Abstract
To examine the in vivo functions of protein kinase N (PKN), one of the effectors of Rho small GTPases, we used the nematode Caernorhabditis elegans as a genetic model system. We identified a C. elegans homologue (pkn-1) of mammalian PKN and confirmed direct binding to C. elegans Rho small GTPases. Using a GFP reporter, we showed that pkn-1 is mainly expressed in various muscles and is localized at dense bodies and M-lines. Over-expression of the PKN-1 kinase domain and loss-of-function mutations by genomic deletion of pkn-1 resulted in a loopy Unc phenotype, which has been reported in many mutants of neuronal genes. The results of mosaic analysis and body wall muscle specific expression of PKN-1 kinase domain suggests that this loopy phenotype is due to the expression of PKN-1 in body wall muscle. The genomic deletion of pkn-1 also showed a defect in force transmission. These results suggest that PKN-1 functions as a regulator of muscle contraction-relaxation and as a component of the force transmission mechanism.
Keywords: C. elegans, muscle, Rho GTPase, PKN, contraction
Introduction
The small GTPases have two convertible forms; GDP bound inactivated and GTP bound activated1. The two states are regulated by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs). The GEFs have been shown to accelerate release of GDP from GTPases, resulting in binding to GTP in the cell, thus activating the GTPase2. Rho family small GTPases include Rho, Cdc42 and Rac. Each of these GTPases, in their GTP bound forms, preferentially interact with specific effector proteins. So far, many effectors of Rho GTPases have been identified3. Among them, protein kinase N (PKN) was the first identified effector of Rho GTPase4. PKN was identified as novel protein kinase containing a protein kinase domain similar to protein kinase C (PKC)5. It has been reported that PKN is involved in many physiological functions, such as cytoskeletal regulation, cell adhesion, vesicle transport, apoptosis, and cell cycle regulation6. In Drosophia, a PKN homologue has been reported to be involved in a signaling cascade that regulates dorsal closure and development7, 8. Nevertheless, the mechanisms by which PKN participates in these various cellular functions are not well understood. We used C. elegans as a genetic model for investigation of in vivo functions of PKN.
Biochemical characterization of a C. elegans PKN ortholog, PKN-1
We identified F46F6.2 as the single C. elegans ortholog of mammalian Protein Kinase N (PKN) by an homology search using the WormBase database (http://www.wormbase.org/). WormBase predicts two isoforms, F46F6.2a and b, by alternative splicing. We confirmed the splicing pattern of F46F6.2a by sequencing of EST cDNA clones (Figure 1A). We named the gene pkn-1 (C. elegans protein kinase N). The C. elegans PKN-1 protein shares structural features with mammalian PKN; three Hr1 repeats at the N-terminus, a C2-like domain in the middle, and a serine/threonine kinase domain at the C-terminus (Figure 1B). The human and nematode PKN proteins are 38% identical in overall protein sequence and 71% identical within the protein kinase domains. Since mammalian PKN is one of the effectors of Rho GTPase, we examined whether PKN-1 could bind to C. elegans Rho GTPases in vitro. We prepared GST fusions of human RhoA and C. elegans Rho type GTPases (RHO-1 as RhoA, CED-10 and MIG-2 as Rac, and CDC-42 as cdc42) with GTPγS (a non-hydrolyzable form of GTP) and GDP, and an MBP fusion of PKN-1 N-terminus (residues 1-291) containing the three Hr1 repeats, a region which has been reported to be essential for the interaction of mammalian PKN with RhoA4. By using those GST fusion proteins (Figure 1D), MBP-PKN-1 N-terminus showed binding to GST-RHO-1 and GST-RhoA, and weaker binding to GST-CED-10 and GST-MIG-2, but no binding to GST-CDC-42 (Figure 1C). Binding was much higher with GTPγS than with GDP. These results suggest that C. elegans PKN-1 is also one of the effectors of RhoA type GTPases. It has been reported that PKN can bind to Rac type GTPases in mammalian cells9 and Drosophila8. However, in C. elegans, pkn-1 is expressed mainly in muscle (see below) and ced-10 and mig-2 are not expressed in muscle based on anatomical expression data. Thus, we suggest that PKN-1 is only regulated by RHO-1 GTPase in muscle cells.
Figure 1. Direct binding between PKN-1 and Rho type small GTPases.
A: Genomic structure of pkn-1. The exon-intron structure of pkn-1a (F46F6.2a) is depicted, below which is shown the location of the intragenic deletion ok1673, and the location of the region used for the promoter-GFP fusion construct (pKK44) expressed in transgenic worms (Figure 2A). The red box denotes the trans-spliced leader RNA SL1, the grey boxes denote the 5’ and 3’ UTR sequences, and the blue boxes denote exons.
B: Schematic showing organization of domains within PKN-1 (F46F6.2a). PKN-1 has three Hr1 repeats, a C2-like domain, and a serine/threonine kinase domain, similar to the human PKN protein in the types, numbers and organization of domains. Percentages under each domain denote the identity between the C. elegans and human PKN proteins. Lower brackets indicate regions used for binding assay (Figure 1C) and for overexpression (Figure 4).
C: Regulatory region of PKN-1 can bind to C. elegans Rho-type GTPases. Purified MBP-PKN-1 N-terminus (residues 1-295) was mixed with glutathione beads coated with GST fusions of Rho-type GTPases (GST-RHO-1, GST-CED-10, GST-MIG-2, GST-CDC-42, and GST-RhoA (human)) loaded with GTPγS or GDP. After extensive washes, binding proteins were eluted with glutathione, subjected to SDS-PAGE separation, and visualized by Coomassie Blue staining4. The MBP-PKN-1 N-terminus binds to GTP forms of GST-RHO-1 and GST-RhoA, and GTP forms of GST-CED-10 and GST-MIG-2 more weakly. All proteins were expressed in E. coli and purified using glutathione agarose (for GST fusions) or amylose resin (for MBP fusion). Expression plasmids were constructed by cloning of corresponding cDNA fragments amplified by PCR from a cDNA library (RB2) into pGEX-4T-1 (GE Healthcare) or pMAL-c2 (New England Biolab) bacterial expression vectors28.
D: Coomassie Blue staining of an SDS-PAGE containing the GST-RHO-1, GST-CED-10, GST-MIG-2, GST-CDC-42, GST-RhoA (Hs), and MBP-PKN-1 N-terminus used in this assay. 2 µg proteins were loaded on 10% SDS-PAGE gel, and stained with Coomassie Brilliant Blue.
Expression pattern of the pkn-1 gene and localization of the PKN-1 protein
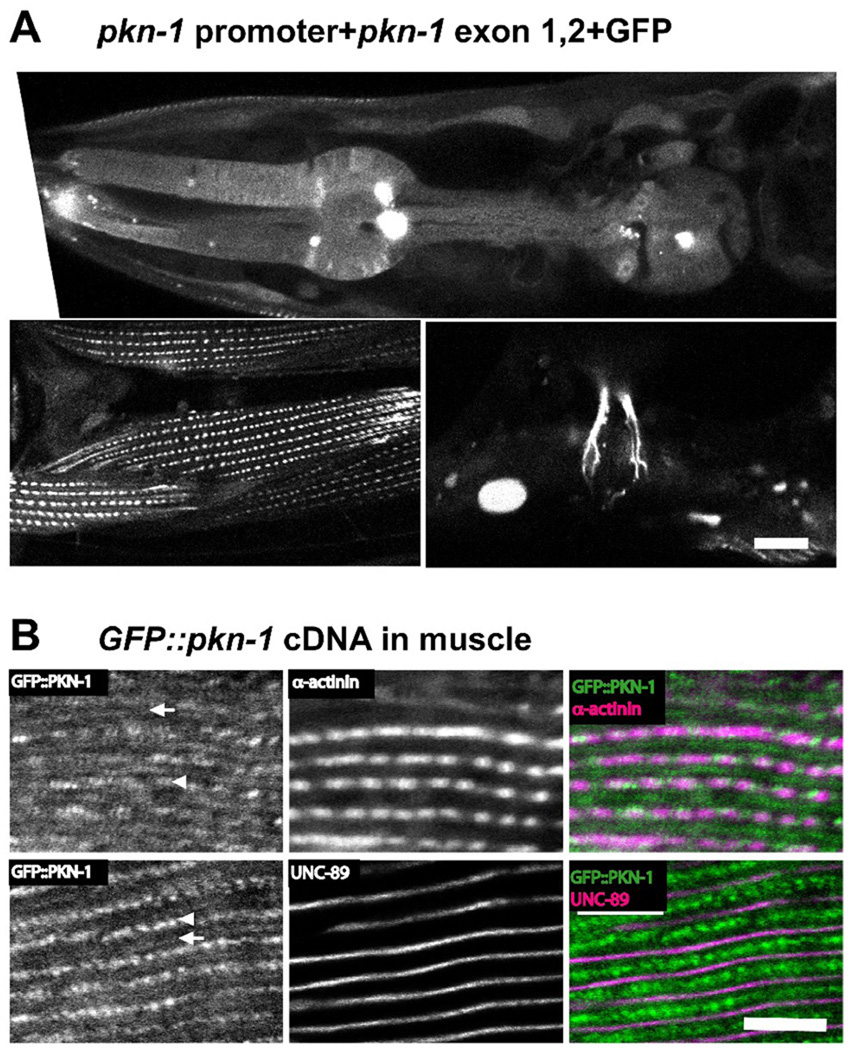
To examine the expression pattern of pkn-1, we used GFP fused to the pkn-1 promoter region (Figure 1A). GFP fusion containing 5 Kb upstream and first and second exons of pkn-1 showed expression in various muscle cells; pharyngeal muscle, body wall muscle, vulval muscle (Figure 2A), and some other tissues including neurons (data not shown). To characterize the localization of PKN-1 in body wall muscle, we expressed a GFP fusion of full-length pkn-1 cDNA under the control of a heat shock promoter and immunostained with anti-GFP and antibodies to proteins of known sarcomeric location. GFP::PKN-1 co-localized with α-actinin10 and UNC-8911, indicating localization at dense bodies (arrowheads in Figure 2B) and M-lines (arrows in Figure 2B). In the major striated muscle of C. elegans, residing in the body wall, thick filaments are organized around M-lines, and thin filaments are attached at dense bodies, equivalent to Z-disks (lines) in vertebrate striated muscle. Many proteins are localized to both M-lines and dense bodies (e.g. PAT-3 (β-integrin), UNC-112 (Kindlin) etc.), whereas others are localized only to M-lines (e.g. UNC-89 (obscurin), UNC-98, UNC-96), and others are localized only to dense bodies (e.g. DEB-1 (vinculin), ATN-1 (α-actinin), ALP-1 (ALP/Enigma) etc.)12, 13. Since GFP fusion of the first and second exons of pkn-1 also localized to dense bodies and M-lines in body wall muscle (Figure 2A, bottom left), the N-terminal 138 residues of PKN-1a might be sufficient for its localization to dense bodies and M-lines. Muscle expression of pkn-1 is supported by SAGE data available on WormBase (http://www.wormbase.org/). SAGE data showed that the pkn-1 also expresses in neurons, hypodermis, pharynx, gonad, and gut cells. It has been reported that rho-1 and unc-89 (encoding a giant protein that includes a RhoA GEF domain, and is localized to M-lines), function in muscle in the regulation of sarcomere organization14. Therefore, it is possible that at sarcomeric M-lines, the RHO-1 / PKN-1 interaction is a part of this UNC-89 / RHO-1 pathway in C. elegans striated muscle. A major component of nematode muscle dense bodies is α-actinin15, 16. Therefore, the observed dense body localization of PKN-1 is consistent with the known interaction of vertebrate PKN with α-actinin17. Whether nematode PKN-1 interacts with α-actinin will require additional experiments.
Figure 2. The expression and localization of PKN-1 in muscle.
A: GFP expressed by the pkn-1 promoter was observed in many types of muscle. We injected a mixture of pKK44 plasmid (5 Kb of upstream putative promoter, the first and second exons, and the first intron of pkn-1 was cloned into pPD95.75, a gift from Dr. Fire) and a lin-15 rescuing plasmid into lin-15 (n765) (temperature sensitive multinulva (Muv)) worms. To obtain transgenic animals, we picked non-Muv worms at 25°C. In transgenic animals, GFP driven by pkn-1 promoter was observed in pharynx including pharyngeal muscle (upper panel), body wall muscle (lower left panel), and vulva muscle (lower right panel). Scale bar, 10 µm.
B: GFP tagged PKN-1 is localized at dense bodies (arrowheads) and M-lines (arrows). We prepared transgenic worms harboring an extrachromosomal array containing both a plasmid expressing GFP tagged full-length pkn-1 cDNA under the control of heat shock promoter, and a plasmid for the rol-6 (roller) dominant marker. Worms were fixed by the Nonet method29. Antibody staining with anti-GFP (Invitrogen A11122, 1/200 dilution), and MH35 (1/200 dilution; α-actinin) or MH42 (1/200 dilution; UNC-89) was performed as described previously28. All images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope using an Apochromat 63x/1.4 oil objective, in 2.5x zoom mode. The color balances of the images were adjusted by using Adobe Photoshop. Scale bar, 10 µm.
Activation and inactivation of pkn-1 in C. elegans
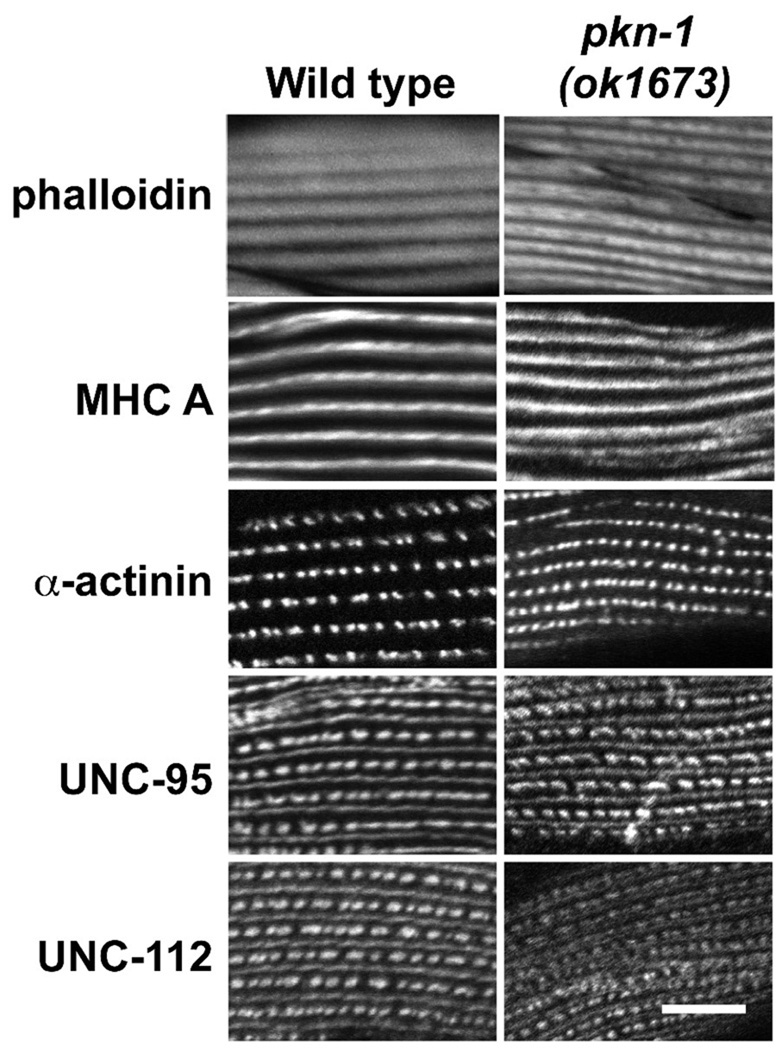
We examined sarcomeric organization in an intragenic deletion of pkn-1 (pkn-1 (ok1673)) using a battery of antibodies to sarcomeric proteins. The ok1673 is a 1064 bp deletion and 18 bp insertion beginning in the 4th exon and ending in the 7th exon (Figure 1A), resulting in a premature stop inside of C2-like domain. Thus, if the truncated protein from ok1673 were stable, it would lack the kinase domain. Compared to wild type, we observed no defects in the organization of thin filaments, thick filaments, and in both the deep and surface regions of muscle attachment structures (dense bodies and M-lines) (Figure 3).
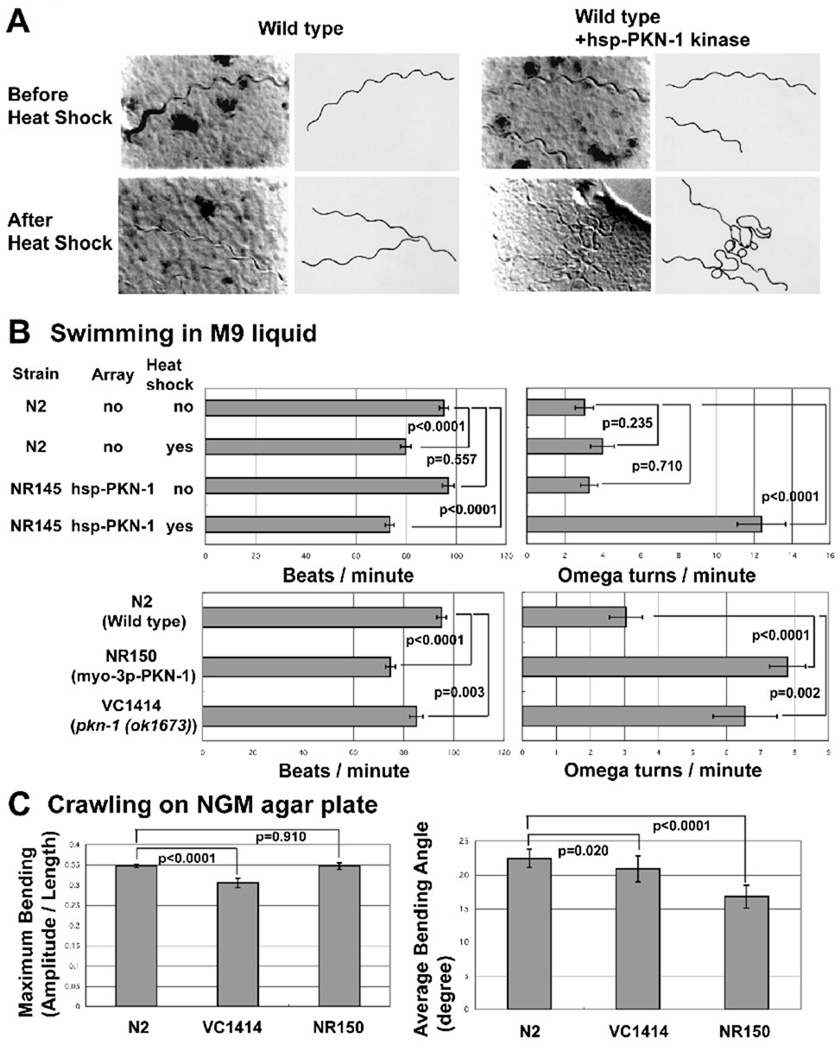
Next, we examined the phenotype of overexpressing of PKN-1 kinase domain as a type of gain of function mutation for pkn-1. We prepared transgenic animals harboring PKN-1 kinase domain expressed by the control of a heat shock promoter (hsp). After heat shock, animals with hsp-PKN-1 kinase showed locomotion with more exaggerated bending (“loopy”)(Figure 4A). Heat shock itself had no effect on animal movement (Figure 4A). To characterize this loopy Unc phenotype quantitatively, we counted swimming or thrashing (beats per minute) and omega turn frequency (omega turn per minute) in M9 liquid. An omega turn is a deep ventral bend in the animal that is shaped like the Greek letter omega. It usually is at the end of a large reversal (defined as more than two heads wings backwards) and the animal changes direction of locomotion when it completes an omega turn18. With heat shock, both wild type and PKN-1 kinase over-expressed worms showed a slight reduction in motility (Figure 4B). Most significantly, we observed that omega turn frequency increased after heat shock of worms harboring hsp-PKN-1 kinase domain (Figure 4B). Query of WormBase database indicates that there are many mutants that have a “loopy” phenotype: baf-1 (encoding novel protein), eat-16 (encoding RGS protein, regulator of hetrotrimetic G protein), egl-8 (encoding phospholipase C beta), flp-1 (encoding FMRFamide-related peptides), gpb-2 (encoding beta subunit of heterotrimeric G protein), gsa-1 (encoding alpha subunit of heterotrimeric G protein), kin-2 (encoding cAMP-dependent protein kinase), rab-3 (encoding rab GTPase), syc-3 (not yet identified), unc-43 (encoding Ca2+-Calmodulin dependent protein kinase), unc-77 (not yet identified), and unc-117 (not yet identified). Since almost all of these mutants are reported to have defects in neuronal functions (for example, described previously19) and some pkn-1 is expressed in neurons, we conducted two types of experiments to determine whether expression of pkn-1 in muscle or neurons is required for display of the loopy Unc phenotype. The first experiment was mosaic analysis using a transgenic animal harboring an extrachromosomal array containing hsp-PKN-1 kinase and sur-5::nls::gfp20. In C. elegans, as development proceeds, at a low frequency, extrachromosomal arrays are randomly lost when cells divide, resulting in mosaic expression of each gene on the array21. In our experiment, the maintenance of the extrachromosomal array was scored by the existence of sur-5::nls::gfp in each tissue’s nuclei. After heat shock of transgenic animals, we picked 42 “loopy” worms and 6 non-“loopy” worms and determined whether sur-5::nls::gfp was present in neurons or muscle cells. In all 42 “loopy” animals, sur-5::nls::gfp was maintained in muscle cells, but was missing in some neurons (such as those in the neural ring). In all 6 non-“loopy” worms, sur-5::nls::gfp was missing in muscle, but present in neuronal cells. Therefore, the loopy phenotype is exactly correlated with overexpression PKN-1 in muscle. Second, we prepared transgenic worms harboring the PKN-1 kinase domain driven by a muscle specific promoter, from the gene myo-3, and examined worm locomotion. When the PKN-1 kinase domain was expressed by the myo-3 promoter (strain NR150), transgenic worms showed a loopy phenotype with reduced motility and increased frequency of omega turn (Figure 4B). When PKN-1 kinase domain was overexpressed by myo-3, muscle structure was normal, as assessed by polarized light microscopy (data not shown). Taken together, we conclude that overexpression of the PKN-1 kinase domain in the muscle cells causes the loopy Unc phenotype. Although intragenic deletion of pkn-1, ok1673, showed normal muscle structure (Figure 3), ok1673 (strain VC1414) also displayed a weak loopy Unc phenotype, including reduced motility and increased frequency of omega turns (Figure 4B). This weak loopy phenotype was also observed in worms subjected to RNAi mediated knockdown of pkn-1 (data not shown).
Figure 3. Muscle structure in a pkn-1 loss of function mutant.
pkn-1 intragenic deletion mutant (pkn-1 (ok1673)) and wild type worms were stained with phalloidin (actin thin filaments), anti-MHC A (5–6, myosin thick filaments and deep part of M-lines30), anti-α-actinin (MH35, deep part of dense bodies10), anti-UNC-95 (a LIM domain protein31 located at the membrane proximal portion of dense bodies and M-lines28), and anti-UNC-112 (mammalian Kindlin ortholog32 located at the membrane proximal portion of dense bodies and M-lines33). As shown, the organization of all these sarcomeric components is normal. Worms were fixed by the Nonet method29 and immunostaining was performed as described previously28. All images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope using an Apochromat 63x/1.4 oil objective, in 2.5x zoom mode. The color balances of the images were adjusted by using Adobe Photoshop. Scale bar, 10 µm.
Figure 4. Either gain of function or loss of function of pkn-1 results in a “loopy Unc phenotype”.
A: Images (left) together with tracings (right) of worm tracks made along a bacterial lawn, from wild type, or wild type with hsp (heat shock promoter)-PKN-1 kinase before or after heat shock (30°C for 2 hrs). Without heat shock, wild type and wild type with hsp-PKN-1 kinase domain worms showed normal movement. After heat shock, only wild type with hsp-PKN-1 kinase domain worms showed more loopy tracks on the bacterial lawn surface. Transgenic worms harboring an extrachromosomal array consisting of hsp-PKN-1 kinase domain and sur-5::nls::gfp was constructed by injection34 of DNA mixture of pPD49.78-PKN-1 kinase, pPD49.83-PKN-1 kinase, and pTG96 (sur-5::nls::gfp marker plasmid) into N2 wild type, and selecting progeny showing GFP expression. The extrachromosomal array was integrated by UV irradiation35. To construct pPD49.78-PKN-1 kinase and pPD49.83-PKN-1 kinase, PCR amplified pkn-1 cDNA containing the kinase domain (see Figure 1A) was cloned into pPD49.78 or pPD49.83.
B: Bar graphs showing the results of two assays of adult worm swimming in M9 liquid: beats per minute (left) and omega turns per minute (right), each with means and standard errors (SEs) (n=20). Pair-wise comparisons for statistical significance are indicated with brackets and p values. These comparisons were made with one-way ANOVA using the post hoc Tukey HSD test, on StatPlus (AnalystSoft, Inc.). NR145 (harboring hsp-PKN-1 kinase) with heat shock, NR150 (harboring myo-3p-PKN-1 kinase), VC1414 (pkn-1 genomic deletion) show increased omega turn frequency. NR150 and VC1414 show reduced beats per minute. NR150 is an integrated line of the myo-3p-PKN-1 kinase array. The myo-3 promoter is reported to be expressed only in muscle36. To construct the plasmid expressing the PKN-1 kinase domain under the control of the myo-3 promoter (pPD95.86-PKN-1 kinase), a PCR amplified cDNA fragment for pkn-1 kinase domain was cloned into pPD95.86. Transgenic worms harboring extrachromosomal arrays were obtained by injection34 of myo-3p-PKN-1 kinase plasmid with pTG96 (sur-5::nls::gfp) into wild type worms. The extrachromosomal array was integrated by UV irradiation35. We obtained VC1414 from Caenorhabditis Genetics Center.
C: Bar graphs showing the maximum bending amplitude during an induced reversal (left) and the average bending angle during normal forward locomotion (right) of N2 (wild type), VC1414 (pkn-1 genomic deletion), and NR150 (integrated myo-3p-PKN-1 kinase), each with means and SEs. For maximum bending amplitude, n=20 except for N2 (n=65). For average bending angle, n=15 for each strain. Pair-wise comparisons for statistical significance are indicated with brackets and p values. These comparisons were made with one-way ANOVA using the post hoc Bonferroni test for the graph on the left (Maximum Bending), the post hoc Tukey HSD test for the graph on the right (Average Bending Angle), on StatPlus (AnalystSoft, Inc.). Each assay was performed on a blank NGM agar plate without bacteria. The average bending angle is the average of the absolute values of the angles of the worm when it is divided into 12 equal segments (13 points). The angle is between consecutive segments and relative to 180 degrees. The deeper the bend, the higher the angle. The numbers shown are averages over a 6–15 second interval when the animals were moving forward. VC1414 shows a reduction of maximum bending ability. NR150 shows a reduction of average bending angle. The methods for these measurements were described previously22, 37, 38.
In addition to characterizing worm behavior in liquid, we characterized worm crawling on NGM agar plates using a recently established video recording and analysis system. We characterized precise movement of wild type (N2), pkn-1 genomic deletion mutant (VC1414), and constitutively over-expressed PKN-1 kinase domain in body wall muscle worm (NR150). We found that the maximum bending amplitude was reduced in the pkn-1 deletion mutant, as compared to wild type or worms overexpressing PKN-1 kinase in muscle (Figure 4C), similar to loss of function of atn-1 (α-actinin)22. This reduction in maximal bending suggests a role for PKN-1 in the transmission of force, and is consistent with the fact that PKN-1 co-localizes with ATN-1 (α -actinin) at dense bodies (Figure 2B). We also found that the average bending angle of worms overexpressing PKN-1 kinase in muscle was reduced as compared to wild type and genomic deletion (Figure 4C). This defect might explain the slow motility of this strain (Figure 4B). Theoretically the “loopy” phenotype of pkn-1 mutants might result from worms making more deeper turning angles or changing direction more frequently. The later possibility is more likely since: (1) pkn-1 mutants display normal or reduced maximum bending and average bending angles, and (2) pkn-1 mutants display more frequent omega turns.
We found that both gain of function and loss of function of pkn-1 in muscle cells results in a loopy phenotype. The loopy phenotype that has been reported for loss of function for many neuronal genes has been explained as too much contraction caused by de-regulated neurotransmitter release19. In the case of pkn-1, we propose two hypotheses to explain this loopy phenotype. First, the PKN-1 kinase could phosphorylate and modify the function of regulators of muscle contraction-relaxation, resulting in too much bending of the worm and thus a loopy Unc phenotype. Such regulators, phosphorylatated by PKN-1, are likely to be located at sarcomeric dense bodies and M-lines, the sites of PKN-1 localization. In mammalian cells, it has been reported that PKN can phosphorylate a regulatory subunit of myosin phosphatase and that this phosphorylation results in a decrease in the activity of myosin phosphatase23. Reduced myosin phosphatase activity could result in increased phosphorylation of regulatory myosin light chains, and thus increased myosin activity and contraction. This is plausible for nematode body wall muscle which has both thin filament and myosin based calcium regulation24. Second, PKN-1 could be involved in retrograde signaling from muscle to neurons to enhance the release of neurotransmitter. It has been reported that muscle expression aex-1, a gene involved in neuronal function, is essential for its neuronal function25.
That both loss of function and gain of function mutations in pkn-1 result in motility defects suggests that a tightly controlled level of PKN-1 activity is required for proper regulation of muscle activity. One explanation for sharing the same phenotype for both gain-of-function and loss-of-function mutations, is some defect in the cycling of two interconvertible states. For example, mutations in small GTPase activators (GEFs) and inactivators (GAPs) show the same phenotype26, 27. In the muscle cells, it is possible that the defect in contraction-relaxation cycle might be caused by unbalanced contraction or relaxation, resulting in a loopy phenotype. Thus, we prefer the first hypothesis. Further examination of the mechanism of loopy phenotype caused by mutation of pkn-1 through the identification of genetic interactors and PKN-1 substrates may provide insight into the role of PKN-1 in regulating muscle contraction-relaxation.
Acknowledgement
We thank Dr. Alan Coulson (Wellcome Trust Genome Campus) for cosmids clones; Dr. Yuji Kohara (National Institute of Genetics) for EST cDNA clones; Andy Fire (Stanford University School of Medicine) for GFP and ectopic expression vectors; Dr. Min Han (University of Colorado) for pTG96 plasmid; Dr. Robert Barstead (Oklahoma Medical Research Foundation) for C. elegans cDNA library RB2; Dr. Kristy J. Wilson (Emory University) for making the worm tracings for Fig. 4A; and Dr. Shinya Kuroda (University of Tokyo) for helpful discussion. We also thank Dr. Ken Norman (Albany Medical College) for help with initial characterization of confocal microscopic images, and Dr. Carlos Moreno (Emory University) for advice on statistical analysis. Some of the strains used in this work were provided by the Caenorhabditis elegans Genetics Center, which is funded by the National Institutes of Health (NIH) Center for Research Resources. HQ, MA, and KK were supported by grants-in-aid for Scientific Research from the Ministry of Education, Science, and Culture, Japan, by the Japan Society of the Promotion of Science Research for the Future, by the Human Frontier Science Program. GMB was supported by NIAMS / NIH grant AR051466. HL is supported by NIH (NIBIB-R21EB012803, NIA-R01AG035317), NSF (CBET/CAREER-0954578), and the Alfred P. Sloan Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu. Rev. Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 2.Cherfils J, Chardin P. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 3.Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu. Rev. Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- 4.Amano M, Mukai H, Ono Y, Chihara K, Matsui T, Hamajima Y, Okawa K, Iwamatsu A, Kaibuchi K. Identification of a putative target for Rho as the serine-threonine kinase protein kinase N. Science. 1996;271:648–650. doi: 10.1126/science.271.5249.648. [DOI] [PubMed] [Google Scholar]
- 5.Mukai H, Ono Y. A novel protein kinase with leucine zipper-like sequences: its catalytic domain is highly homologous to that of protein kinase C. Biochem. Biophys. Res. Commun. 1994;199:897–904. doi: 10.1006/bbrc.1994.1313. [DOI] [PubMed] [Google Scholar]
- 6.Mukai H. The structure and function of PKN, a protein kinase having a catalytic domain homologous to that of PKC. J. Biochem. 2003;133:17–27. doi: 10.1093/jb/mvg019. [DOI] [PubMed] [Google Scholar]
- 7.Betson M, Settleman J. A rho-binding protein kinase C-like activity is required for the function of protein kinase N in Drosophila development. Genetics. 2007;176:2201–2212. doi: 10.1534/genetics.107.072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Y, Settleman J. The Drosophila Pkn protein kinase is a Rho/Rac effector target required for dorsal closure during embryogenesis. Genes Dev. 1999;13:1168–1180. doi: 10.1101/gad.13.9.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vincent S, Settleman J. The PRK2 kinase is a potential effector target of both Rho and Rac GTPases and regulates actin cytoskeletal organization. Mol. Cell. Biol. 1997;17:2247–2256. doi: 10.1128/mcb.17.4.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benian GM, Tinley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qadota H, Benian GM. Molecular structure of sarcomere-to-membrane attachment at M-Lines in C. elegans muscle. J. Biomed. Biotechnol. 2010;2010:864749. doi: 10.1155/2010/864749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lecroisey C, Segalat L, Gieseler K. The C. elegans dense body: anchoring and signaling structure of the muscle. J. Muscle Res. Cell Motil. 2007;28:79–87. doi: 10.1007/s10974-007-9104-y. [DOI] [PubMed] [Google Scholar]
- 14.Qadota H, Blangy A, Xiong G, Benian GM. The DH-PH region of the giant protein UNC-89 activates RHO-1 GTPase in Caenorhabditis elegans body wall muscle. J. Mol. Biol. 2008;383:747–752. doi: 10.1016/j.jmb.2008.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barstead RJ, Kleiman L, Waterston RH. Cloning, sequencing, and mapping of an alpha-actinin gene from the nematode Caenorhabditis elegans. Cell Motil. Cytoskeleton. 1991;20:69–78. doi: 10.1002/cm.970200108. [DOI] [PubMed] [Google Scholar]
- 17.Mukai H, Toshimori M, Shibata H, Takanaga H, Kitagawa M, Miyahara M, Shimakawa M, Ono Y. Interaction of PKN with alpha-actinin. J. Biol. Chem. 1997;272:4740–4746. doi: 10.1074/jbc.272.8.4740. [DOI] [PubMed] [Google Scholar]
- 18.Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U S A. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, Meyer BJ. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci. 1997;17:8061–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman RK. Mosaic analysis. Methods Cell Biol. 1995;48:123–146. [PubMed] [Google Scholar]
- 22.Moulder GL, Cremona GH, Duerr J, Stirman JN, Fields SD, Martin W, Qadota H, Benian GM, Lu H, Barstead RJ. alpha-actinin is required for the proper assembly of Z-disk/focal-adhesion-like structures and for efficient locomotion in Caenorhabditis elegans. J. Mol. Biol. 2010;403:516–528. doi: 10.1016/j.jmb.2010.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamaguchi T, Ito M, Feng J, Seko T, Koyama M, Machida H, Takase K, Amano M, Kaibuchi K, Hartshorne DJ, Nakano T. Phosphorylation of CPI-17, an inhibitor of myosin phosphatase, by protein kinase N. Biochem. Biophys. Res. Commun. 2000;274:825–830. doi: 10.1006/bbrc.2000.3225. [DOI] [PubMed] [Google Scholar]
- 24.Harris HE, Tso MY, Epstein HF. Actin and myosin-linked calcium regulation in the nematode Caenorhabditis elegans. Biochemical and structural properties of native filaments and purified proteins. Biochemistry. 1977;16:859–865. doi: 10.1021/bi00624a008. [DOI] [PubMed] [Google Scholar]
- 25.Doi M, Iwasaki K. Regulation of retrograde signaling at neuromuscular junctions by the novel C2 domain protein AEX-1. Neuron. 2002;33:249–259. doi: 10.1016/s0896-6273(01)00587-6. [DOI] [PubMed] [Google Scholar]
- 26.Powers S, Gonzales E, Christensen T, Cubert J, Broek D. Functional cloning of BUD5, a CDC25-related gene from S. cerevisiae that can suppress a dominant-negative RAS2 mutant. Cell. 1991;65:1225–1231. doi: 10.1016/0092-8674(91)90017-s. [DOI] [PubMed] [Google Scholar]
- 27.Park HO, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- 28.Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol. Biol. Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- 30.Miller DM, 3rd, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- 31.Broday L, Kolotuev I, Didier C, Bhoumik A, Podbilewicz B, Ronai Z. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in. C. elegans. J. Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogalski TM, Mullen GP, Gilbert MM, Williams BD, Moerman DG. The UNC-112 gene in Caenorhabditis elegans encodes a novel component of cell-matrix adhesion structures required for integrin localization in the muscle cell membrane. J. Cell Biol. 2000;150:253–264. doi: 10.1083/jcb.150.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hikita T, Qadota H, Tsuboi D, Taya S, Moerman DG, Kaibuchi K. Identification of a novel Cdc42 GEF that is localized to the PAT-3-mediated adhesive structure. Biochem. Biophys. Res. Commun. 2005;335:139–145. doi: 10.1016/j.bbrc.2005.07.068. [DOI] [PubMed] [Google Scholar]
- 34.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 35.Mitani S. Genetic regulation of mec-3 gene expression impricated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Dev. Growth & Diff. 1995;37:551–557. doi: 10.1046/j.1440-169X.1995.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- 36.Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geng W, Cosman P, Berry CC, Feng Z, Schafer WR. Automatic tracking, feature extraction and classification of C elegans phenotypes. IEEE Trans. Biomed. Eng. 2004;51:1811–1820. doi: 10.1109/TBME.2004.831532. [DOI] [PubMed] [Google Scholar]
- 38.Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput. Biol. 2008;4:e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]