Abstract
BACKGROUND & AIMS
In a genome-wide association study of patients being treated for chronic hepatitis C, 2 functional variants in ITPA that cause inosine triphosphatase (ITPase) deficiency were shown to protect against ribavirin (RBV)-induced hemolytic anemia during early stages of treatment. We aimed to replicate this finding in an independent cohort from the Study of Viral Resistance to Antiviral Therapy of Chronic Hepatitis C and to investigate the effects of these variants beyond week 4.
METHODS
Genetic material was available from 318 patients. The ITPA variants, rs1127354 (exon 2, P32T) and rs7270101 (intron 2, splice altering), were genotyped and tested for association with hemoglobin (Hb) reduction at week 4. An ITPase deficiency variable was defined that combined both ITPA variants according to documented effect on ITPase activity. We investigated the impact of ITPA variants on Hb levels over the course of therapy and on the need for RBV dose reduction.
RESULTS
The final analysis included 304 patients with genotype 1 hepatitis C virus (167 white patients and 137 black patients). The polymorphisms rs1127354 and rs7270101 were associated with Hb reduction at week 4 (P = 3.1 × 10−13 and 1.3 × 10−3, respectively). The minor alleles of each variant protected against Hb reduction. Combining the variants into the ITPase deficiency variable strengthened the association (P = 2.4 × 10−18). The ITPase deficiency variable was associated with lower rates of anemia over the entire treatment period (48 weeks), as well as a lower rate of anemia-related RBV dose reduction (hazard ratio, 0.52; P = .0037). No association with sustained virological response was observed.
CONCLUSIONS
Two polymorphisms that cause ITPase deficiency are strongly associated with protection from RBV-induced hemolytic anemia and decrease the need for RBV dose reduction.
Keywords: Pharmacogenomics, Genome-Wide Association Study, Polymorphism, Single Nucleotide Polymorphism, HCV, Adverse Event
Therapy for chronic hepatitis C is difficult to tolerate and associated with significant morbidity. Among the adverse effects of treatment, ribavirin (RBV)-induced hemolytic anemia (HA) is of particular importance because it is common and can necessitate early dose reductions that may affect efficacy. In the 2 phase 3 registration trials of pegylated interferon alfa (pegIFN-α) and RBV, dose modification for anemia was required in 9% to 22% of patients.1,2
A genome-wide association study of participants in the IDEAL study recently identified a strong association between quantitative hemoglobin (Hb) reduction at week 4 of treatment and the single nucleotide polymorphism rs6051702.3 Genotyping of known functional variants showed that the association signal was entirely explained by 2 functional variants in the ITPA gene (encoding inosine triphosphatase, ITPase) on chromosome 20: a missense variant in exon 2 (rs1127354, P32T) and a splice-altering single nucleotide polymorphism in intron 2 (rs7270101). Both polymorphisms had previously been well characterized and validated as functional variants in studies of patients with ITPase deficiency, a benign inherited enzymopathy in which inosine triphosphate (the substrate for ITPase) accumulates in red blood cells.4–7 Although the causal variants responsible for protection against RBV hemolysis were identified, the mechanism whereby ITPase deficiency confers protection is yet to be explained. The impact of these genetic variants on anemia beyond week 4 of therapy and the association between the variants and need for RBV dose reduction has not been defined.
We therefore sought to confirm and extend our understanding of this recently reported genetic association in an independent cohort of patients who participated in the Study of Viral Resistance to Antiviral Therapy of Chronic Hepatitis C (ViraHep-C).8 Specifically, we have replicated the association analysis between the functional ITPA variants and anemia at week 4 and investigated the impact on Hb levels over the course of therapy. Finally, we have explored whether knowledge of these genetic polymorphisms might affect clinical decision making by assessing the relationship between these variants and the need for RBV dose reduction.
Patients and Methods
Patients
Patients in this study were participants in ViraHep-C (clinicaltrials.gov, NCT00038974). This study was designed to compare the rates of response to pegIFN and RBV combination therapy for genotype 1 hepatitis C virus (HCV) infection between white patients and black patients.8 Patients consented to the provision of genetic material as part of the primary protocol, and a genetic biorepository linked to the clinical dataset was created at the National Institute for Diabetes and Digestive and Kidney Diseases (www.niddkrepository.org/niddk/home.do) using samples from 318 of the 401 participants. Official permission for access to both the clinical dataset and genetic material was obtained. Inclusion/exclusion criteria, as well as laboratory and viral testing protocols, have been described in detail previously.8 All patients were treated with pegIFN-α-2a (180 µg/wk) and RBV (1200/1000 mg/day according to body weight of 75 kg or less), and virological responders continued to 48 weeks of treatment.8 Patients in whom a qualitative serum HCV RNA test result was positive at week 24 were considered nonresponders, and therapy was stopped. Specific dose adjustment guidelines for pegIFN and RBV were included in the study protocol. In brief, RBV dose reduction was from 1200 to 800 mg/day or 1000 to 600 mg/day for Hb levels of 8.5 to 10.0 g/dL. When the Hb level decreased to less than 8.5 g/dL, RBV therapy was stopped permanently. For nonhematologic RBV toxicity, stepwise dose reduction of RBV was performed in decrements of 200 mg/day to the lowest dose of 400 mg/day. Protocol-specified pegIFN dose reductions were mandated if neutropenia occurred.8 Physician-prescribed changes to study medication doses throughout the treatment period, as well as patient discontinuation from treatment, were recorded as part of the original study. Growth factors were not part of the study protocol. Two patients included in this study were treated with erythropoietin off-protocol at the discretion of individual investigators. Growth factor was started after week 12 in both patients, following the first dose reduction. They were therefore included in the analysis of week 4 and week 12 Hb reduction, as well as time to first RBV dose reduction.
Genotyping
A total of 318 patients were genotyped at polymorphic sites rs1127354 and rs7270101 on chromosome 20 using the ABI TaqMan allelic discrimination kit and the ABI7900HT Sequence Detection System (Applied Biosystems, Carlsbad, CA) as previously described.3 The possible genotypes for each biallelic polymorphism are as follows: rs1127354: C/C, A/C, A/A (minor allele = A); rs7270101: A/A, A/C, C/C (minor allele = C). The cohort was also tested for the IL28B polymorphism rs12979860, recently identified to be associated with pegIFN/RBV treatment response.9
Definition of an ITPase Deficiency Variable According to rs1127354/rs7270101 Genotypes
Severity of ITPase deficiency was defined according to previous studies3–7 (Table 1).
Table 1.
ITPase Deficiency Variable
| rs1127354 | rs7270101 | Predicted ITPase activity |
Predicted ITPase deficiency |
|---|---|---|---|
| Wild-type (C/C) | Wild-type (A/A) | 100% | − |
| Wild-type (C/C) | Heterozygosity (A/C) | 60% | + |
| Heterozygosity (C/A) | Wild-type (A/A) | 30% | + + |
| Wild-type (C/C) | Homozygosity (C/C) | 30% | + + |
| Heterozygosity (C/A) | Heterozygosity (A/C) | 10% | + + + |
| Homozygosity (A/A) | Wild-type (A/A) | <5% | + + + |
NOTE. Definition of an ITPase deficiency variable according to rs1127354/rs7270101 genotypes: severity of ITPase deficiency was predicted as absent, representing wild-type activity (−) and mild (+), moderate (+ +), or severe (+ + +) deficiency according to previous studies (Supplementary Table 1). ITPase activity was defined by the accumulated presence of minor alleles at the respective polymorphic sites. Note that only 3 haplotypes are observed (the minor allele for both single nucleotide polymorphisms never occurs together on the same chromosome).3
Definition of Clinical End Points
For replication of the primary genome-wide association study discovery, we analyzed Hb reduction at week 4. Two primary end points were considered: (1) absolute Hb reduction as a continuous trait and (2) Hb reduction >3 g/dL. Secondary end points were (1) Hb reduction over the course of 48 weeks of therapy, defined both qualitatively and quantitatively (reduction >3 g/dL); (2) the need for RBV dose reduction because of anemia; and (3) rate of sustained virological response (SVR).
Statistical Analysis
For descriptive statistics, continuous variables were summarized as median (25th to 75th percentiles). Categorical variables were described as frequency and percentage. Comparisons between groups for demographic, clinical, and virological data were performed using a Wilcoxon test for skewed continuous data or the χ2 test/Fisher exact test for categorical data. Significance was defined at P < .05. Association between the individual polymorphisms/ITPase deficiency variable and the anemia phenotypes was tested using single-marker genotype trend tests of association in linear or logistic regression models. Multivariable regression models with backward selection were used to identify independent predictors of anemia. A significance level of .05 was used for removal from the model. Covariates included age, sex, weight (kg), METAVIR fibrosis stage on pretreatment liver biopsy, baseline Hb level, baseline serum creatinine level, and RBV dose (1000/1200 mg). The relationship between the ITPase-deficient phenotype and the need for RBV dose reduction was tested in a multivariable survival analysis. Each predictor was first tested by univariable analysis, considering the tests of equality across strata to select the predictors in the final model. For the categorical variables, the log-rank test of equality across strata was used. For the continuous variables, we used a univariable Cox proportional hazards model. The predictor was included in the final survival analysis if the univariable test had a P value of <.2. All statistical analyses were performed in SAS version 9.2 (SAS Institute, Cary, NC); Stata version 10.1 (StataCorp LP, College Station, TX) was used to generate survival curves.
Results
Patient Characteristics
The clinical characteristics of the study population are described in Table 2. A total of 304 patients had DNA available and complete clinical datasets (ITPA genotyping was not successful in 2 patients; week 4 Hb data were missing in 12 patients). A total of 167 (55%) were white, and 137 (45%) were black. Black patients in this cohort were heavier, had a lower baseline Hb level, and had a higher baseline creatinine level compared with white patients. Black patients had lower baseline alanine aminotransferase levels but had similar liver histology scores for METAVIR inflammatory grade and fibrosis stage. HCV RNA levels were similar in the 2 ethnic groups at baseline.
Table 2.
Clinical Characteristics of the Study Population
| Overall | White patients | Black patients | P value (white patients vs black patients) | |
|---|---|---|---|---|
| n | 304 | 167 | 137 | |
| Male sex, n (%) | 198 (65) | 111 (67) | 87 (64) | .5896 |
| Age (y) | 48.5 (44–53) | 48 (43–52) | 49 (46–53) | .1242 |
| Baseline weight (kg) | 85 (75–98) | 83 (73–95) | 89 (79–102) | .0009 |
| Body mass index (kg/m2) | 28.4 (25.2–32.6) | 27.6 (24.5–31.6) | 28.8 (26.6–34.2) | .0013 |
| Hb (g/dL) | 15.1 (14.2–15.9) | 15.4 (14.5–16.1) | 14.8 (13.8–15.5) | 2.9 × 10−5 |
| Serum creatinine (mg/dL) | 0.9 (0.8–1.0) | 0.9 (0.8–1.0) | 1.0 (0.8–1.1) | .0003 |
| Alanine aminotransferase (IU/mL) | 66 (46–105) | 74 (52–139) | 59.5 (41–83) | 6.2 × 10−5 |
| HCV RNA level (log10IU/mL) | 6.5 (5.7–6.8) | 6.5 (5.6–6.8) | 6.4 (5.7–6.7) | .2973 |
| METAVIR activity, n (%) | ||||
| A1 | 61 (20) | 36 (22) | 25 (18) | .7578 |
| A2 | 83 (27) | 44 (26) | 39 (28) | |
| A3 | 160 (53) | 87 (52) | 73 (53) | |
| METAVIR fibrosis, n (%) | ||||
| F0–2 | 250 (82) | 137 (82) | 113 (82) | .9194 |
| F3–4 | 54 (18) | 30 (18) | 24 (18) |
NOTE. Values are expressed as median (25th to 75th percentiles) unless otherwise specified.
Population Distribution of ITPA Gene Variants
The 2 polymorphisms, rs1127354 and rs7270101, were genotyped in the study population (Table 3). The minor allele (A) frequency for rs1127354 was 0.07, and the population distribution was in Hardy–Weinberg equilibrium (P = .22). The minor allele frequency for rs7270101 was 0.09, again in Hardy–Weinberg equilibrium (P = .73). Genotype distributions were not significantly different between white and black patients for either polymorphism. The 2 genotypes were then used to define the ITPase deficiency variable; a majority (70%) of patients were predicted to have wild-type ITPase activity, but a significant minority was predicted to have reduced ITPase activity (Table 3). There was no significant difference in population distribution of the ITPase deficiency variable between white and black patients.
Table 3.
Population Distribution of the ITPA Variants
| Overall | White patients | Black patients |
P value (white patients vs black patients) |
||||
|---|---|---|---|---|---|---|---|
| rs1127354 | n | % | n | % | n | % | |
| CC | 264 | 86.8 | 142 | 85.0 | 122 | 89.0 | .3201 |
| CA | 40 | 13.2 | 25 | 15.0 | 15 | 11.0 | |
| AA | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| rs7270101 | |||||||
| AA | 251 | 82.6 | 133 | 79.6 | 118 | 86.1 | .1823 |
| AC | 51 | 16.8 | 32 | 19.2 | 19 | 13.9 | |
| CC | 2 | 0.7 | 2 | 1.2 | 0 | 0.0 | |
| Predicted ITPase deficiency | |||||||
| − | 212 | 69.7 | 109 | 65.3 | 103 | 75.2 | .2250 |
| + | 50 | 16.5 | 31 | 18.6 | 19 | 13.9 | |
| + + | 41 | 13.5 | 26 | 15.1 | 15 | 11 | |
| + + + | 1 | 0.3 | 1 | 0.6 | 0 | 0 | |
NOTE. The table shows rs1127354 and rs7270101 genotype prevalence within the study population and population distribution of predicted level of ITPase deficiency according to ITPA haplotype (Table 1).
ITPase Deficiency Protected Against Anemia at Week 4
The ITPA variants (rs1127354 and rs7270101) were both strongly associated with quantitative Hb reduction at week 4 in multivariable linear regression analysis, with the minor allele having a protective effect in each case (Tables 4 and 5). Furthermore, each functional variant was strongly and independently associated with protection in regression models that included the other ITPA variant, as observed by Fellay et al (Tables 4 and 5).3 The combined ITPase deficiency variable was also tested as a predictor of quantitative Hb reduction. The combined variable had the strongest association with protection against anemia at week 4 (Tables 4 and 5) and was estimated to explain 19% of the variability in quantitative Hb reduction at week 4 (comparing favorably with the study by Fellay et al, suggesting minimal “winner’s curse” effect in the original description [Supplementary Table 2]3). The composite variable was also most strongly associated with Hb reduction >3 g/dL (Tables 4 and 5).
Table 4.
ITPA Variants and ITPase Deficiency Are Associated With Reduction in Week 4 Hb Level
| Estimate | Standard error |
P value | Adjusted estimatea |
Standard error |
Adjusted P valuea |
|
|---|---|---|---|---|---|---|
| ITPA variantb | ||||||
| rs1127354 | −1.64 | 0.21 | 3.12 × 10−13 | −1.77 | 0.21 | 1.53 × 10−15 |
| rs7270101 | −0.63 | 0.19 | 1.28 × 10−3 | −0.83 | 0.18 | 4.22 × 10−6 |
| ITPase deficiency variable | −0.89 | 0.09 | 2.35 × 10−18 |
NOTE. ITPA variants were strongly and independently associated with protection from quantitative Hb reduction at week 4 of therapy in multivariable linear regression analyses. The composite ITPase deficiency variable was more strongly associated with protection from week 4 quantitative Hb reduction than either individual ITPA variant in a multivariable linear regression model.
Adjusted estimates and P values were calculated in models in which the other functional variant was already included.
Covariables rs1127354 and/or rs7270101, or ITPase defciency variable, plus ethnicity (white vs black), age (years), sex, baseline Hb level (g/dL), baseline creatinine level (mg/dL), METAVIR F3–4 vs F0–2, and RBV starting dose (mg/kg) were considered. Backward selection was used to remove variables from the model. A P value of .05 was used to remove variables from the final model.
Table 5.
The Composite ITPase Deficiency Variable Was Strongly Associated With Protection From Hb Reduction >3 g/dL at Week 4 of Therapy in a Multivariable Logistic Regression Model
| OR | 95% CI | P value | Adjusted ORa | 95% CI | Adjusted P valuea | |
|---|---|---|---|---|---|---|
| ITPA variantb | ||||||
| rs1127354 | 0.03 | 0.004–0.22 | .0006 | 0.02 | 0.003–0.18 | .0003 |
| rs7270101 | 0.54 | 0.28–1.06 | .07 | 0.38 | 0.19–0.76 | .006 |
| ITPase deficiency variable | 0.26 | 0.15–0.43 | 2.7 × 10−7 |
NOTE. The 2 ITPA variants are shown to be strongly and independently associated with protection from Hb reduction >3 g/dL at week 4. The strength of association with the ITPase deficiency variable is shown to be greater than for either ITPA variant alone. Supplementary Table 2 presents the analyses stratified by ethnicity.
Adjusted OR and independent P values were calculated in models in which the other functional variant was already included.
Covariables rs1127354 and/or rs7270101, or ITPase defciency variable, plus ethnicity (white vs black), age (years), sex, baseline Hb level (g/dL), baseline creatinine level (mg/dL), METAVIR F3–4 vs F0–2, and RBV starting dose (mg/kg) were considered. Backward selection was used to remove variables from the model. A P value of .05 was used to remove variables from the final model.
There was no association between the ITPA variants or the ITPase deficiency variable and baseline Hb level (data not shown). No significant association was observed between either the ITPA variants or the ITPase deficiency variable and week 4 reductions in neutrophil or platelet count (data not shown).
ITPase Deficiency Was Associated With Protection Against Anemia Throughout the Course of Anti-HCV Therapy
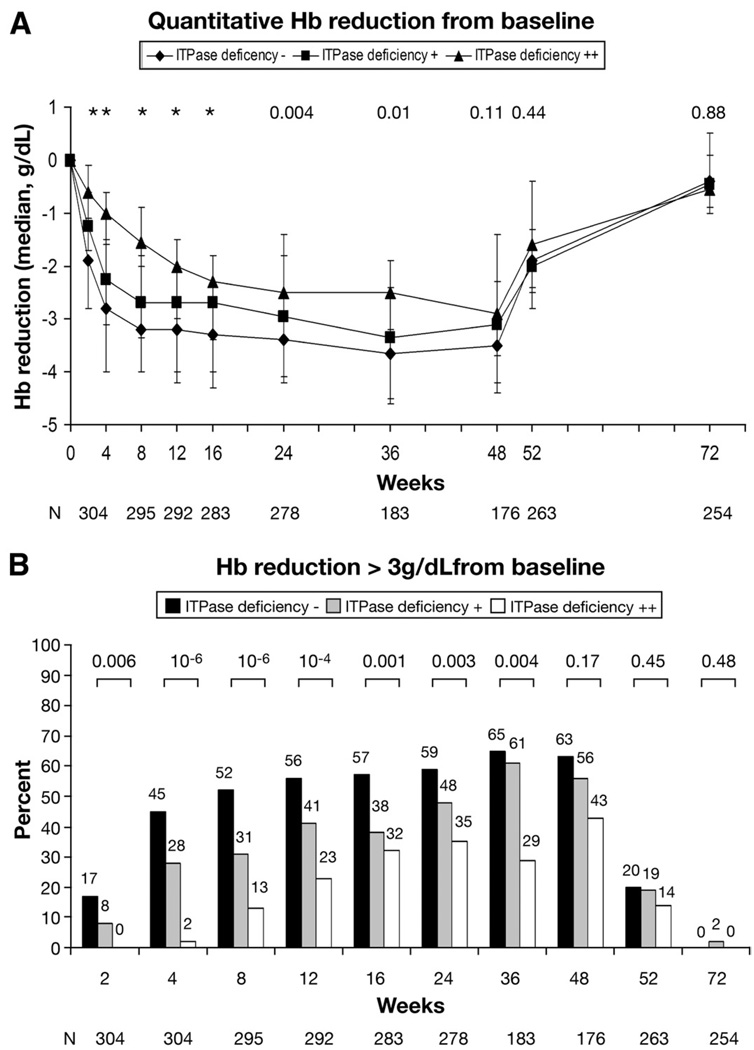
The association between the ITPase deficiency variable and anemia was evaluated throughout the course of 48 weeks of treatment. ITPase deficiency was associated with delayed kinetics of Hb decline during the first 12 weeks and less absolute reduction in Hb over the full course of therapy until 48 weeks (Figure 1A). This translated to significant differences in the numbers of patients experiencing Hb reduction >3 g/dL (Figure 1B). Notably, 95 of 212 patients (45%) predicted to have wild-type ITPase activity had experienced >3 g/dL reduction of Hb by week 4 of therapy, compared with only 1 of 41 patients (2%) with predicted moderate ITPase deficiency (P = 1.4 × 10−6). The frequency of clinically significant anemia increased over time to week 24, reaching 59%, 48%, and 35% in high-, medium-, and low-risk patients, respectively (P = .003; Figure 1B), before significant patient drop out due to virological nonresponse.
Figure 1.
(A) Quantitative Hb reduction from baseline. The ITPase deficiency variable was associated with less absolute reduction in median Hb level at all time points on treatment. There was only 1 patient with severe (+ + +) ITPase deficiency, and this patient was excluded from the analysis. Data are presented as median and interquartile range. P values are listed above the relevant time points (*P ≤ .001). (B) Hb reduction >3 g/dL from baseline. The ITPase deficiency variable was protective against clinically significant reductions (>3 g/dL) in Hb throughout the course of pegIFN plus RBV combination therapy. There was only 1 patient with severe (+ + +) ITPase deficiency, and this patient was excluded from the analysis. P values for each time point are listed above the columns. ViraHep-C protocol involved a week 24 stopping rule for virological nonresponders, explaining the decreased cohort size after 24 weeks.
ITPase Deficiency Was Associated With Lower Rates of RBV Dose Reduction
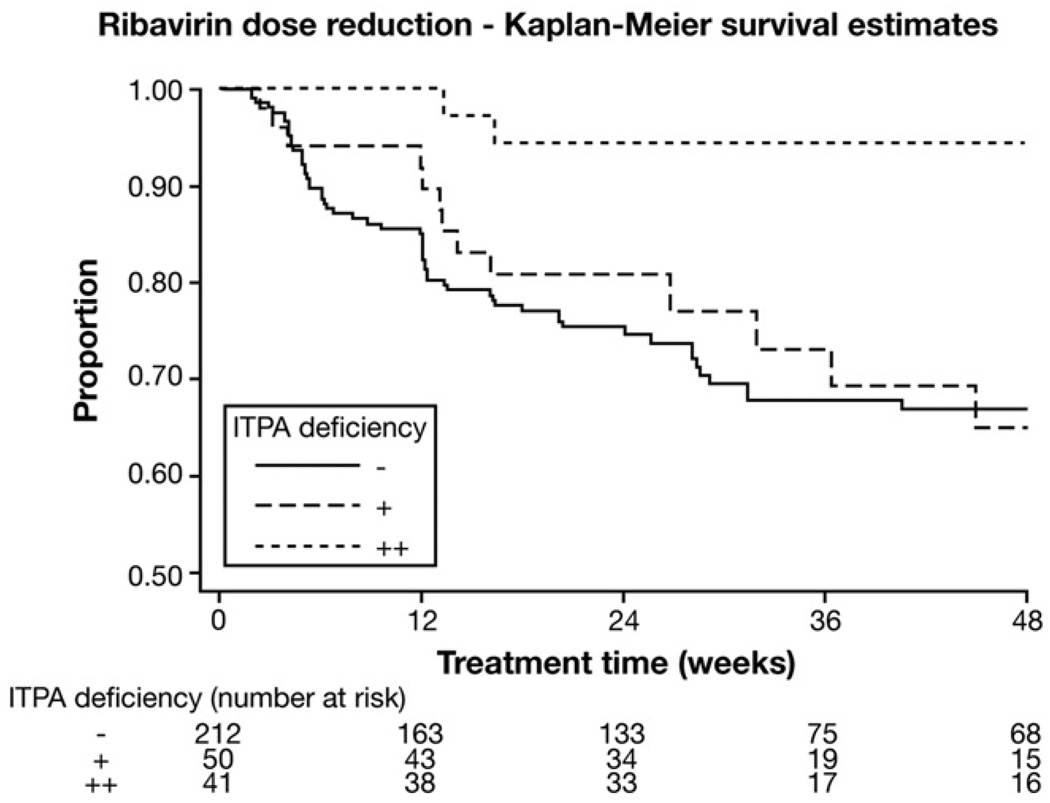
We first considered RBV dose reduction, defined as any patient in whom RBV dosing was decreased (step-down or discontinuation). Dose reduction was common in the ViraHep-C cohort (140/304; 46%). Anemia was the indication for dose reduction in only 73 of 140 patients (52%). In contrast, 37 (27%) had dose reduction because of other adverse events and 30 (21%) per the patient’s preference (as per the ViraHep-C database). When the relationship between the ITPase deficiency variable and RBV dose reduction for any reason (at any time in the study) was considered, a protective trend only was noted (odds ratio, 0.74; 95% confidence interval [CI], 0.53–1.01). However, there was a significant protective benefit when anemia was the indication for dose reduction (odds ratio, 0.53; 95% CI, 0.33– 0.84; P = .006). The association between the ITPase deficiency variable and time to first anemia-related RBV dose reduction was explored in greater detail by survival analysis (Figure 2), showing that RBV dose reductions were less frequent and delayed in the setting of ITPase deficiency. The Cox proportional hazards model confirmed an independent relationship with the risk for anemia-related RBV dose reduction over time, after adjustment for possible confounders including ethnicity (white vs black), age, baseline weight, baseline Hb and creatinine levels, METAVIR fibrosis stage, and RBV starting dose (ITPase deficiency variable: hazard ratio, 0.52; 95% CI, 0.34 –0.81; P = .0037; Table 6). Finally, we also considered the association between ITPase deficiency and whether patients received at least 80% of the planned maximal RBV dose, recognized as a key threshold for maximizing treatment response.10 In the overall cohort, adjusting for all RBV dose modifications, patients with moderate to severe predicted ITPase deficiency were significantly more likely to receive >80% of the planned RBV dose (82% vs 64%; P = .0281).
Figure 2.
Survival analysis of ribavirin dose reduction. Survival analysis of time to first dose reduction (indication = anemia) according to ITPase deficiency variable, showing that mild or moderate ITPase deficiency protects against the need for RBV dose reduction. Censor criteria: 1 = dose reduction for anemia, 0 = last follow-up or date of dose reduction for an indication not related to anemia.
Table 6.
Multivariable Cox Proportional Hazards Model for Time to Anemia-Related RBV Dose Reduction
| Variable | Parameter estimate | Standard error | P value | Hazard ratio | Hazard ratio 95% CI |
|---|---|---|---|---|---|
| ITPase deficiency variable | −0.65 | 0.22 | .0037 | 0.521 | 0.34–0.81 |
| Age (y) | 0.04 | 0.01 | .0104 | 1.037 | 1.01–1.07 |
| RBV starting dose (mg/kg) | 0.18 | 0.06 | .0032 | 1.194 | 1.06–1.34 |
| Baseline Hb (g/dL) | −0.74 | 0.10 | <.0001 | 0.478 | 0.40–0.58 |
NOTE. Multi-variable Cox proportional hazards model considering the following covariables: ITPase deficiency variable, baseline Hb level (g/dL), sex, age (years), and RBV starting dose (mg/kg). Sex was removed by backward selection (a P value of .05 was used for removal from the model). Ethnicity, baseline serum creatinine level (mg/dL), and fibrosis stage (METAVIR F3–4 vs F0–2) were not significantly associated with time to first RBV dose reduction in univariable analyses.
When dose discontinuation alone was considered, of the 51 patients who prematurely discontinued RBV, only 12 stopped treatment due to anemia; 17 stopped treatment for other adverse events, and 22 stopped per the patient’s preference. It was notable that all 12 patients had either wild-type ITPase activity (n = 7) or mild ITPase deficiency (n = 5); no patients with moderate to severe ITPase deficiency (0/42) had treatment stopped because of anemia. The low event number limited any statistical analysis of the relationship between the ITPase deficiency variable and RBV discontinuation. There was no significant association between the ITPA variants or the ITPase deficiency variable and pegIFN dose reduction or discontinuation (data not shown).
ITPase Deficiency and Treatment Outcome
Rs1127354, rs7270101, and the combined variable were tested for association with treatment outcome, both SVR and week 4 rapid virological response, in logistic regression models that included age, sex, ethnicity, baseline HCV RNA level, baseline alanine aminotransferase level, METAVIR fibrosis stage, and RBV dose per kilogram body weight. No association with either SVR or rapid virological response was observed (Supplementary Tables 3 and 4). This was true both for the overall cohort and a subset analysis that excluded patients in whom RBV was dose reduced for indications other than anemia. The regression analyses were repeated, including the IL28B single nucleotide polymorphism rs12979860 as a covariate, recently identified to be an important predictor of viral clearance in response to pegIFN and RBV therapy.9,11,12 Again, the ITPA variants did not associate with treatment outcome in the overall cohort or when stratified analyses were run in subgroups according to IL28B genotype (data not shown). IL28B genotype was strongly and independently associated with SVR in this cohort, as recently reported (data not shown).13 Finally, the ITPA variants were not associated with SVR in models stratified according to rapid virological response.
Discussion
To our knowledge, this is the first study to replicate the recent discovery of ITPA genetic variants that protect against RBV-induced HA in an independent cohort of patients from ViraHep-C. We provide the first description of a strong association between ITPase deficiency and Hb reduction over the entire 48-week course of pegIFN and RBV therapy for genotype 1 HCV. We also address the clinical implications of these findings by reporting the protective benefit of the ITPA variants against RBV dose reduction.
Almost one-third of patients (30%) were identified to have moderate to severe ITPase deficiency and to be relatively protected from the hemolytic toxicity of RBV. Patients with moderate to severe ITPase deficiency were least likely to become anemic. The effect was most evident early in treatment but persisted throughout. Further, this protective effect translated into fewer RBV dose reductions and greater cumulative RBV exposure. Indeed, the greater RBV dose reduction in the patients with normal ITPase activity likely explains the convergence of the Hb curves over time. The use of erythropoietin was not allowed by the ViraHep-C protocol, so the issue of whether these variants predicted need for growth factor support could not be directly addressed, but it is logical to expect that ITPase deficiency would be very relevant.
It was interesting that despite this strong protective effect against anemia, and less need for RBV dose reduction, no benefit in terms of treatment outcome was observed. There are a number of possible explanations. Firstly, it is the minor alleles that are protective and only a minority of patients are protected against anemia. Thus, we were most likely underpowered to observe an effect on SVR. Secondly, there was a high incidence of RBV dose reduction and discontinuation for reasons other than anemia in ViraHep-C, which may have obscured any effect. Thirdly, we are assuming that RBV dose reduction is the only mechanism whereby ITPA variants might increase the rate of SVR. However, if these genetic variants affect RBV pharmacodynamics outside the erythrocyte, it may be that ITPase deficiency also reduces antiviral efficacy, despite protecting against anemia. This would be consistent with the fact that no increase in SVR rate was observed, despite maintenance of RBV dosing, and with the recent observation that early-onset anemia (likely to lead to dose reduction) was associated with an increased rate of SVR in a large study that permitted the use of erythropoietin.14 Finally, it has also been suggested that the impact of RBV dose reduction on the likelihood of SVR may be negligible, as long as RBV is not discontinued.15,16 Therefore, the relationship among anemia, ITPA variants, and SVR is complicated, and there may be opposing mechanisms involved that now require further investigation.
ITPase deficiency causes the accumulation of inosine triphosphate within red blood cells and is generally believed to be a benign condition,4–7 although it has been associated with thiopurine toxicity.17,18 RBV is metabolized within the red cells to RBV triphosphate, and it is the accumulation of RBV triphosphate that is believed to lead to oxidative damage to the red cell membrane and erythrophagocytic extravascular destruction.19 Whether high levels of inosine triphosphate restrict the conversion and accumulation of RBV triphosphate within red blood cells is not known, and the mechanism whereby ITPase deficiency, or inosine triphosphate accumulation, protects against RBV hemolysis is yet to be resolved. Direct investigation of the relationship between red cell ITPase activity, using functional assays, and accumulation of RBV/RNB metabolites, and hemolysis, will be required.
There are many genetic variants that have been shown to influence the pharmacokinetics of drugs but that nevertheless fail to have any well-documented impact on clinical decision making, even at the level of dose reduction.20 In contrast, the ITPA variants described here are shown to be strongly associated with differential sensitivity to RBV-induced anemia as well as the need for dose reduction during treatment, meaning that they join a small set of pharmacogenetic variants that have been documented to influence clinical management.21,22
This finding has the potential to inform clinical decision making. Patients with normal ITPase activity require particularly vigilant monitoring of Hb levels early in therapy. Indeed, it has been shown that a decrease in Hb of 1.5 g/dL during the first 2 weeks of treatment is predictive of the development of severe anemia.23 Early intervention in this group with RBV dose reduction and/or growth factor support may be indicated to maximize safety and minimize premature discontinuation of RBV. Conversely, the smaller subgroup of patients, protected from anemia, could be monitored less frequently. Prospective studies will be required to investigate the clinical utility, as well as the cost-effectiveness, of these approaches. Patients who are ITPase deficient may also be candidates for more aggressive RBV dose escalation strategies, because higher-dose RBV has been associated with higher rates of SVR.24 This will require evaluation in randomized studies. Separation of patient subgroups based on the predisposition to develop RBV-induced anemia may also allow clinicians to consider therapy despite comorbidities that were previously considered relative contraindications, such as renal impairment or mild to moderate coronary artery disease (where the risks of developing significant anemia and its effect on the underlying disease are of concern). Whether genetic testing can expand treatment options for these complex patients must be prospectively investigated. Finally, a number of the new HCV direct antiviral drugs that are currently under investigation in combination regimens with pegIFN and RBV have been associated with increased rate of anemia (telaprevir, boceprevir) and frequent need for erythropoietin use (boceprevir).25,26 A specific role for ITPase function in mediating the anemia associated with these direct antivirals will need to be explored. As a final note, functional testing of ITPase activity is possible and will need to be explored and compared with ITPA genotyping for the prediction of anemia risk.
In conclusion, functional variants of the ITPA gene, associated with ITPase deficiency, are protective against RBV-induced hemolytic anemia, reducing the need for RBV dose reduction, and maintaining cumulative RBV dosage. ITPA genotype was not associated with SVR, although the sample size of this cohort may have been too small to detect a modest effect. ITPA genotyping, or functional ITPase assays, may help guide clinical decision making in patients considering antiviral therapy for HCV, especially those at high risk for hemolytic anemia or anemia-related morbidity.
Supplementary Material
Acknowledgments
The ViraHep-C study was conducted by the ViraHep-C Study Group and supported by the National Institute of Diabetes and Digestive and Kidney Diseases. This report was not prepared in collaboration with the investigators of the ViraHep-C Study Group and does not necessarily reflect the opinions or views of the ViraHep-C Study Group or the National Institute of Diabetes and Digestive and Kidney Diseases. However, the authors gratefully acknowledge their efforts to make this valuable biorepository and clinical dataset publicly available.
Funding
A.J.T. received funding support from the Duke Clinical Research Institute, a generous research gift from the Richard B. Boebel Family Fund, the National Health and Medical Research Council of Australia, the Gastroenterology Society of Australia, and the Royal Australasian College of Physicians. M.W.F. is funded in part by grant K24 DK066144.
Abbreviations used in this paper
- CI
confidence interval
- HA
hemolytic anemia
- Hb
hemoglobin
- ITPase
inosine triphosphatase
- pegIFN
pegylated interferon
- RBV
ribavirin
- SVR
sustained virological response
- ViraHep-C
Study of Viral Resistance to Antiviral Therapy of Chronic Hepatitis C
Footnotes
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi:10.1053/j.gastro.2010.06.016.
Conflicts of interest
The authors disclose the following: A.J.T., J.F., D.G., T.J.U., K.V.S., D.B.G., and J.G.M. are coinventors of a patent application based on the ITPA discovery. K.P., H.L.T., S.N., A.J.M., M.W.F., and N.H.A. disclose no conflicts.
References
- 1.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 2.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 3.Fellay J, Thompson AJ, Ge D, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405–408. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 4.Sumi S, Marinaki AM, Arenas M, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet. 2002;111:360–367. doi: 10.1007/s00439-002-0798-z. [DOI] [PubMed] [Google Scholar]
- 5.Maeda T, Sumi S, Ueta A, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency in the Japanese population. Mol Genet Metab. 2005;85:271–279. doi: 10.1016/j.ymgme.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Shipkova M, Lorenz K, Oellerich M, et al. Measurement of erythrocyte inosine triphosphate pyrophosphohydrolase (ITPA) activity by HPLC and correlation of ITPA genotype-phenotype in a Caucasian population. Clin Chem. 2006;52:240–247. doi: 10.1373/clinchem.2005.059501. [DOI] [PubMed] [Google Scholar]
- 7.Atanasova S, Shipkova M, Svinarov D, et al. Analysis of ITPA phenotype-genotype correlation in the Bulgarian population revealed a novel gene variant in exon 6. Ther Drug Monit. 2007;29:6–10. doi: 10.1097/FTD.0b013e3180308554. [DOI] [PubMed] [Google Scholar]
- 8.Conjeevaram HS, Fried MW, Jeffers LJ, et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470–477. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 10.McHutchison JG, Manns M, Patel K, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123:1061–1069. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1009. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 12.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 13.Howell CD, Thompson AJ, Ryan K, et al. IL28B genetic variation is associated with early viral kinetics and SVR in HCV genotype 1: the VIRAHEP-C Study. Proceedings of the 45th Annual Meeting of the European Association for the Study of the Liver (EASL); Vienna, Austria. 2010. [Google Scholar]
- 14.Sulkowski M, Shiffman M, Afdhal N, et al. Hemoglobin decline is associated with SVR among HCV genotype 1-infected persons treated with peginterferon (PEG)/ribavirin(RBV): analysis from the IDEAL study (A126) J Hepatol. 2009;50 Suppl 1:S51. [Google Scholar]
- 15.Shiffman ML, Ghany MG, Morgan TR, et al. Impact of reducing peginterferon alfa-2a and ribavirin dose during retreatment in patients with chronic hepatitis C. Gastroenterology. 2007;132:103–112. doi: 10.1053/j.gastro.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Reddy KR, Shiffman ML, Morgan TR, et al. Impact of ribavirin dose reductions in hepatitis C virus genotype 1 patients completing peginterferon alfa-2a/ribavirin treatment. Clin Gastroenterol Hepatol. 2007;5:124–129. doi: 10.1016/j.cgh.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Stepchenkova EI, Tarakhovskaya ER, Spitler K, et al. Functional study of the P32T ITPA variant associated with drug sensitivity in humans. J Mol Biol. 2009;392:602–613. doi: 10.1016/j.jmb.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bierau J, Lindhout M, Bakker JA. Pharmacogenetic significance of inosine triphosphatase. Pharmacogenomics. 2007;8:1221–1228. doi: 10.2217/14622416.8.9.1221. [DOI] [PubMed] [Google Scholar]
- 19.De Franceschi L, Fattovich G, Turrini F, et al. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology. 2000;31:997–1004. doi: 10.1053/he.2000.5789. [DOI] [PubMed] [Google Scholar]
- 20.Grossman I, Sullivan PF, Walley N, et al. Genetic determinants of variable metabolism have little impact on the clinical use of leading antipsychotics in the CATIE study. Genet Med. 2008;10:720–729. doi: 10.1097/GIM.0b013e3181863239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 22.Mallal S, Nolan D, Witt C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 23.Reau N, Hadziyannis SJ, Messinger D, et al. Early predictors of anemia in patients with hepatitis C genotype 1 treated with peginterferon alfa-2a (40KD) plus ribavirin. Am J Gastroenterol. 2008;103:1981–1988. doi: 10.1111/j.1572-0241.2008.01957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiffman ML, Salvatore J, Hubbard S, et al. Treatment of chronic hepatitis C virus genotype 1 with peginterferon, ribavirin, and epoetin alpha. Hepatology. 2007;46:371–379. doi: 10.1002/hep.21712. [DOI] [PubMed] [Google Scholar]
- 25.McHutchison JG, Everson GT, Gordon SC, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 26.Kwo P, Lawitz EJ, McCone J, et al. HCV SPRINT-1 final results: SVR 24 from a phase II study of boceprevir plus peginterferon alfa-2b/ribavirin in treatment-naive subjects with genotype 1 chronic hepatitis C. J Hepatol. 2009;50:S4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.