a tightly organized assembly of multiple different viral proteins, with many proteins providing a level of redundancy, helping to ensure successful budding. The activities of HA, NA, M1 and M2 may serve to sequentially modify membrane curvature, with each protein furthering the process of virus assembly and budding. Thus, for influenza virus, budding is not a solo performance, but a carefully orchestrated symphony with each component functioning at precisely defined points in space and time. By better understanding this orchestration it may be possible to alter these relationships, causing a failure of budding and inhibiting the replication of influenza viruses.
Acknowledgments
We thank Andrew S. Pekosz for useful discussions. Work in the authors’ laboratory was supported by a grant from the National Institutes of Health, R01 AI-20201. J.S.R. is an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ada GL, Perry BT, Abbot A. Biological and physical properties of the Ryan strain of filamentous influenza virus. J Gen Microbiol. 1958;19:23–39. doi: 10.1099/00221287-19-1-23. [DOI] [PubMed] [Google Scholar]
- Ali A, Avalos RT, Ponimaskin E, Nayak DP. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J Virol. 2000;74:8709–8719. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzt S, Baudin F, Barge A, Timmins P, Burmeister WP, Ruigrok RW. Combined results from solution studies on intact influenza virus M1 protein and from a new crystal form of its N-terminal domain show that M1 is an elongated monomer. Virology. 2001;279:439–446. doi: 10.1006/viro.2000.0727. [DOI] [PubMed] [Google Scholar]
- Arzt S, Petit I, Burmeister WP, Ruigrok RW, Baudin F. Structure of a knockout mutant of influenza virus M1 protein that has altered activities in membrane binding, oligomerisation and binding to NEP (NS2) Virus Res. 2004;99:115–119. doi: 10.1016/j.virusres.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Barman S, Adhikary L, Chakrabarti AK, Bernas C, Kawaoka Y, Nayak DP. Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza a virus neuraminidase in raft association and virus budding. J Virol. 2004;78:5258–5269. doi: 10.1128/JVI.78.10.5258-5269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S, Nayak DP. Lipid raft disruption by cholesterol depletion enhances influenza A virus budding from MDCK cells. J Virol. 2007;81:12169–12178. doi: 10.1128/JVI.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourmakina SV, Garcia-Sastre A. Reverse genetics studies on the filamentous morphology of influenza A virus. J Gen Virol. 2003;84:517–527. doi: 10.1099/vir.0.18803-0. [DOI] [PubMed] [Google Scholar]
- Brock SC, Goldenring JR, Crowe JE., Jr Apical recycling systems regulate directional budding of respiratory syncytial virus from polarized epithelial cells. Proc Natl Acad Sci USA. 2003;100:15143–15148. doi: 10.1073/pnas.2434327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- Bruce EA, Digard P, Stuart AD. The Rab11 pathway is required for influenza a virus budding and filament formation. J Virol. 2010;84:5848–5859. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Medcalf L, Crump CM, Noton SL, Stuart AD, Wise HM, Elton D, Bowers K, Digard P. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology. 2009;390:268–278. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh LM, Calder LJ, Skehel JJ, Steinhauer DA. Influenza a viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes. J Virol. 2005;79:1262–1270. doi: 10.1128/JVI.79.2.1262-1270.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnet FM, Lind PE. Studies on filamentary forms of influenza virus with special reference to the use of dark-ground-microscopy. Arch Gesamte Virusforsch. 1957;7:413–422. doi: 10.1007/BF01241959. [DOI] [PubMed] [Google Scholar]
- C.D.C.; CfD Control. Emerging Infections Program. Atlanta, GA: 2009. CDC estimates of 2009 H1N1 cases and related hospitalizations and deaths from April–October 17, 2009, by age group. [Google Scholar]
- Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci USA. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- Cary LA, Cooper JA. Molecular switches in lipid rafts. Nature. 2000;404:945–947. doi: 10.1038/35010257. [DOI] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol. 2008;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Takeda M, Lamb RA. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J Virol. 2005;79:13673–13684. doi: 10.1128/JVI.79.21.13673-13684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TK, Guan Y, Ng SS, Chen H, Wong CH, Peiris JS, Poon LL. Generation of recombinant influenza A virus without M2 ion-channel protein by introduction of a point mutation at the 5′ end of the viral intron. J Gen Virol. 2005;86:1447–1454. doi: 10.1099/vir.0.80727-0. [DOI] [PubMed] [Google Scholar]
- Choppin PW. Multiplication of two kinds of influenza A2 virus particles in monkey kidney cells. Virology. 1963;21:342–352. doi: 10.1016/0042-6822(63)90195-8. [DOI] [PubMed] [Google Scholar]
- Choppin PW, Murphy JS, Stoeckenius W. The surface structure of influenza virus filaments. Virology. 1961;13:548–550. doi: 10.1016/0042-6822(61)90287-2. [DOI] [PubMed] [Google Scholar]
- Choppin PW, Murphy JS, Tamm I. Studies of two kinds of virus particles which comprise influenza A2 virus strains. III Morphological characteristics: Independence of morphological and functional traits. J Exp Med. 1960;112:945–952. doi: 10.1084/jem.112.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CM, Dawson IM, Elford WJ. Filamentous forms associated with newly isolated influenza virus. Lancet. 1949;1:602. doi: 10.1016/s0140-6736(49)91699-2. [DOI] [PubMed] [Google Scholar]
- Donaldson JG. Arfs and membrane lipids: sensing, generating and responding to membrane curvature. Biochem J. 2008;414:e1–2. doi: 10.1042/BJ20081438. [DOI] [PubMed] [Google Scholar]
- Eierhoff T, Hrincius ER, Rescher U, Ludwig S, Ehrhardt C. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 2010;6:e1001099. doi: 10.1371/journal.ppat.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman CJ, Barclay WS. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology. 2004;321:144–153. doi: 10.1016/j.virol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Elster C, Fourest E, Baudin F, Larsen K, Cusack S, Ruigrok RWH. A small percentage of influenza virus M1 protein contains zinc but zinc does not influence in vitro M1-RNA interaction. J Gen Virol. 1994;75:37–42. doi: 10.1099/0022-1317-75-1-37. [DOI] [PubMed] [Google Scholar]
- Epand RM, Shai Y, Segrest JP, Anantharamaiah GM. Mechanisms for the modulation of membrane bilayer properties by amphipathic helical peptides. Biopolymers. 1995;37:319–338. doi: 10.1002/bip.360370504. [DOI] [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Cell-surface expression of influenza haemagglutinin from a cloned DNA copy of the RNA gene. Nature. 1981;293:620–625. doi: 10.1038/293620a0. [DOI] [PubMed] [Google Scholar]
- Gomez-Puertas P, Albo C, Perez-Pastrana E, Vivo A, Portela A. Influenza virus matrix protein is the major driving force in virus budding. J Virol. 2000;74:11538–11547. doi: 10.1128/jvi.74.24.11538-11547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouttenoire J, Castet V, Montserret R, Arora N, Raussens V, Ruysschaert JM, Diesis E, Blum HE, Penin F, Moradpour D. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J Virol. 2009;83:6257–6268. doi: 10.1128/JVI.02663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham ML, Stewart SM, Lalime EN, Pekosz A. Tyrosines in the influenza A virus M2 protein cytoplasmic tail are critical for production of infectious virus particles. J Virol. 2010;84:8765–8776. doi: 10.1128/JVI.00853-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Forouhar F, Qiu S, Sha B, Luo M. The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology. 2001;289:34–44. doi: 10.1006/viro.2001.1119. [DOI] [PubMed] [Google Scholar]
- Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- Holsinger LJ, Lamb RA. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- Horvath CA, Vanden Broeck D, Boulet GA, Bogers J, De Wolf MJ. Epsin: inducing membrane curvature. Int J Biochem Cell Biol. 2007;39:1765–1770. doi: 10.1016/j.biocel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Hughey PG, Compans RW, Zebedee SL, Lamb RA. Expression of the influenza A virus M2 protein is restricted to apical surfaces of polarized epithelial cells. J Virol. 1992;66:5542–5552. doi: 10.1128/jvi.66.9.5542-5552.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey PG, Roberts PC, Holsinger LJ, Zebedee SL, Lamb RA, Compans RW. Effects of antibody to the influenza A virus M2 protein on M2 surface expression and virus assembly. Virology. 1995;212:411–421. doi: 10.1006/viro.1995.1498. [DOI] [PubMed] [Google Scholar]
- Hui EK, Barman S, Tang DH, France B, Nayak DP. YRKL sequence of influenza virus M1 functions as the L domain motif and interacts with VPS28 and Cdc42. J Virol. 2006a;80:2291–2308. doi: 10.1128/JVI.80.5.2291-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hui EK, Barman S, Yang TY, Nayak DP. Basic residues of the helix six domain of influenza virus M1 involved in nuclear translocation of M1 can be replaced by PTAP and YPDL late assembly domain motifs. J Virol. 2003;77:7078–7092. doi: 10.1128/JVI.77.12.7078-7092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hui EK, Barman S, Yang TY, Tang DH, France B, Nayak DP. Retraction: “Basic residues of the helix six domain of influenza virus M1 involved in nuclear translocation of M1 can be replaced by PTAP and YPDL late assembly domain motifs” and “YRKL sequence of influenza virus M1 functions as the L domain motif and interacts with VPS28 and Cdc42”. J Virol. 2006b;80:10289. [Google Scholar]
- Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, Ogasawara K, Kawaoka Y. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki-Horimoto K, Horimoto T, Noda T, Kiso M, Maeda J, Watanabe S, Muramoto Y, Fujii K, Kawaoka Y. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J Virol. 2006;80:5233–5240. doi: 10.1128/JVI.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Leser GP, Zhang J, Lamb RA. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J, Prekeris R. Polarized endocytic transport: the roles of Rab11 and Rab11-FIPs in regulating cell polarity. Histol Histopathol. 2009;24:1171–1180. doi: 10.14670/hh-24.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbourne ED, Murphy JS. Genetic studies of influenza viruses. I Viral morphology and growth capacity as exchangeable genetic traits Rapid in ovo adaptation of early passage asian strain isolates by combination with PR8. J Exp Med. 1960;111:387–406. doi: 10.1084/jem.111.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JC, Chan WW, Kien F, Nicholls JM, Peiris JS, Garcia JM. Formation of virus-like particles from human cell lines exclusively expressing Influenza neuraminidase. J Gen Virol. 2010;91:2322–2330. doi: 10.1099/vir.0.019935-0. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Pinto LH. The proton selective ion channels of influenza A and B viruses. In: Kawaoka Y, editor. Contemporary Topics in Influenza Virology. Horizon Scientific Press; Wymondham, Norfolk, UK: 2005. pp. 65–93. [Google Scholar]
- Lamb RA, Zebedee SL, Richardson CD. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- Latham T, Galarza JM. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J Virol. 2001;75:6154–6165. doi: 10.1128/JVI.75.13.6154-6165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- McCown MF, Pekosz A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J Virol. 2005;79:3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol. 2006;80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min CK, Bang SY, Cho BA, Choi YH, Yang JS, Lee SH, Seong SY, Kim KW, Kim S, Jung JU, Choi MS, Kim IS, Cho NH. Role of amphipathic helix of a herpesviral protein in membrane deformation and T cell receptor downregulation. PLoS Pathog. 2008;4:e1000209. doi: 10.1371/journal.ppat.1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Rose HM, Moore DH. Structure and development of viruses observed in the electron microscope III. Influenza virus. J Exp Med. 1956;104:171–182. doi: 10.1084/jem.104.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima N, Hata S, Sato Y, Tobiume M, Katano H, Kaneko K, Nagata N, Kataoka M, Ainai A, Hasegawa H, Tashiro M, Kuroda M, Odai T, Urasawa N, Ogino T, Hanaoka H, Watanabe M, Sata T. The first autopsy case of pandemic influenza (A/H1N1pdm) virus infection in Japan: detection of a high copy number of the virus in type II alveolar epithelial cells by pathological and virological examination. Jpn J Infect Dis. 2010;63:67–71. [PubMed] [Google Scholar]
- Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PA, Soto CS, Polishchuk A, Caputo GA, Tatko CD, Ma C, Ohigashi Y, Pinto LH, DeGrado WF, Howard KP. pH-induced conformational change of the influenza M2 protein C-terminal domain. Biochemistry. 2008;47:9934–9936. doi: 10.1021/bi801315m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J Gen Virol. 2007;88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Shaw ML. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- Pattnaik AK, Brown DJ, Nayak DP. Formation of influenza virus particles lacking hemagglutinin on the viral envelope. J Virol. 1986;60:994–1001. doi: 10.1128/jvi.60.3.994-1001.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- Roberts PC, Lamb RA, Compans RW. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology. 1998;240:127–137. doi: 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. Influenza virus M2 ion channel protein is cecessary for filamentous virion formation. J Virol. 2010a;84:5078–5088. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Lamb RA. The influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010b;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok R, Baudin F, Petit I, Weissenhorn W. Role of influenza virus M1 protein in the viral budding process. Int Congress Series. 2001;1219:397–404. [Google Scholar]
- Rust MJ, Lakadamyali M, Zhang F, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 2004;11:567–573. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Roth MG, Simons K. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 1997;16:5501–5508. doi: 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, Lamb RA. Influenza virus assembly and budding at the viral budozone. Adv Virus Res. 2005;64:383–416. doi: 10.1016/S0065-3527(05)64012-2. [DOI] [PubMed] [Google Scholar]
- Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C, Heider H, Moncke-Buchner E, Lin TI. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur Biophys J. 2005;34:52–66. doi: 10.1007/s00249-004-0424-1. [DOI] [PubMed] [Google Scholar]
- Schwartz M, Chen J, Lee WM, Janda M, Ahlquist P. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc Natl Acad Sci USA. 2004;101:11263–11268. doi: 10.1073/pnas.0404157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha B, Luo M. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat Struct Biol. 1997;4:239–244. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- Shishkov AV, Goldanskii VI, Baratova LA, Fedorova NV, Ksenofontov AL, Zhirnov OP, Galkin AV. The in situ spatial arrangement of the influenza A virus matrix protein M1 assessed by tritium bombardment. Proc Natl Acad Sci USA. 1999;96:7827–7830. doi: 10.1073/pnas.96.14.7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieczkarski SB, Whittaker GR. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J Virol. 2002;76:10455–10464. doi: 10.1128/JVI.76.20.10455-10464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieczkarski SB, Whittaker GR. Characterization of the host cell entry of filamentous influenza virus. Arch Virol. 2005;150:1783–1796. doi: 10.1007/s00705-005-0558-1. [DOI] [PubMed] [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Skehel JJ. Influenza virus. Amantadine blocks the channel. Nature. 1992;358:110–111. doi: 10.1038/358110b0. [DOI] [PubMed] [Google Scholar]
- Stewart SM, Wu WH, Lalime EN, Pekosz A. The cholesterol recognition/interaction amino acid consensus motif of the influenza A virus M2 protein is not required for virus replication but contributes to virulence. Virology. 2010;405:530–538. doi: 10.1016/j.virol.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue RJ, Hay AJ. Structural characteristics of the M2 protein of the influenza A viruses: Evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Hemgard GV, Susanto SA, Wirth M. Caveolin-1 influences human influenza A virus (H1N1) multiplication in cell culture. Virol J. 2010;7:108. doi: 10.1186/1743-422X-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M. Lipid rafts and assembly of enveloped viruses. Traffic. 2002;3:705–709. doi: 10.1034/j.1600-0854.2002.31002.x. [DOI] [PubMed] [Google Scholar]
- Takeda M, Leser GP, Russell CJ, Lamb RA. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc Natl Acad Sci USA. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaa B, Herrmann A, Veit M. Intrinsic Cytoskeleton-Dependent Clustering of Influenza Virus M2 Protein with Hemagglutinin Assessed by FLIM-FRET. J Virol. 2010;84:12445–12449. doi: 10.1128/JVI.01322-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley TJ, Ducharme NA, Varthakavi V, Shepherd BE, Santangelo PJ, Lindquist ME, Goldenring JR, Crowe JE., Jr Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc Natl Acad Sci USA. 2008;105:10209–10214. doi: 10.1073/pnas.0712144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcarcel J, Portela A, Ortin J. Regulated M1 mRNA splicing in influenza virus-infected cells. J Gen Virol. 1991;72:1301–1308. doi: 10.1099/0022-1317-72-6-1301. [DOI] [PubMed] [Google Scholar]
- Wakefield L, Brownlee GG. RNA-binding properties of influenza A virus matrix protein M1. Nucl Acids Res. 1989;17:8569–8580. doi: 10.1093/nar/17.21.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Harmon A, Jin J, Francis DH, Christopher-Hennings J, Nelson E, Montelaro RC, Li F. The lack of an inherent membrane targeting signal is responsible for the failure of the matrix (M1) protein of influenza A virus to bud into virus-like particles. J Virol. 2010;84:4673–4681. doi: 10.1128/JVI.02306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Lamb RA. Influenza virus budding does not require a functional AAA+ ATPase, VPS4. Virus Res. 2010;153:58–63. doi: 10.1016/j.virusres.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Muller B, Krausslich HG. More than one door - Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebedee SL, Lamb RA. Growth restriction of influenza A virus by M2 protein antibody is genetically linked to the M1 protein. Proc Natl Acad Sci USA. 1989;86:1061–1065. doi: 10.1073/pnas.86.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebedee SL, Richardson CD, Lamb RA. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J Virol. 1985;56:502–511. doi: 10.1128/jvi.56.2.502-511.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lamb RA. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology. 1996;225:255–266. doi: 10.1006/viro.1996.0599. [DOI] [PubMed] [Google Scholar]
- Zhang J, Leser GP, Pekosz A, Lamb RA. The cytoplasmic tails of the influenza virus spike glycoproteins are required for normal genome packaging. Virology. 2000a;269:325–334. doi: 10.1006/viro.2000.0228. [DOI] [PubMed] [Google Scholar]
- Zhang J, Pekosz A, Lamb RA. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J Virol. 2000b;74:4634–4644. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P, Wu F, Lu L, Huang JH, Chen YH. The cytoplasmic domain of influenza M2 protein interacts with caveolin-1. Arch Biochem Biophys. 2009;486:150–154. doi: 10.1016/j.abb.2009.02.001. [DOI] [PubMed] [Google Scholar]