Abstract
Hepatitis C virus (HCV) has a high rate of replication and lacks RNA-proofreading capabilities, thereby leading to variant or mutant viruses circulating within the host as a quasispecies. Previous work in our laboratory identified viral variants that emerged in a class-II immunodominant epitope NS3358–375 of the non-structural-3 (NS3) protein region of HCV, the sequence of which is based on genotype 1A, the most prevalent genotype in the U.S. population. Further work suggested that positive immune selection pressure was driving viral variation. Paradoxically, viral variants account for only a small percentage of the circulating virus in humans and chimpanzees, suggesting that passive evasion is not the only means of escape by HCV. This observation suggests a unique pathogenesis for HCV as it persists in the host. In the current study, we hypothesize that viral variants are acting as altered peptide ligands (APLs). To test this hypothesis, we used cloned T-cells specific for NS3358–375 peptide, which demonstrated attenuated T-cell and IFNγ responses to individual variant peptides, when compared to the NS3358–375 stimulated T-cell clones. Furthermore, such variants could act as altered peptide ligands (APLs), based on their ability to antagonize the IFNγ proliferative responses of clones specific for NS3358–375. In addition, MHC class II tetramer staining demonstrated that variant peptide-MHC complexes were able to specifically bind to NS3358–375 T-cell clones and that both the variant and NS3358–375 tetramers were able to bind to the same CD4+ T cells. Taken together, the results suggest that viral variants may act as APL to effectively blunt the T-cell response to an important HCV epitope.
Keywords: Hepatitis C virus, human CD4 T cells, altered peptide ligands, MHC class II tetramer, antagonism
Introduction
T cell activation occurs when a T cell receptor (TCR) binds to cognate or specific peptide bound to major histocompatibility complex (pMHC) molecules on the surface of antigen presenting cells (APCs) [1]. The engagement of the TCR to pMHC is necessary for the activation of CD4+ and CD8+ T cells, thereby leading to an effective adaptive immune response against an invading pathogen [2]. Single amino acid substitution in the cognate peptide, termed altered peptide ligands (APLs), can elicit a gradient of effector function changes in a specific T cell [3].
The recognition of these segments of a pathogen or epitope(s) presented by the pMHC to T cells is critical for the clearance of viruses [4]. As in the case of hepatitis C virus (HCV) infection, clearance of the virus has been found to be dependent upon the quality of the CD4+ T cell response in up to 30% of individuals infected [5–8]. Mueller et al. [9] have detected virus-specific CD4+ T cells in chronic HCV patients, suggesting a novel mechanism of viral persistence [10–14].
Previous work in our laboratory identified viral variants that emerged in an MHC class-II immunodominant epitope NS3358–375 of the nonstructural-3 (NS3) protein region of HCV [15]. Further work suggested that positive immune selection pressure was driving viral variation [16]. In contradiction to viral escape models, HCV variants account for only a small percentage of the circulating virus in humans and chimpanzees [17, 18]. Also, HCV is able to modulate the immune response specifically towards viral persistence, indicating that HCV may be able to exploit a different mechanism in place of a more passive evasion strategy, leading to non-detection of the HCV pathogen by the immune system [16, 19].
One such mechanism that HCV may be able to exploit for persistence includes APLs, which have been observed within the hypervariable region 1 [20, 21], and in other pathogens, such as HBV [4, 22], HIV [23, 24] and plasmodium falciparium [25]. Our previous studies identified and tracked viral variants arising in a single HCV chronic individual and further determined the effect these variants had on the in vitro immune response using polyclonal assays [15, 16, 18]. Although HCV circulates in the host as a quasispecies, it appears that such variation does not lead to viral escape, but rather that some variants are able to suppress the immune response in an antigen-specific manner.
The importance of T cells in HCV clearance leads to the supposition that fully activated HCV specific T cells correlate with HCV clearance; however, the impact of viral variants on TCR activation in conjunction with the cognate peptide has not been fully elucidated in HCV pathogenesis. There are several non-mutually exclusive factors that lead to a fully activated T cell, such as the quantity of pMHC presented on the surface of APCs and TCR avidity. Previous work demonstrated an attenuated T cell proliferative response to the cognate peptide in polyclonal assays when PBMC from a chronic HCV patient were pre-incubated with the variant peptides [16, 19]. A question that arises from the previous polyclonal work is whether the variants that arise in this important Th1- viral clearing epitope (NS3358–375) are antagonizing the same CD4+ T cell or inducing a subset of CD4+ T cells to suppress the cognate T cell response. To test the hypothesis that viral variants could act as APLs leading to a partially activated T cell and not the induction of another subset of T cells that can suppress the immune response, in this study we used cloned T cells specific for the MHC class II epitope NS3358–375 peptide. The APL peptides alone stimulated low T cell and cytokine responses in comparison to cognate peptide in polyclonal assays, all of which has been documented elsewhere [16]. Prepulsing T cell clones with variant peptides revealed that variants were able to antagonize proliferative and IFNγ responses to cognate NS3358–375 peptide. MHC class II tetramers, loaded with variant peptides, are able to bind specifically to NS3358–375 T cell clones. Further, both variant S370P and NS3358–375 are able to bind to the same NS3358–375 specific CD4+ T cells. Thus, viral epitope variants are able to blunt the responses of the very T cells that should help with the elimination of virus. Our results also help to explain observations that CD4+ T cell responses to HCV antigens seem attenuated or missing in chronically infected patients; they also bear on the findings that CD8+ killer T cells are ineffective at eliminating HCV infected target cells [26, 27].
Materials and Methods
Blood Samples
These studies have been reviewed and approved by the University of Utah and the Medical College of Wisconsin Institutional Review Boards. Isolation of lymphocytes and subsequent HLA typing was previously described [28].
Synthetic Peptides
In vitro PBMC and T cell clones were stimulated with synthetic peptides representing one human leukocyte antigen DRB1*1501 restricted-epitope surrounding HCV NS3 amino acids 358–375 (a.a. 1384–1401 of the HCV polyprotein) as previously described [16, 28]. The three single amino acid variants were identified in a chronic HCV patient (P.B3019) [16, 28]. Peptide sequences were as follows: cognate 358–375 (NS3358–375), VIKGGRHLIFCHSKKKCD; variant H369R, VIKGGRHLIFCRSKKKCD; variant S370P, VIKGGRHLIFCHPKKKCD; variant K371E, VIKGGRHLIFCHSEKKCD.
T Cell Proliferation Assay
To measure proliferative responses of PBMC and T cell clones following stimulation with cognate peptide NS3358–375 and variants peptides, cells were plated at 1 × 105/ well in round-bottom 96 well plates and incubated at 37°C, 5% CO2 at indicated times and peptide doses. The cells were pulsed overnight with 1 µCi/well of titrated thymidine (3H-TdR) (Perkin Elmer, Waltham, Massachusetts) and harvested onto glass fiber filters (Perkin Elmer) for measurement of radiolabel incorporation by scintillation counter (Perkin Elmer).
T Cell Clones
Specific helper T cell clones, towards the cognate HCV NS3 peptide 358–375 (NS3358–375), were generated from a resolved DRB1*1501/1301 hemophiliac individual (PH1127) that had resolved their infection. As described previously [28], PH1127 was PCR negative but antibody positive for HCV. Further, PH1127 has the same MHC class II (DRB1*1501) allele as the chronically infected patient PB3019, in which the viral variants were identified, and PH1127 was able to respond to NS3358–375 peptide in a dose dependent manner. PH1127 PBMC were re-suspended and plated into 96 well flat bottom plates (200µl/well) at 2 × 106 cells/ml. Cells were stimulated with NS3358–375 at 5µM/ml at 37°C, 5% CO2 incubator. Cells were provided 10 U/ml rhIL-2 (BD bioscience). On day 14, cells were tested for responsiveness to NS3358–375. Functional cells were further subcloned by limiting dilution at cell densities of 10, 3, 1, and 0.3 cells/well in 96 flat bottom plates, which contained 105 irradiated (3000 rad) autologous PBMC. T cells from 0.3 cells/well were moved to 24 well plates and cultured with 10µM/ml NS3358–375 peptide and 10U/ml of rhIL-2 in complete media. Culture of PBMC and T cell clones were cultured in complete media: RPMI 1640 tissue culture medium (BioWhittaker, Walkersville, ME) supplemented with 25 mM HEPES, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 1 mM sodium pyruvate, 5 µg/ml gentamycin (all from Mediatech Cellgro, Herndon, VA), 10 U/ml heparin sodium (Fisher Scientific, Pittsburgh, PA) and 10% pure human serum (Atlanta Biologicals, Lawrenceville, GA). T cell clones were tested for responsiveness to NS3358–375 and variant peptides presented by autologous PBMC for each T cell clone experiment. Cells were cultured in a 37°C, 5% CO2 incubator at indicated times.
Cytokine ELISA
Cell culture supernatants IFNγ levels were determined by a commercial ELISA kit from Pharmingen, Inc. (San Diego, CA, USA) according to the manufacturer’s instructions. Experimental values were determined by comparing the optical densities with a standard curve derived from recombinant IFNγ. Negative controls consisted of background levels of APC cultured with T cell clones and complete media. Each experiment was done in triplicate.
Antagonism Assay
Antigen presenting cells (APCs) were prepulsed 2 hrs. with NS3358–375 peptide at 1µM, washed (3×) in complete media + 10%PHS and irradiated at 3000 Rad. APCs were split (2 × 105 cells/well) into 96 well trays containing variant peptides at indicated concentrations and T cells were added at 1 × 104 cells/well. Cells were incubated at 37°C for 48 hrs. Then 100µl of supernatant was removed for ELISA screening and 3[H]-thymidine was added for 18 hrs. before harvesting and measuring 3[H]-thymidine incorporation.
Anergy Assay
APCs were prepulsed for 3 hrs. with variant peptides at 1µM, washed, and irradiated (3000 rad). The APCs were split into wells containing NS3358–375 peptide at 0.1, 1, or 10µM concentrations. T cells were subsequently added at 1 × 104 cells/well and incubated at 37°C for 48 hrs. 100µl of supernatant was saved for ELISA screening and 3[H]-thymidine was added for 18 hrs. before harvesting and measuring 3[H]-thymidine incorporation.
Tetramer Staining
All tetramers were obtained from the NIH tetramer facility at Emory University. T cell clones were stained either with 358–375- Phycoerythrin (PE), or variant H369R-Allophycocyanin (APC); variant S370P-APC; variant K371E –APC tetramers for 1hr at 37°C. 7-AAD (BD bioscience), cell viability probe, CD4-pacific blue (BD bioscience), CD3-Amcyan (BD bioscience), CD8-FITC (eBioscience) were added for 15 min. at 4°C, washed with flow stain buffer (BD bioscience) and analyzed on a BD FACSCanto II. Negative controls consisted of staining cells with nonspecific peptide, CLIP-DR15 tetramer, labeled with either –PE label or –APC respectively. Further, T cell clone HA306–318 was stained with each tetramer (data not shown). Results were compared using Student’s t-test and considered significant if the p<0.05. Flow cytometry data analysis was performed using Flow Jo software (Tree Star).
CD4+ T Cell Line
NS3358–375 antigen-specific CD4+ CFSE T cells were derived from PH1127 and PB3019. PBMC from PH1127 and PB3019 were labeled with 0.5µM Carboxyfluorescein succinimidyl ester (CFSE) and stimulated with NS3358–375 (5µM) synthetic peptide. On day 7, the cells were stained with anti-CD4-APC (Invitrogen, Carlsbad, CA) and cell sorted at the University of Utah cell sorting core facility. The CD4+ T cells were sorted by either CFSEhigh (non-proliferating) or CFSElow (proliferating). The cells were washed and expanded by CD3/CD28 dynabeads (Invitrogen) with 10U/ml of human rIL-2 (BD Pharmingen, San Diego, CA) and the CD4+ CFSElow T cells were used for confocal imaging. As a control, Tropomyosin-specific CD4+ T cells were selected and expanded using the same method as the CD4+ NS3358–375 T cell lines.
Fluorescent Microscopy
Images were obtained at the University of Utah School of Medicine Cell Imaging Facility. The CD4+ T cells were fixed in BD cytofix for 1hr at 4°C. The cells were washed 1× with BD pharmingen’s Stain Buffer [Dulbecco’s Phosphate Buffered Saline (DPBS), pH 7.4, 0.2% Bovine Serum Albumin, containing 0.09% Sodium azide]. CD4+ T cells (1 × 105 cells/50µl) were stained with 10µg/ml of MHC class II tetramers for 2 hrs. at 4°C, washed 2× in Stain Buffer, and imaged. Fluorescent images were obtained using a Nikon AR1 system with a 60× oil objective. The Emissions wavelength (EmW) and Excitation wavelength (ExW) for -PE (EmW/ ExW) was 595/561.2 and -APC (EmW/ ExW) was 700/635.8.
Results
Attenuated T Cell Proliferative Responses to Epitope Variants
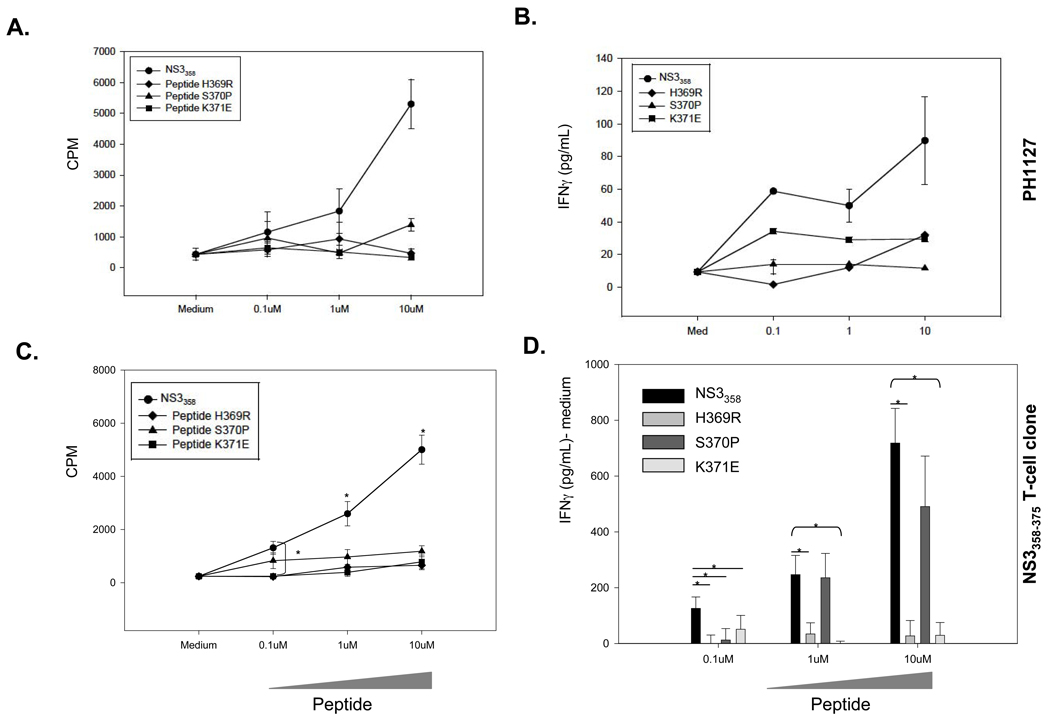
To determine if naturally occurring epitope variants are acting as APL and thus antagonizing or anergizing potential helper T cell responses, we used both PBMC polyclonal assays (Figure 1A,B) and T cell clones specific for NS3358–375. The T cell clones specific for NS3358–375 (Figure 1C,D) were derived from a resolved HCV patient PH1127 (HLA-DR15) who is viral load negative but antibody positive and recognizes a previously characterized immunodominant NS3358–375 epitope (Figure 1A, B) [31, 32]. Furthermore, previous work has demonstrated that the variants are able to bind to HLA-DR15 molecules [16, 33]. T cell clones derived from PH1127 (Figure 1A,B) are labeled as NS3358–375 T cell clones and these T cell clones have an attenuated T cell proliferative and IFNγ response to the variant peptides (Figure 1C, D). The inability of variant peptides to activate T cell proliferation and IFNγ secretion when incubated with variant peptides in both polyclonal and T cell clone assays led us to investigate if these variants were antagonizing the cognate T cell response.
Figure 1. Viral variants H369R, S370P, and K371E attenuate T cell proliferative and IFNγ responses.
A,C. NS3358–375 peptide has a dose-dependent T cell response and the variant peptides have an attenuated T cell response. Medium alone was used as a negative control. Results are shown in mean counts per minute (CPM) +/− standard error of triplicate cultures. B,D. Corresponding IFNγ response for polyclonal (B, PH1127) and NS3358–375 T cell clones (D). Results are shown pg/ml- medium +/− standard error of triplicate cultures. D. Medium was subtracted from the peptide-stimulated cultures for each sample. PBMC from PH1127 were cultured for 5 days and T cell clones were incubated for 72 hrs with irradiated (1 × 105 cells/well) PBMCs used as APCs at indicated peptide concentrations. *, P<0.05 as determined by student’s t-test. D. Results are representative of 3 independent experiments.
Variant Peptides Act as Antagonists
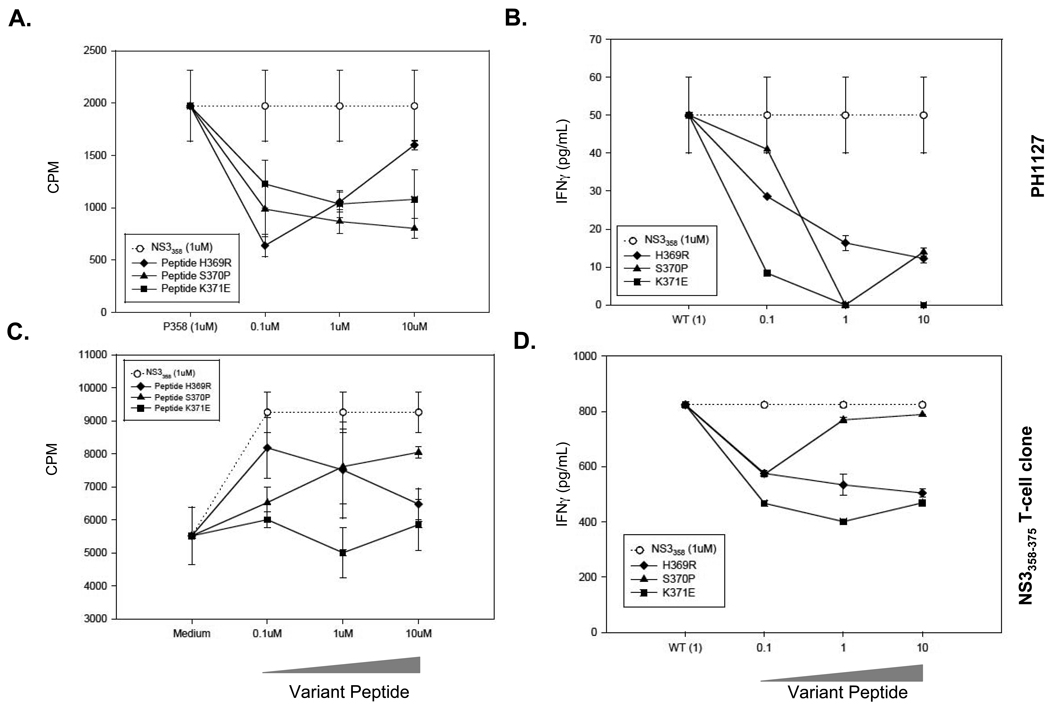
To determine if the variants could serve as antagonists, cells were prepulsed with 1µM NS3358–375 (open circle) and co-cultured in a dose-dependent manner with variant peptides (Figure 2). The variants effectively antagonize the T cell response in both the polyclonal (Figure 2A,B) and T cell clone assays (Figure 2C,D) in a dose-dependent manner (Figure 2). Furthermore, the IFNγ cytokine levels are attenuated in a similar fashion as the proliferative response (Figure 2B,D). Taken together, the viral variants act as an antagonist due to the lack of both proliferative and IFNγ response, when both variant and cognate peptides are present.
Figure 2. Variant peptides act as antagonists.
A, B. Polyclonal assays using PBMC from PH1127. A. PBMC were incubated with 1µM NS3358–375 for 3 hrs. and then washed. NS3358–375 variant peptides were added to the culture at indicated doses and incubated for 5 days. B. Supernatants from the polyclonal assays were collected at 48 hrs. from the cell culture and IFNγ levels were measured. C,D. T cell clone proliferative and INFγ responses. Antigen-presenting cells were prepulsed with NS3358–375 peptide for 3 hrs. washed, irradiated and cultured with NS3358–375 T cell clones with variant peptide for 72 hrs with irradiated autologous PBMCs used as APCs. The dotted line with open circle represents proliferation by NS3358–375 peptide prepulse alone. D. The supernatants from T358 were collected at 48 hrs. and IFNγ levels were measured. Each experiment was performed in triplicate.
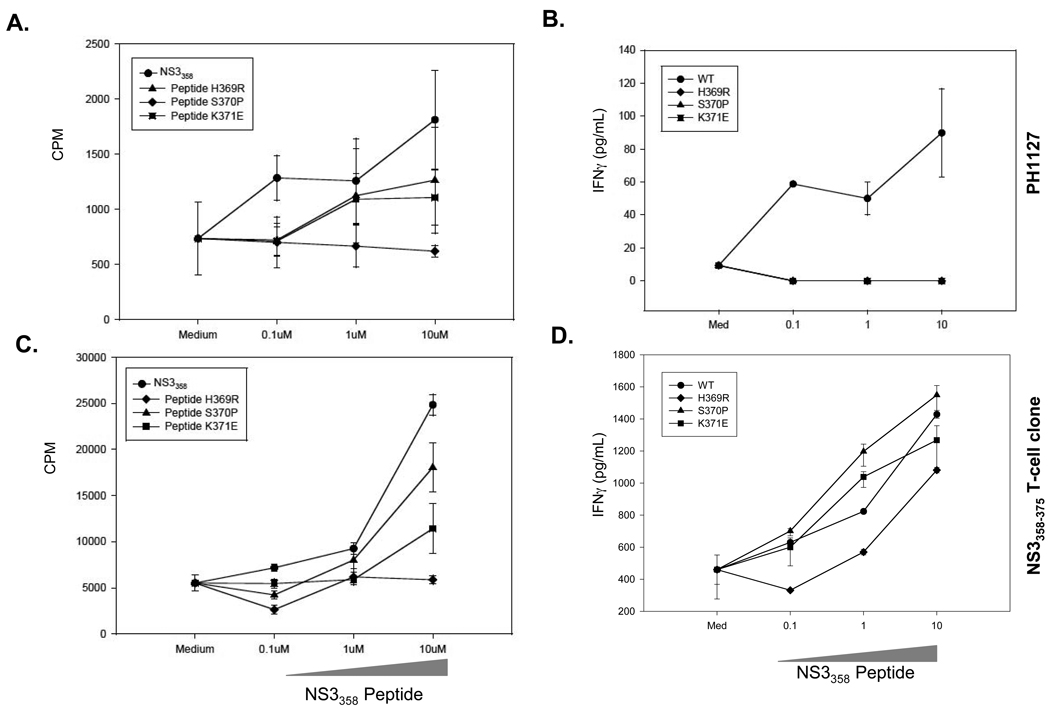
To assess if the variant peptides are anergizing the cognate T cell response as found in polyclonal assays (Figure 3A,B), APCs were prepulsed with 1µM variant peptides for 3 hrs. washed, irradiated, and cultured with T cell clones and cognate NS3358–375 (Figure 3C,D). H369R blunts both T cell proliferative and IFNγ response, but variants S370P and K371E act as partial-antagonists (Figure 3C,D). These results suggest that one variant is able to anergize T cells and variants S370P and K371E act as partial-antagonists.
Figure 3. Variants antagonize NS3358–375 specific T cells.
A–B. PBMC from PH1127 were prepulsed with indicated variant peptide and subsequently stimulated with NS3358–375 peptide in a dose dependent manner. C–D. APC cells were incubated with variant peptide for 3 hrs., washed, irradiated and added to NS3358–375 T cell clones with varying concentration of the cognate peptide. B,D. Supernatants were collected at 48 hrs. and the IFNγ levels were measured. Each experiment was performed in triplicate.
Viral Variant Tetramers bind to NS3358–375 T Cell Clones
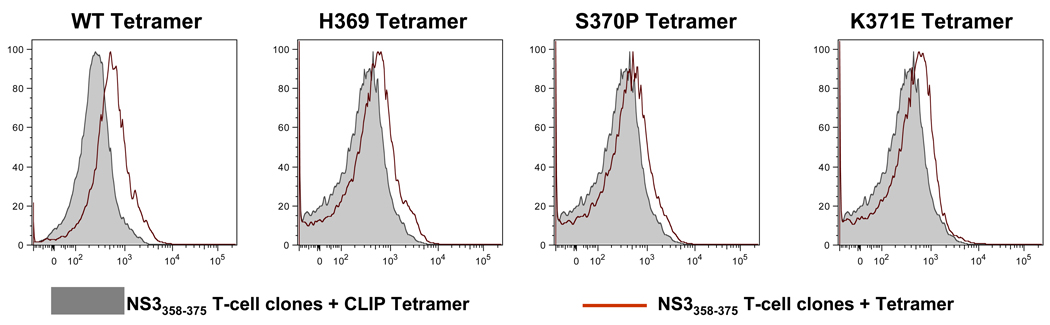
To establish viral variants capacity to bind to NS3358–375 T cells, T cell clones were stained with MHC class II tetramers (Figure 4). Tetramers loaded with each individual peptide were used to stain the NS3358–375 T cell clone. All three variant tetramers were able to bind to the NS3358–375 T cell clone (Figure 4, red histogram). The ability of these MHC class II tetramers to bind to the same T cell clones as the NS3358–375 suggests that these variants are able to not only bind to the same TCR, but also to antagonize antigen-specific CD4+ T cells.
Figure 4. Variant MHC class II tetramers bind to NS3358 T cell clones.
Histograms of MHC class II tetramer (red) staining. CLIP loaded DR15 tetramer was used as a control (gray). The control for the variant tetramer was performed at the same time for the variant stains.
MHC II S370P Tetramer Stains the Same Cell as MHC II NS3358–375 Tetramer
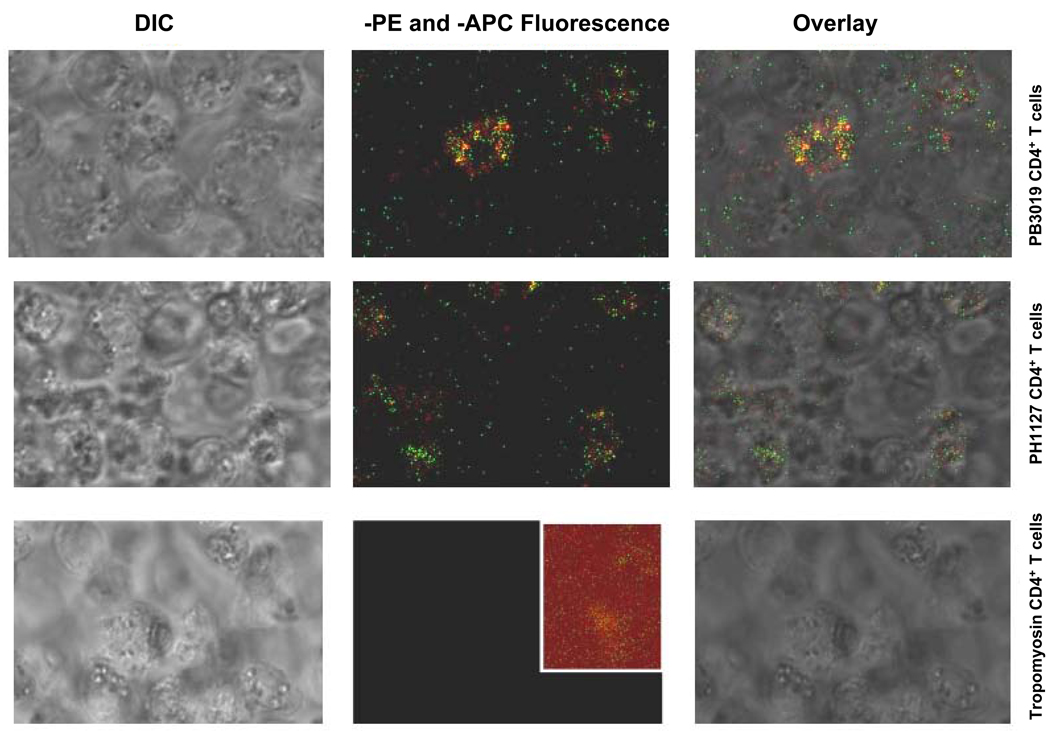
To solidify if variant S370P was binding to the same T cells as the NS3358–375 tetramer, we performed MHC class II tetramer staining on NS3358–375-specific CD4+ T cells. NS3358–375 and S370P tetramers were added to the indicated CD4+ T cell line in tandem (Figure 5). The NS3358–375 tetramer appears to stain the majority of T cells but the S370P tetramer does not individually stain the cells. Instead the variant co-localizes with the NS3358–375 tetramer (Figure 5). CD4+ Tropomyosin-specific T cells were used as a control for the tetramers (Figure 5, last column). The CD4+ Tropomyosin-specific T cells were stained at the same time and with the same tetramers as PH1127 and PB3019 T cells (Figure 5, third row). These results show that variant S370P is able to bind to the same TCR as the cognate peptide, albeit at a lower avidity in comparison to the NS3358–375 tetramer.
Figure 5. NS3358–375 and S370P MHC class II tetramers are able to bind to CD4+ NS3358–375 T cells.
The fluorescent images were all taken using identical confocal settings including laser power, emission filters, pinhole, and scan speed. NS3358–375 specific CD4+ T cells (Materials and Methods) were stained with both NS3358–375-PE (green) and S370PAPC (red) MHC class II tetramers at 10µg/ml. NS3358–375 specific CD4+ T cells from PB3019 and PH1127 are double positive for NS3358–375 and S370P tetramers (rows one and two). CD4+ Tropomyosin-specific T cells were used as a control (third row). The inset (row 3, panel 2) has been multiplied by three to reveal the level of background for comparison to the experimental panels shown in the same column.
Discussion
We demonstrate that variant epitopes modulate in vitro immune responses to the cognate peptide. Our results suggest that naturally occurring variants within a protective immunodominant epitope may act as APLs, leading to changes in the quality of T cell responses, which could allow for viral persistence.
This viral persistence seems relevant considering that HCV is able to modulate the immune response in an antigen-specific manner. This was shown by the fact that the variants were able to attenuate the response to NS3358–375 in antagonism assays. Each variant showed unique effects on the cognate NS3358–375 response that ranged from rendering the T cells unresponsive or anergic by H369R to partial-antagonism by S370P and K371E. By comparison, work by Scotta et al. [21] studied the effects of hypervariable region 1 variants in a CD4+ T cell epitope and found that the antagonists were able to induce apoptosis. We observed no evidence of induced programmed cell death, as Annexin-V/PI staining in polyclonal assays showed no difference in cultures treated with variants alone or in combination with cognate NS3358–375 (data not shown). Interestingly, our lab had shown that variant S370P was found to be stable in a chronic HCV patient for over 2 years, consistent with selection and fixation of this variant [15]. In the current study, S370P was still able to induce a proliferative and IFNγ response, albeit at lower levels than the NS3358–375 peptide, and was able to bind specifically to T cell clones specific for cognate peptide in an individual who was able to clear HCV. It seems that S370P is able to act on the in vitro response to HCV through varied suppressive mechanisms [34, 35]. One such mechanism of suppression is Tregs, in that, preincubating PBMC with S370P increases phenotypic markers for Tregs [19]. Expanding these findings to a larger cohort of subjects, the Treg markers are found at significantly higher levels in PBMC from both resolved and chronically infected patients; we were able to show the antigen-specific induction of Tregs [19]. Taken together, it appears that Treg induction in the periphery of HCV patients could be the result of viral variants inhibiting viral clearing T cells from reaching a level of activation that is necessary to maintain a sustained CD4+ T cell viral clearing response. Similarly, Gottschalk et al. [36] demonstrates an upregulation of Foxp3 in vivo with variant peptide and low doses of cognate peptide and they determined that by decreasing the “density” of the TCR ligand leads to a partially activated T cell and induction of Foxp3. The role of H369R and K371E has not been determined but recently we have demonstrated that these two variants do not increase Foxp3 expression in PBMC stimulated cultures in comparison to S370P stimulated PBMC and further work needs to be done in order to determine the avidity of these variants to the TCR [19].
A defining feature of APLs is the ability of the antagonist peptide, coupled to pMHC, to bind to the TCR. In the current study, we used MHC class II tetramers to determine whether variant peptides coupled to MHC were able to bind to T cell clones specific for NS3358–375 and possess the same TCR. The variants were able to bind to the T cell clones, suggesting that this has an effect on the biological outcome of the T cell. These results do not clearly demonstrate the kinetics of the pMHC-TCR and is currently being investigated. Although tetramer staining addresses the ability of the variants to bind to the same TCR as the NS3358–375 tetramer, further investigation needs to be performed to determine if markers of antagonism are upregulated, such as the phosphatase SHP-1 [37].
Using cloned T cells specific for the MHC class II epitope NS3358–375 peptide and previously identified naturally occurring APLs within a protective immunodominant epitope, suggests that changes in TCR activation may be used by HCV to deviate or blunt an HCV-specific T cell response [16]. These results provide an explanation for the attenuated or missing CD4+ T cells in chronically infected patients [26, 27]. Although these results suggest that HCV is exploiting TCR signaling for viral persistence, further investigation into APLs effect on the differentiation of these antigen-specific CD4+ T cells is necessary to understand HCV pathogenesis.
Acknowledgements
This work was supported by the National Institutes of Health [5R01AI047347-11]. The authors thank Drs Matthew Williams, Robert Fujinami, Curt Hagedorn, and Matthew Mulvey for helpful discussions along with Jane Libbey for critical feedback of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by alpha beta T cell receptors. Annual Review of Immunology. 1998;16:523. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 2.McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci U S A. 1995;92(11):5042. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sloan-Lancaster J, Allen PM. Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu Rev Immunol. 1996;14:1. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Zajac AJ, Murali-Krishna K, Blattman JN, Ahmed R. Therapeutic vaccination against chronic viral infection: the importance of cooperation between CD4+ and CD8+ T cells. Curr Opin Immunol. 1998;10(4):444. doi: 10.1016/s0952-7915(98)80119-2. [DOI] [PubMed] [Google Scholar]
- 5.Gerlach JT, Diepolder HM, Jung MC, Gruener NH, Schraut WW, Zachoval R, Hoffmann R, Schirren CA, Santantonio T, Pape GR. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology. 1999;117(4):933. doi: 10.1016/s0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 6.Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302(5645):659. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 7.Day CL, Lauer GM, Robbins GK, McGovern B, Wurcel AG, Gandhi RT, Chung RT, Walker BD. Broad specificity of virus-specific CD4+ T-helper-cell responses in resolved hepatitis C virus infection. J Virol. 2002;76(24):12584. doi: 10.1128/JVI.76.24.12584-12595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckels DD, Wang H, Bian TH, Tabatabai N, Gill JC. Immunobiology of hepatitis C virus (HCV) infection: the role of CD4 T cells in HCV infection. Immunol Rev. 2000;174:90. doi: 10.1034/j.1600-0528.2002.017403.x. [DOI] [PubMed] [Google Scholar]
- 9.Mueller M, Spangenberg HC, Kersting N, Altay T, Blum HE, Klenerman P, Thimme R, Semmo N. Virus-specific CD4+ T cell responses in chronic HCV infection in blood and liver identified by antigen-specific upregulation of CD154. J Hepatol. 2010;52:800. doi: 10.1016/j.jhep.2009.12.038. [DOI] [PubMed] [Google Scholar]
- 10.Gruner NH, Gerlach TJ, Jung MC, Diepolder HM, Schirren CA, Schraut WW, Hoffmann R, Zachoval R, Santantonio T, Cucchiarini M, Cerny A, Pape GR. Association of hepatitis C virus-specific CD8+ T cells with viral clearance in acute hepatitis C. Journal of Infectious Diseases. 2000;181(5):1528. doi: 10.1086/315450. [DOI] [PubMed] [Google Scholar]
- 11.Chang KM, Thimme R, Melpolder JJ, Oldach D, Pemberton J, Moorhead-Loudis J, McHutchison JG, Alter HJ, Chisari FV. Differential CD4(+) and CD8(+) T-cell responsiveness in hepatitis C virus infection. Hepatology. 2001;33(1):267. doi: 10.1053/jhep.2001.21162. [DOI] [PubMed] [Google Scholar]
- 12.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. Journal of Experimental Medicine. 2001;194(10):1395. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes E, Lauer G, Walker B, Klenerman P. T cell failure in hepatitis C virus infection. Viral Immunology. 2002;15(2):285. doi: 10.1089/08828240260066233. [DOI] [PubMed] [Google Scholar]
- 14.Lauer GM, Barnes E, Lucas M, Timm J, Ouchi K, Kim AY, Day CL, Robbins GK, Casson DR, Reiser M, Dusheiko G, Allen TM, Chung RT, Walker BD, Klenerman P. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology. 2004;127(3):924. doi: 10.1053/j.gastro.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Eckels DD. Mutations in immunodominant T cell epitopes derived from the nonstructural 3 protein of hepatitis C virus have the potential for generating escape variants that may have important consequences for T cell recognition. J Immunol. 1999;162(7):4177. [PubMed] [Google Scholar]
- 16.Wang JH, Layden TJ, Eckels DD. Modulation of the peripheral T-Cell response by CD4 mutants of hepatitis C virus: transition from a Th1 to a Th2 response. Hum Immunol. 2003;64(7):662. doi: 10.1016/s0198-8859(03)00070-3. [DOI] [PubMed] [Google Scholar]
- 17.Fuller MJ, Shoukry NH, Gushima T, Bowen DG, Callendret B, Campbell KJ, Hasselschwert DL, Hughes AL, Walker CM. Selection-driven immune escape is not a significant factor in the failure of CD4 T cell responses in persistent hepatitis C virus infection. Hepatology. 2009 doi: 10.1002/hep.23319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Bian T, Merrill SJ, Eckels DD. Sequence variation in the gene encoding the nonstructural 3 protein of hepatitis C virus: evidence for immune selection. J Mol Evol. 2002;54(4):465. doi: 10.1007/s00239-001-0037-6. [DOI] [PubMed] [Google Scholar]
- 19.Cusick MF, Schiller JJ, Gill JC, Eckels DD. Hepatitis C virus induces regulatory T cells by naturally occurring viral variants to suppress T cell responses. Clin Dev Immunol. 2011:806061. doi: 10.1155/2011/806061. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frasca L, Del Porto P, Tuosto L, Marinari B, Scotta C, Carbonari M, Nicosia A, Piccolella E. Hypervariable region 1 variants act as TCR antagonists for hepatitis C virus-specific CD4+ T cells. J Immunol. 1999;163(2):650. [PubMed] [Google Scholar]
- 21.Scotta C, Tuosto L, Masci AM, Racioppi L, Piccolella E, Frasca L. Hypervariable region 1 variant acting as TCR antagonist affects hepatitis C virus-specific CD4+ T cell repertoire by favoring CD95-mediated apoptosis. J Leukoc Biol. 2005;78(2):372. doi: 10.1189/jlb.0804456. [DOI] [PubMed] [Google Scholar]
- 22.Bertoletti A, Sette A, Chisari FV, Penna A, Levrero M, De Carli M, Fiaccadori F, Ferrari C. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. 1994;369(6479):407. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 23.Klenerman P, Rowland-Jones S, McAdam S, Edwards J, Daenke S, Lalloo D, Koppe B, Rosenberg W, Boyd D, Edwards A, et al. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature. 1994;369(6479):403. doi: 10.1038/369403a0. [DOI] [PubMed] [Google Scholar]
- 24.Norris PJ, Stone JD, Anikeeva N, Heitman JW, Wilson IC, Hirschkorn DF, Clark MJ, Moffett HF, Cameron TO, Sykulev Y, Stern LJ, Walker BD. Antagonism of HIV-specific CD4+ T cells by C-terminal truncation of a minimum epitope. Mol Immunol. 2006;43(9):1349. doi: 10.1016/j.molimm.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert SC, Plebanski M, Gupta S, Morris J, Cox M, Aidoo M, Kwiatkowski D, Greenwood BM, Whittle HC, Hill AV. Association of malaria parasite population structure, HLA, and immunological antagonism. Science. 1998;279(5354):1173. doi: 10.1126/science.279.5354.1173. [DOI] [PubMed] [Google Scholar]
- 26.Cox AL, Mosbruger T, Lauer GM, Pardoll D, Thomas DL, Ray SC. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42(1):104. doi: 10.1002/hep.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rehermann B, Chang KM, McHutchison JG, Kokka R, Houghton M, Chisari FV. Quantitative analysis of the peripheral blood cytotoxic T lymphocyte response in patients with chronic hepatitis C virus infection. J Clin Invest. 1996;98(6):1432. doi: 10.1172/JCI118931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckels DD, Tabatabai N, Bian TH, Wang H, Muheisen SS, Rice CM, Yoshizawa K, Gill J. In vitro human Th-cell responses to a recombinant hepatitis C virus antigen: failure in IL-2 production despite proliferation. Human Immunology. 1999;60(3):187. doi: 10.1016/s0198-8859(98)00111-6. [DOI] [PubMed] [Google Scholar]
- 31.Diepolder HM, Gerlach JT, Zachoval R, Hoffmann RM, Jung MC, Wierenga EA, Scholz S, Santantonio T, Houghton M, Southwood S, Sette A, Pape GR. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71(8):6011. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabatabai NM, Bian TH, Rice CM, Yoshizawa, Gill J, Eckels DD. Functionally distinct T-cell epitopes within the hepatitis C virus non-structural 3 protein. Hum Immunol. 1999;60(2):105. doi: 10.1016/s0198-8859(98)00102-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhu F, Yang M, Eckels DD. Interactions between helper T-cell epitopes of hepatitis C virus. Vaccine. 2005;23(27):3572. doi: 10.1016/j.vaccine.2005.01.157. [DOI] [PubMed] [Google Scholar]
- 34.MacDonald AJ, Duffy M, Brady MT, McKiernan S, Hall W, Hegarty J, Curry M, Mills KH. CD4 T helper type 1 and regulatory T cells induced against the same epitopes on the core protein in hepatitis C virus-infected persons. J Infect Dis. 2002;185(6):720. doi: 10.1086/339340. [DOI] [PubMed] [Google Scholar]
- 35.Groux H, O'Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE, Roncarolo MG. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389(6652):737. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 36.Gottschalk RA, Corse E, Allison JP. TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. J Exp Med. 2010;207(8):1701. doi: 10.1084/jem.20091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wasserman HA, Beal CD, Zhang Y, Jiang N, Zhu C, Evavold BD. MHC variant peptide-mediated anergy of encephalitogenic T cells requires SHP-1. J Immunol. 2008;181(10):6843. doi: 10.4049/jimmunol.181.10.6843. [DOI] [PubMed] [Google Scholar]