Abstract
In C. elegans, mosaic analysis is a powerful genetic tool for determining in which tissue or specific cells a gene of interest is required. For traditional mosaic analysis, a loss-of-function mutant and a genomic fragment that can rescue the mutant phenotype are required. Here we establish an easy and rapid mosaic system using RNAi (RNA mediated interference), using a rde-1 mutant that is resistant to RNAi. Tissue-specific expression of the wild type rde-1 cDNA in rde-1 mutants limits RNAi sensitivity to a specific tissue. We established hypodermal- and muscle-specific RNAi systems by expressing rde-1 cDNA under the control of the lin-26 and hlh-1 promoters, respectively. We confirmed tissue-specific RNAi using two assays: (1) tissue-specific knockdown of GFP expression, and (2) phenocopy of mutations in essential genes that were previously known to function in a tissue-specific manner. We also applied this system to an essential gene, ajm-1, expressed in hypodermis and gut, and show that lethality in ajm-1 mutants is due to loss of expression in hypodermal cells. Although we demonstrate tissue-specific RNAi in hypodermis and muscle, this method could be easily applied to other tissues.
Keywords: muscle, hypodermis, mutant, mosaic, phenotype
1. Introduction
The expression pattern of a gene of interest implicates the function of that gene in specific cells or tissues. In the case of a gene that is expressed in a complicated or broad pattern, however, it is difficult to determine the specific requirements for that gene in a particular tissue or subset of cells, due to potential defects arising from neighboring tissues.
In C. elegans, mosaic analysis is a powerful genetic tool for determining in which tissues or specific cells a gene of interest is required (Hedgecock and Herman, 1995; Herman, 1995). Originally, before the use of transgenic lines, a chromosomal duplication was used for mosaic analysis. More recently, however, a transgenic approach has been taken to produce mosaics. In this procedure, a genomic fragment rescuing a loss-of-function mutation is injected into mutant worms. The injected DNA forms an extrachromosomal array, which is heritably transmitted to subsequent generations. Occasionally, this extrachromosomal array is not segregated into one of the daughter cells during mitosis, resulting in the production of daughter cells that lack the rescuing array. Using this property of extrachromosomal arrays, mosaic animals can be obtained. Several markers for mosaic analysis exist, such as ncl-1 (Hedgecock and Herman, 1995) and sur-5::NLS::GFP (Yochem et al., 1998). In the case of the ncl-1 system, the genotype of each cell can be assessed by the size of nucleoli, since ncl-1 mutants have enlarged nucleoli. In the case of sur-5::NLS:GFP, the presence of the extrachromosomal array in a given cell can be easily assessed by nuclear GFP fluorescence. In both systems, a loss-of-function mutant and a genomic fragment that can rescue the mutant phenotype are required. As an alternative to these methods, rescue of a mutant phenotype can be attempted by driving expression of the wild-type gene using cell/tissue specific promoters. Even in this system, a mutant in the gene of interest and a cloned genomic fragment are required.
Here we describe a mosaic system using RNAi (RNA-mediated interference) and a rde-1 mutant that is resistant to RNAi (Tabara et al., 1999). In C. elegans, RNAi is widely used for rapid and convenient knockdown of most genes of interest (Fire et al., 1998). We show that tissue-specific expression of wild-type rde-1 in rde-1 mutant worms confers RNAi sensitivity only in the tissues expressing rde-1. Since many tissue-specific promoters are available for C. elegans, this system will be applicable to many tissues or selected cells.
2. Materials and Methods
2.1 Strains and worm procedure
Nematodes were grown at 20°C on NGM agar plates with Escherichia coli strain OP50 (Brenner, 1974). We used Bristol N2 as wild type and WM27 (rde-1 (ne219) V) and KW1309 (rde-1 (ne219) dpy-11 (e224) V) as the rde-1 mutant (Tabara et al., 1999). The genotypes of strains used in this study are:
NR220 (kzIs7[pKK1260(lin-26p::nls::gfp), pRF4(rol-6 marker)]);
NR221 (rde-1(ne219) V; kzIs8[pKK1260(lin-26p::nls::gfp), pRF4(rol-6 marker)]);
NR222 (rde-1(ne219) V; kzIs9[pKK1260(lin-26p::nls::gfp), pKK1253(lin-26p::rde-1), pRF4(rol-6 marker)]);
NR230 (kzIs17[pPD93.97(myo-3p::nls::gfp), pRF4(rol-6 marker)]);
NR225 (rde-1(ne219) V; kzIs12[pPD93.97(myo-3p::nls::gfp), pRF4(rol-6 marker)]);
NR229 (rde-1(ne219) V; kzIs16[pPD93.97(myo-3p::nls::gfp), pKK1253(lin-26p::rde-1), pRF4(rol-6 marker)]);
NR320 (rde-1 (ne219) V; kzEx320[pDM#715(hlh-1p::rde-1), pTG96(sur-5p::nls::gfp)]);
NR321 (rde-1(ne219) V; kzEx321[pKK1253(lin-26p::rde-1), pTG96(sur-5p::nls::gfp)]);
NR350 (rde-1(ne219) V; kzIs20[pKK1253(lin-26p::rde-1), pTG96(sur-5p::nls::gfp)]).
NR220 and NR221 have an integrated array containing pKK1260 (lin-26p::nls::gfp, 10 μg/ml) and pRF4 (rol-6 dominant marker, 100 μg/ml) (Mello et al., 1991) in the wild-type or rde-1 mutant background, respectively. NR222 has an integrated array containing pKK1253 (lin-26p::rde-1, 10 μg/ml), pKK1260 (lin-26p::nls::gfp, 10 μg/ml) and pRF4 (rol-6 dominant marker, 100 μg/ml) in the rde-1 mutant background. NR230 and NR225 have an integrated array containing pPD93.97 (myo-3::nls::gfp, 10 μg/ml) and pRF4 (rol-6 dominant marker, 100 μg/ml) in the wild-type or rde-1 mutant background, respectively. NR229 has an integrated array containing pKK1253 (lin-26p::rde-1, 10 μg/ml), pPD93.97 (myo-3::nls::gfp, 10 μg/ml) and pRF4 (rol-6 dominant marker, 100 μg/ml) in the rde-1 mutant background. NR320 and NR321 have extrachromosomal arrays containing pDM#715 (hlh-1p::rde-1, 10 μg/ml) or pKK1253 (lin-26p::rde-1, 10 μg/ml) with pTG96 (sur-5p::nls::gfp, 100 μg/ml) as a marker in the rde-1 mutant background. NR350 have integrated arrays containing pDM#715 (hlh-1p::rde-1, 10 μg/ml) with pTG96 (sur-5p::nls::gfp, 100 μg/ml) as a marker in the rde-1 mutant background. Injection of plasmids into worms was performed as described (Mello and Fire, 1995). The dominant Rol marker (pRF4) (Mello and Fire, 1995) or GFP marker (pTG96) (Yochem et al., 1998) were used as transformation markers. Integration of extrachromosomal arrays was done by the UV irradiation method (Mitani, 1995).
2.2 Molecular biology
RNAi was performed as described (Fire et al., 1998). The procedure for feeding RNAi was described previously (Mercer et al., 2006). To synthesize double-stranded RNA (dsRNA) for RNAi experiments, we used the Large-scale T7 RNA Transcription Kit (Novagen Inc, Wisconsin). We used PCR products derived from plasmids harboring cDNAs as templates for synthesizing dsRNA. For synthesizing dsRNA for GFP, hlh-1, and ajm-1, we used PCR products amplified from pKK1351 (GFP cDNA in pBluescript KS+), yk523e12 (cDNA EST clone containing hlh-1), and yk28c11 (cDNA EST clone containing ajm-1) (yk clones are gifts from Dr. Yuji Kohara) using primers CMo24 (5′-TTG TAA AAC GAC GGC CAG-3′) and CMo422 (5′-GCG TAA TAC GAC TCA CTA TAG GGA ACA AAA GCT GGA GCT-3′). dsRNA for lin-26 was produced by using PCR product amplified from H13-4 (RB2 cDNA clone containing lin-26) (Tsuboi et al., 2002) with primers T7-GAD (5′-GCG TAA TAC GAC TCA CTA TAG GGC AAA CCC AAA AAA AGA GAT C-3′) and GAD-T7 (5′-CGC TAA TAC GAC TCA CTA TAG GGG TTG AAG TGA ACT TGC GCG G-3′).
To make pKK1351, the Sac I/EcoR I fragment (GFP cDNA) of pPD95.67 (a plasmid for construction of GFP fusion, a gift from Dr. Andy Fire) was cloned into pBluescript SK+. pKK1260 contains 3.7 kb upstream sequence of lin-26 (den Boer et al., 1998) derived from the F18A1 cosmid and NLS-GFP cDNA from pPD95.67. For making pKK1253, rde-1 cDNA derived from yk296b10 (cDNA EST clone containing rde-1, a gift from Dr. Yuji Kohara) was cloned downstream of the 3.7 kb upstream promoter sequence of lin-26. For expression of NLS-GFP in muscle cells, we used pPD93.97 (a gift from Dr. Andy Fire). pDM#715 was made by cloning of a fragment of yk296b10 (rde-1 cDNA) into pPD52.99 (a plasmid for expression under the control of hlh-1 promoter, a gift from Dr. Andy Fire). A plasmid for RNAi by feeding for the unc-98 gene was described previously (Mercer et al., 2006).
2.3 Staining and microscopy
Immunostaining was performed as described (Soto et al., 2002) using MH27 monoclonal antibodies, which specifically recognizes the AJM-1 protein (Francis and Waterston, 1985; Koppen et al., 2001). Estimation of GFP fluorescence and phenotype of embryos was performed by microscopy 24 hrs after injection of each dsRNA. The procedure for polarized light microscopy was described previously (Mercer et al., 2006).
3. Results
3.1 Tissue specific expression of rde-1
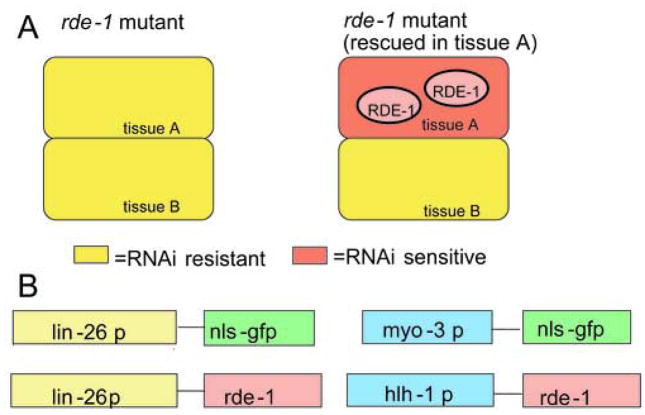
rde-1 mutant worms are reported to be resistant to RNAi, and rde-1 functions in a cell autonomous fashion (Tabara et al., 1999). We reasoned that expression of wild-type rde-1 in a specific tissue in rde-1 mutants should rescue susceptibility to RNAi only in that tissue (Fig. 1A). Initially, we chose hypodermis as a model tissue. For hypodermal expression of rde-1, we used the lin-26 promoter (den Boer et al., 1998). LIN-26 is an essential transcription factor required for the differentiation of hypodermis and is expressed specifically in the hypodermis beginning in mid embryogenesis and continuing to adulthood. We expressed the rde-1 cDNA under the control of a lin-26 hypodermal-specific promoter element (Labouesse et al., 1996) in rde-1 mutant worms (four constructs are shown in Fig. 1B). Worms harboring an extrachromosomal array containing this construct displayed no defects in growth or movement. To avoid mosaic expression of the lin-26p::rde-1 construct, we prepared worms containing integrated arrays. To test hypodermis-specific rescue of rde-1, we chose muscle as a negative control tissue (see sections 3.2 and 3.3).
Fig. 1.
Strategy of tissue-specific RNAi using the rde-1 mutant
A: Yellow colour shows RNAi resistant cells. Red colour shows RNAi sensitive cells. Expression of rde-1 in tissue A should result in RNAi sensitivity in tissue A, but not tissue B.
B: Plasmids used in this study. The lin-26 promoter (3.7 kb 5′ sequence) was placed upstream of NLS-GFP or rde-1, resulting in hypodermis-specific NLS-GFP or rde-1 expression. The myo-3 promoter (2.4 kb 5′ sequence) was fused to NLS-GFP, resulting in a muscle-specific NLS-GFP expressing plasmid (pPD93.97). The hlh-1 promoter (3.0 kb 5′ sequence) was fused to rde-1, resulting in a muscle-specific rde-1 expressing plasmid.
3.2 Examination of tissue specificity using GFP markers
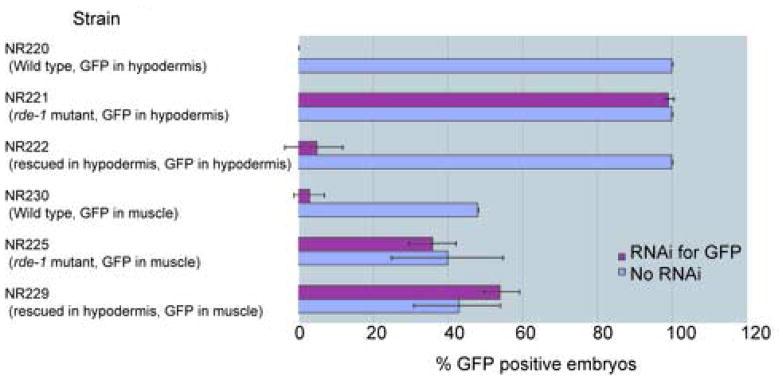
To examine the tissue specificity of RNAi in rde-1 mutant worms expressing the wild type rde-1 gene in the hypodermis, we conducted two assays. The first assay utilized GFP markers expressed specifically in either hypodermis or muscle using the plasmids shown in Fig. 1B. These plasmids contain NLS-GFP under the control of the lin-26 promoter for hypodermal expression (den Boer et al., 1998) or the myo-3 promoter for muscle expression (Okkema et al., 1993). We confirmed hypodermal or muscle-specific GFP expression in the nucleus in both cases (data not shown). We co-injected lin-26p::rde-1 DNA together with either lin-26p::nls::gfp or myo-3p::nls::gfp plasmids, created transgenic lines, and then integrated these extrachromosomal arrays into the genome. Worm strains used in this assay (Fig. 2) have the following characteristics. In the NR220, NR221, and NR222 strains, the GFP marker is expressed in the hypodermis, and in NR230, NR225, and NR229, the GFP marker is expressed in muscle. NR220 and NR230 are wild type strains, in which RNAi is effective in all tissues, including hypodermis and muscle. NR221 and NR225 are rde-1 mutant strains, in which RNAi is inactive in all tissues including hypodermis and muscle. NR222 and NR229 are hypodermis-specific RNAi strains, in which the rde-1 mutation is rescued by the rde-1 cDNA solely in the hypodermis. As shown in Fig. 2, gfp(RNAi) eliminated GFP fluorescence in the hypodermis and muscle in wild type strains (i.e., NR220 and NR230), and these effects were suppressed by the rde-1 mutation (i.e., NR221 and NR225) in both hypodermis and muscle. In the hypodermis-specific RNAi strain expressing GFP in the hypodermis, gfp(RNAi) effectively eliminated GFP fluorescence. However, in the hypodermis-specific RNAi strain expressing GFP in muscle, gfp(RNAi) had no effect on GFP fluorescence (Fig. 2). These results show that expression of the rde-1 cDNA in hypodermis can rescue the rde-1 mutation only in hypodermis, not in muscle, and that as a result, RNAi is effective only in the hypodermis.
Fig. 2.
Estimation of tissue specific RNAi system using GFP markers
“Strain” shows the worm strain used for each assay with its properties. Purple bar represents percentage of GFP positive embryos after RNAi for GFP. Blue bar represents percentage of GFP positive embryos after no RNAi. Double sided T-bar shows standard deviation.
3.3 Examination of tissue specificity using embryonic lethal genes
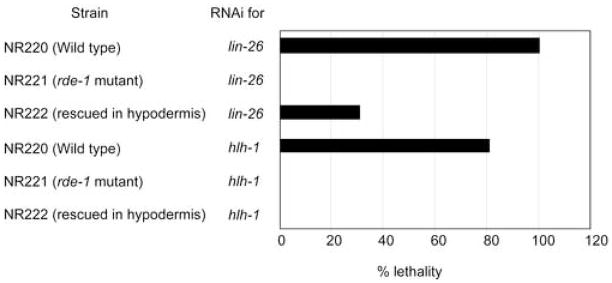
As a second assay to confirm tissue specificity, we chose to test two genes that are essential for embryonic development. One is lin-26 (Labouesse et al., 1994; Labouesse et al., 1996), which is expressed only in the hypodermis; lin-26(RNAi) causes embryonic lethality. The other is hlh-1 (Krause et al., 1994), which is expressed only in muscle; hlh-1(RNAi) causes embryonic and larval lethality. Both LIN-26 and HLH-1 are essential for tissue differentiation and function in a cell-autonomous manner. Double stranded RNA prepared from lin-26 and hlh-1 cDNAs were injected into worms with the following genotypes: wild type, rde-1 mutant, and rde-1 mutant rescued in the hypodermis. lin-26(RNAi) in wild type showed 100% lethality, in contrast to lin-26 (RNAi) in the rde-1 mutant, in which 0% lethality was observed. In the case of rde-1 rescued in the hypodermis, lin-26(RNAi) resulted in 31% lethality (Fig. 3). RNAi of hlh-1 caused 81% lethality in wild type, but 0% in the rde-1 mutant and 0% in the hypodermal rescued rde-1 (Fig. 3), again demonstrating that the RNAi effect in muscle is not rescued by rde-1 expressed in the hypodermis. From these results, we conclude that the expression of rde-1 under the control of the lin-26 promoter can rescue the rde-1 mutation solely in the hypodermis and that as a result, RNAi is only effective in the hypodermis in these worms.
Fig. 3.
Estimation of the efficacy of the tissue specific RNAi system using embryonic lethal genes “Strain” shows the worm strain used for each assay with its properties. “RNAi for” shows which gene was used in the assay. Black bar represents percentage of embryos that showed lethality.
3.4 Application to other tissues
To verify our “tissue-specific RNAi” system, we used the hlh-1 promoter for muscle-specific expression (Krause, 1995). We established two lines in which rde-1 mutants harbor the rde-1 cDNA under control of either the lin-26 or hlh-1 promoter (Table 1). For these two lines, we carried out lin-26(RNAi) and hlh-1(RNAi). We scored worms showing lethal phenotypes from GFP positive worms containing extrachromosomal arrays in each case (Table 1). hlh-1(RNAi) resulted in lethality only in the strain in which rde-1 was rescued in muscle. Similarly, lin-26(RNAi) only resulted in lethality in the strain rescued for rde-1 function in the hypodermis (Table 1). These results demonstrate that as with the lin-26 promoter in the hypodermis, the hlh-1 promoter can be used to induce muscle-specific RNAi.
Table 1.
Muscle- and hypodermis-specific RNAi system
| strain | genotype | RNAi | Lethal phenotype (%) |
|---|---|---|---|
| NR320 | rde-1 (ne219) V; kzEx320[pDM#715(hlh-1p::rde-1), pTG96(sur-5p::nls::gfp)] | lin-26 | 0 |
| NR320 | rde-1 (ne219) V; kzEx320[pDM#715(hlh-1p::rde-1), pTG96(sur-5p::nls::gfp)] | hlh-1 | 23.5 |
| NR321 | rde-1(ne219) V; kzEx321[pKK1253(lin-26p::rde-1), pTG96(sur-5p::nls::gfp)] | lin-26 | 84.5 |
| NR321 | rde-1(ne219) V; kzEx321[pKK1253(lin-26p::rde-1), pTG96(sur-5p::nls::gfp)] | hlh-1 | 0 |
3.5 Tissue-specific analysis of ajm-1 function
We applied the hypodermis-specific RNAi system to a gene expressed in multiple tissues. We chose ajm-1, which is expressed in hypodermis, pharynx, and gut (Koppen et al., 2001). The ajm-1 gene encodes a protein that localizes to apical junctions in epithelial cells, and is required for junctional integrity. ajm-1(RNAi) causes embryonic lethality (Koppen et al., 2001). To determine whether loss of function of ajm-1 in hypodermis but not other tissues causes lethality, we carried out ajm-1(RNAi) in embryos rescued for RNAi only in the hypodermis. ajm-1(RNAi) in wild type showed 100% lethality; in contrast, ajm-1(RNAi) in the rde-1 mutant yielded 0% lethality (Table 2), confirming previous results. ajm-1(RNAi) in the hypodermis-rescued rde-1 strain showed a high percentage of lethality (Table 2). RNAi of ajm-1 in a hypodermis-rescued rde-1strain also eliminated AJM-1 immunostaining using MH27, a monoclonal antibody specific for AJM-1 (Francis and Waterston, 1985; Koppen et al., 2001), in the hypodermis, but not in gut (Fig. 4), confirming that RNAi is effective only in the hypodermis. These results suggest that ajm-1 expression in hypodermal cells is essential for its function, and further confirm the tissue specificity of RNAi using tissue-specific promoters to rescue rde-1 function in rde-1 mutants.
Table 2.
Application of hypodermis-specific RNAi system to the ajm-1 gene
| strain | genotype | Lethal phenotype (%) | No phenotype (%) | N |
|---|---|---|---|---|
| NR220 | kzIs7[pKK1260(lin-26p::nls::gfp), pRF4(rol-6 marker)] | 100 | 0 | 23 |
| NR221 | rde-1(ne219) V; kzIs8[pKK1260(lin-26p::nls::gfp), pRF4(rol-6 marker)] | 0 | 100 | 64 |
| NR222 | rde-1(ne219) V; kzIs9[pKK1260(lin-26p::nls::gfp), pKK1253(lin-26p::rde-1), pRF6(rol-6 marker)] | 56 | 42 | 115 |
Fig. 4.
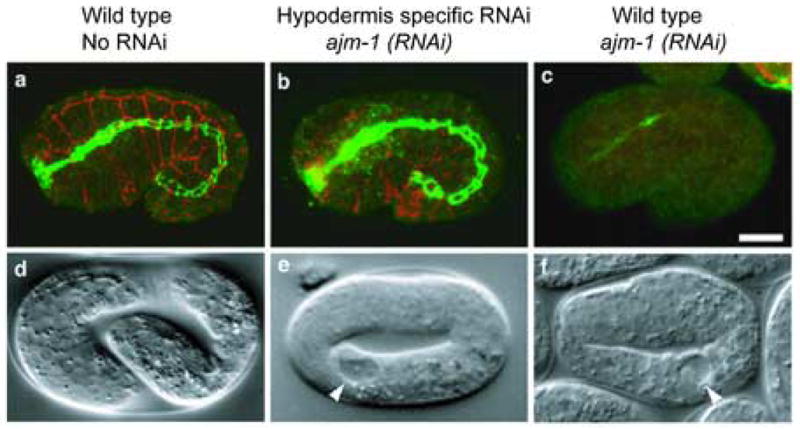
Tissue-specific depletion of AJM-1 via ajm-1(RNAi) in wild type and hypodermis-rescued rde-1 embryos.
a–c, MH27 (a monoclonal antibody specific to AJM-1) staining of wild type, ajm-1(RNAi); rde-1 [plin-26::rde-1] and ajm-1(RNAi) embryos. Hypodermal MH27 signal is shown in red (a projection of three confocal sections of the surface of the embryo), and pharyngeal and intestinal signal is shown in green (a projection of two central confocal sections). In the wild type, MH27 staining is bright and even in all epithelia (a). ajm-1(RNAi) in the strain rescued for rde-1 in the hypodermis (b) results in marked reduction of MH27 staining in the hypodermis, but no effect in the pharynx and intestine. ajm-1(RNAi) in a wild-type embryo causes almost complete loss of MH27 signal from all embryonic epithelia (c). d–f, Nomarski phenotypes of wild type, ajm-1(RNAi); rde-1 [plin-26::rde-1] and ajm-1(RNAi) embryos. A wild type embryo is shown at the 3-fold stage (d). ajm-1(RNAi) causes a nearly identical 2-fold arrest phenotype in both the strain rescued for rde-1 in hypodermis (e) and in the wild type (f). However, in the case of the strain rescued for rde-1 in hypodermis, embryonic lethality is only observed in 50% of the embryos, while 100% lethality is observed in a wild-type background. Scale bar represents 10 μm. Arrowheads indicate vacuoles characteristic of the ajm-1 loss of function phenotype.
3.6 Postembryonic muscle-specific RNAi by the feeding method
To determine whether our tissue-specific RNAi system might also work postembryonically, we performed RNAi by feeding for unc-98. The unc-98 gene is expressed primarily in adults and unc-98 mutants show a characteristic birefringent needle phenotype by polarized light microscopy (Mercer et al., 2003). Bacteria expressing dsRNA of the unc-98 gene were fed to either wild type, the rde-1 mutant, or the rde-1 mutant carrying an integrated array containing wild type rde-1 cDNA fused to the hlh-1 promoter. The progeny from each strain were subjected to polarized light microscopy. Progeny from wild type showed the needle phenotype as previously reported (Mercer et al., 2003), whereas those from the rde-1 mutant strain did not show the needle phenotype (Fig. 5). Although the percentage of adult worms showing the needle phenotype is not high (20%), the progeny from the muscle-specific RNAi strain showed the needle phenotype (Fig. 5), suggesting that the tissue-specific RNAi system can be applied to a gene which functions at postembryonic stages by the feeding procedure.
Fig. 5.
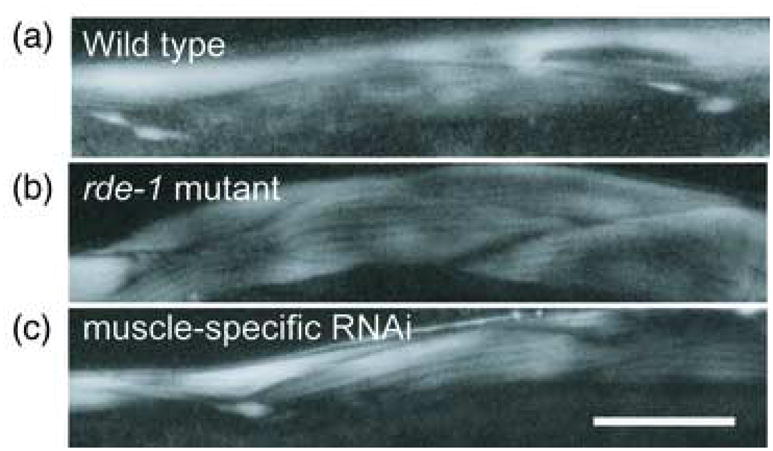
Muscle-specific feeding RNAi for the unc-98 gene in adult worms
All three panels show polarized light microscope images of progeny from worms fed with bacteria expressing dsRNA for unc-98. (a) Wild type strain; (b) the rde-1 mutant strain; (c) muscle-specific RNAi strain. Birefringent needles, which are characteristic of unc-98 mutants, are observed in (a) and (c). Scale bar represents 20 μm.
4. Discussion
4.1 A tissue-specific RNAi system
In this report, we demonstrate a tissue-specific RNAi method in C. elegans, using the hypodermis and muscle as test cases. Based on two lines of evidence, we showed the tissue specificity of this system. First, by using GFP markers expressed in either the hypodermis or muscle, we showed that expression of wild-type rde-1 in the hypodermis can confer RNAi sensitivity only in the hypodermis. GFP is not affected by other cellular functions and is a sensitive reporter of gene activity (Chalfie et al., 1994), and thus confirms the tissue specificity of our technique. Second, we confirmed tissue specificity by using genes essential for embryonic development that are expressed either in the hypodermis (lin-26) (Labouesse et al., 1996) or muscle (hlh-1) (Krause, 1995). Results from functional assays indicate that hypodermis- or muscle-specific RNAi has the desired effect, and can be applied for practical “mosaic” analysis. Furthermore, using this hypodermis-specific RNAi system, we revealed that the expression of ajm-1 in the hypodermis is important for its function during development, thus confirming the usefulness of this tissue-specific RNAi system. A previous report suggested that RDE-1-dependent tissue-specific RNAi can function postembryonically in hypodermis and muscle using promoters different from the ones used in our study (dpy-7 promoter for hypodermis and myo-3 for muscle) to perturb nas-37 function (Suzuki et al., 2004). Although this work did not verify tissue-specific knockdown using protein assays as we have done here, this previous work supports our idea that the RDE-1-dependent tissue-specific RNAi system is useful for the characterization of tissue-specific function of a given gene.
4.2 Efficiency of RNAi in the tissue-specific RNAi system
When we injected lin-26 dsRNA into worms rescued for RNAi only in the hypodermis, only 31% of progeny from injected worms showed a lethal phenotype (Fig. 3). Injection of ajm-1 dsRNA resulted in only 56% lethality (Table 2). In the case of muscle-specific RNAi (Table 1), only 23% of progeny injected with hlh-1 dsRNA showed a lethal phenotype. All three dsRNA injections caused lethality in 81~100% of progeny from wild type worms (Fig. 3, Table 1 and 2), suggesting that rescue of rde-1 in the hypodermis or muscle was not complete. As described previously, maternal and zygotic expression of rde-1 are required for complete rescue of the rde-1 mutant (Tabara et al., 1999). In our tissue-specific RNAi system, rde-1 is expressed from extrachromosomal or integrated arrays. Expression of those arrays is silent in the germline, resulting in a lack of maternal expression. Furthermore, in our tissue-specific RNAi system, the rde-1 gene is regulated by the lin-26 or hlh-1 promoters. These promoters are reported to drive expression beginning during mid-embryogenesis (Krause et al., 1994; Labouesse et al., 1996). Due to the exclusively zygotic expression of rde-1 by the lin-26 or hlh-1 promoters, it is therefore likely that complete rescue of rde-1 in the hypodermis or muscle is not achieved because the hypodermis or muscle lacks maternal rde-1 expression. To overcome this reduced efficiency of RNAi, we attempted to utilize the double mutant, rde-1; rrf-3, since it has been reported that rrf-3 increases RNAi sensitivity (Simmer et al., 2002). However, rrf-3 is also known to cause genome instability and the double mutant genotype was very unstable (soon after we constructed the rde-1; rrf-3 strain, the rde-1 mutation was lost).
4.3 Muscle specific promoters
We used the hlh-1 promoter for muscle-specific expression. Commonly, the myo-3 promoter is used for muscle-specific expression (Okkema et al., 1993). Based on GFP and lacZ expression results (Okkema et al., 1993), the myo-3 promoter does drive gene expression largely in muscle cells, a finding we confirmed using myo-3p::nls::gfp. We therefore tried to establish a muscle-specific RNAi system using the myo-3 promoter, but results of both GFP and functional assays showed that RNAi is effective both in hypodermis and muscle cells in such worms (data not shown), suggesting that the myo-3 promoter may not be sufficiently specific to muscle cells. It is possible that excessive gene expression driven by the myo-3 promoter results in “leaky” expression of rde-1 transcripts in other tissues besides muscle. If the amount of rde-1 function required for rescue of rde-1 loss of function is less than that required for robust GFP or lacZ expression, such a scenario is feasible. One additional possibility is that RDE-1 protein expressed in muscle cells has non- cell autonomous effects on other tissues. However, the result that hypodermis-specific RNAi, using the lin-26 promoter, and muscle-specific RNAi, using the hlh-1 promoter, each yielded tissue-specific effects does not support this possibility. Furthermore, RDE-1 has been reported to be cell-autonomous by others (Tabara et al., 1999). A previous report shows that the myo-3 promoter does function in muscle-specific RNAi (Suzuki et al., 2004). This discrepancy may be due to the difference between integrated arrays (this study) and extrachromosomal arrays (previous study). In addition, the previous study did not verify tissue-specific knockdown via immunostaining. It may be that partial knockdown of hypodermal expression of nas-37 due to “leaky” rescue of rde-1 function driven by the myo-3 promoter is insufficient to yield a noticeable phenotype, whereas our functional assays using lin-26 and ajm-1 are more sensitive to “leaky” RNAi effects.
4.4 Further application of the tissue specific RNAi system
Considerable effort has been expended to determine the pattern of expression of many genes and the cis-acting elements responsible for their expression. In the mouse and Drosophila, a technique for tissue-specific inactivation or knockdown of a specific gene has been established using tissue-specific promoters (Golic, 1991; Herault et al., 1998; Zheng et al., 2000). Our method of tissue-specific RNAi in C. elegans also requires tissue-specific promoters. Presently, data is available for several promoters that drive expression in specific tissues, such as hypodermis (Gilleard et al., 1997; den Boer et al., 1998; Sieburth et al., 1999), muscle (Okkema et al., 1993; Krause et al., 1994; Terami et al., 1999), pharynx (Okkema et al., 1993; Kalb et al., 1998; Anyanful et al., 2001), or gut (Fukushige et al., 1998), and some specific cells, such as distal tip cells (Henderson et al., 1994), sex myoblasts (Harfe et al., 1998), and germline cells (Praitis et al., 2001). Many of these promoters should be useful for extending our tissue-specific RNAi system to other tissues.
By using various promoters to drive the expression of the rde-1 cDNA and a GFP marker, it will be possible to establish additional tissue- or cell-specific RNAi systems in C. elegans. These tissue-specific RNAi systems can be used to rapidly identify tissues or cells in which the function of a gene of interest is essential. In particular, for genes functioning in multiple cells or tissues, such as those encoding cytoskeletal components or other essential proteins, this system should be a powerful tool for examining the function of a given gene in a particular tissue or cell type. We have also found that the feeding RNAi procedure is effective in the tissue-specific RNAi system (Fig. 5) and that this system could apply to the genes functioning both at embryonic (Fig. 4) and postembryonic (Fig. 5) stage. Combined with genome-wide RNAi methods (Fraser et al., 2000; Gonczy et al., 2000), the procedures described here will enable genome-wide estimates of tissue specific functions of genes of interest without the use of the more technically difficult traditional mosaic analysis.
Acknowledgments
We thank Dr. Alan Coulson for cosmids and YAC clones; Dr. Yuji Kohara for cDNA clones; Dr. Andy Fire for vectors; Dr. Robert H. Waterston for MH27 antibody; Drs. Hiroaki Tabara and Craig Mello for the rde-1 strain and cDNA; Fumie Nishimura for technical assistance; Katsuhisa Kasuya for discussions; Dr. Shinya Kuroda and THE TEAM for helpful discussion. We also thank Dr. Ken Norman and Dr. Guy Benian for critical reading of this manuscript. Some of the strains used in this work were provided by the Caenorhabditis elegans Genetics Center, which is funded by the National Institutes of Health (NIH) Center for Research Resources. H. Q., M. A., and K. K. were supported by grants-in-aid for Scientific Research from the Ministry of Education, Science, and Culture, Japan, by the Japan Society of the Promotion of Science Research for the Future, by the Human Frontier Science Program. M.K and J.H. were supported by NIH grant GM058038.
abbreviations
- RNAi
RNA mediated interference
- GFP
green fluorescent protein
- NLS
nuclear localization signal
- dsRNA
double stranded RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anyanful A, Sakube Y, Takuwa K, Kagawa H. The third and fourth tropomyosin isoforms of Caenorhabditis elegans are expressed in the pharynx and intestines and are essential for development and morphology. J Mol Biol. 2001;313:525–37. doi: 10.1006/jmbi.2001.5052. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–5. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- den Boer BG, Sookhareea S, Dufourcq P, Labouesse M. A tissue-specific knock-out strategy reveals that lin-26 is required for the formation of the somatic gonad epithelium in Caenorhabditis elegans. Development. 1998;125:3213–24. doi: 10.1242/dev.125.16.3213. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–49. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–30. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- Gilleard JS, Barry JD, Johnstone IL. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol Cell Biol. 1997;17:2301–11. doi: 10.1128/mcb.17.4.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252:958–61. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, Hannak E, Kirkham M, Pichler S, Flohrs K, Goessen A, Leidel S, Alleaume AM, Martin C, Ozlu N, Bork P, Hyman AA. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–6. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998;12:2623–35. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Herman RK. The ncl-1 gene and genetic mosaics of Caenorhabditis elegans. Genetics. 1995;141:989–1006. doi: 10.1093/genetics/141.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–24. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Herault Y, Rassoulzadegan M, Cuzin F, Duboule D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE) Nat Genet. 1998;20:381–4. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- Herman RK. Mosaic analysis. Methods Cell Biol. 1995;48:123–46. [PubMed] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, Okkema PG, McGhee JD. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–80. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–91. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- Krause M. MyoD and myogenesis in C. elegans. Bioessays. 1995;17:219–28. doi: 10.1002/bies.950170308. [DOI] [PubMed] [Google Scholar]
- Krause M, Harrison SW, Xu SQ, Chen L, Fire A. Elements regulating cell- and stage-specific expression of the C. elegans MyoD family homolog hlh-1. Dev Biol. 1994;166:133–48. doi: 10.1006/dbio.1994.1302. [DOI] [PubMed] [Google Scholar]
- Labouesse M, Hartwieg E, Horvitz HR. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development. 1996;122:2579–88. doi: 10.1242/dev.122.9.2579. [DOI] [PubMed] [Google Scholar]
- Labouesse M, Sookhareea S, Horvitz HR. The Caenorhabditis elegans gene lin-26 is required to specify the fates of hypodermal cells and encodes a presumptive zinc-finger transcription factor. Development. 1994;120:2359–68. doi: 10.1242/dev.120.9.2359. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–82. [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KB, Flaherty DB, Miller RK, Qadota H, Tinley TL, Moerman DG, Benian GM. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol. Biol. Cell. 2003;14:2492–507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KB, Miller RK, Tinley TL, Sheth S, Qadota H, Benian GM. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol. Biol. Cell. 2006;17:3832–47. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani S. Genetic regulation of mec-3 gene expression impricated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Dev Growth & Diff. 1995;37:551–557. doi: 10.1046/j.1440-169X.1995.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–26. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth DS, Sundaram M, Howard RM, Han M. A PP2A regulatory subunit positively regulates Ras-mediated signaling during Caenorhabditis elegans vulval induction. Genes Dev. 1999;13:2562–9. doi: 10.1101/gad.13.19.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–9. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Soto MC, Qadota H, Kasuya K, Inoue M, Tsuboi D, Mello CC, Kaibuchi K. The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 2002;16:620–32. doi: 10.1101/gad.955702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Sagoh N, Iwasaki H, Inoue H, Takahashi K. Metalloproteases with EGF, CUB, and thrombospondin-1 domains function in molting of Caenorhabditis elegans. Biol Chem. 2004;385:565–8. doi: 10.1515/BC.2004.069. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–32. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Terami H, Williams BD, Kitamura S, Sakube Y, Matsumoto S, Doi S, Obinata T, Kagawa H. Genomic organization, expression, and analysis of the troponin C gene pat-10 of Caenorhabditis elegans. J Cell Biol. 1999;146:193–202. doi: 10.1083/jcb.146.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi D, Qadota H, Kasuya K, Amano M, Kaibuchi K. Isolation of the interacting molecules with GEX-3 by a novel functional screening. Biochem Biophys Res Commun. 2002;292:697–701. doi: 10.1006/bbrc.2002.6717. [DOI] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–34. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Sage M, Sheppeard EA, Jurecic V, Bradley A. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol Cell Biol. 2000;20:648–55. doi: 10.1128/mcb.20.2.648-655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]