Abstract
Leukemogenesis requires two classes of mutations, one that promotes proliferation and one that blocks differentiation. The erythroleukemia induced by Friend virus is a multistage disease characterized by an early proliferative stage driven by the interaction of the viral glycoprotein, gp55, with Sf-Stk and the EpoR, and a late block to differentiation resulting from retroviral insertion in the Pu.1 locus. We demonstrate here that activation of Stat3 by Sf-Stk in the early stage of disease is essential for the progression of erythroleukemia in the presence of differentiation signals induced by the EpoR, but is dispensable in the late stages of the disease. Furthermore, we identify Pu.1 as a Stat3 target gene in the early stages of erythroleukemia development. Our results support a model whereby the activation of Stat3 in the early stage of disease plays a pivotal role in regulating differentiation through the upregulation of Pu.1, thus inhibiting differentiation and favoring the expansion of infected erythroblasts and enhancing the pool of progenitors available for the acquisition of additional mutations, including insertional activation of Pu.1, resulting in full leukemic transformation.
Keywords: Stk, Ron, Gab2, Stat3, Pu.1, leukemia
Introduction
There is increasing evidence for a cooperative role of tyrosine kinases that promotes cellular proliferation and transcription factors that regulate cellular differentiation in the progression of leukemia. The erythroleukemia induced by Friend virus (FV) occurs in two distinct stages. The initial stage is characterized by a polyclonal expansion of infected erythroblasts. During the late stage of disease, the acquisition of additional genetic changes results in the clonal expansion of fully transformed cells. The first stage of the disease is driven by the interaction of the viral glycoprotein, gp55, with the EpoR and truncated form of the Stk receptor tyrosine kinase, Sf-Stk (Persons et al., 1999). C57BL/6 mice, which fail to express Sf-Stk, are resistant to Friend disease. However, exogenous expression of Sf-Stk in erythroblasts from C57BL/6 mice, both in vitro and in vivo, restores Epo-independent colony formation and development of erythroleukemia in response to FV infection (Persons et al., 1999; Finkelstein et al., 2002). Alternatively, although activation of the EpoR by gp55 is not required for the erythroblastosis induced by FV, it is required for the development of polycythemia (Zhang et al., 2006).
The ability of gp55 to interact with the EpoR could also explain the difference in disease pathogenesis in mice infected with two different variants of FV. FVP induces polycythemia in addition to erythroleukemia, whereas FVA-induced erythroleukemia is associated with anemia. Although both the viral glycoproteins from FVP (gp55P) and FVA (gp55A) interact with the EpoR (Constantinescu et al., 2003), gp55A is a weak activator of EpoR signaling, due to alterations in the transmembrane domain (Fang et al., 1998). Therefore, although activation of Sf-Stk is required for the polyclonal expansion of infected erythroblasts and the subsequent development of leukemia, activation of the EpoR primarily promotes the differentiation of infected cells.
The essential role played by Sf-Stk in the development of Friend disease led us to investigate the downstream signals required for disease pathogenesis. We have demonstrated that the recruitment of a Grb2/Gab2 complex to Sf-Stk in response to viral infection is both necessary and sufficient to promote the development of leukemia (Finkelstein et al., 2002; Teal et al., 2006). Furthermore, our studies indicated that Gab2, but not the closely related Gab1, can support the Epo-independent growth of FV-infected erythroblasts (Teal et al., 2006), a hallmark of the early stage of the disease. We identified a Stat3 binding site that is unique to Gab2 and demonstrated that this Stat3 binding motif is required for the Epo-independent growth of infected erythroblasts (Ni et al., 2007).
Insertional activation of Pu.1 expression in the second stage of Friend disease plays a critical role in maintaining the undifferentiated state of erythroleukemic cells. Pu.1 inhibits GATA-1 function and erythroid differentiation, and RNA interference of Pu.1 induces terminal differentiation and growth arrest of erythroleukemia cells (Papetti and Skoultchi, 2007). Alternatively, exogenous expression of GATA-1 in erythroleukemia cells reverses tumorigenicity and triggers terminal differentiation of these cells (Choe et al., 2003). In addition to its critical role in the late stage of Friend disease, previous studies have demonstrated that gp55 leads to upregulation of Pu.1 expression, independent of proviral insertion, suggesting that Pu.1 may also contribute to the early polyclonal expansion of infected erythroblasts (Afrikanova et al., 2002).
Here, we set out to examine the function of Stat3 in the development of erythroleukemia induced by FV. We demonstrate for the first time that Stat3 is critical for the development of the early stage of Friend disease in vivo, but appears to be dispensable for the expansion of fully transformed cells in the second stage of the disease. Furthermore, studies comparing the response of these mice to FVP vs FVA suggest that Stat3 plays a critical role in regulating the differentiation of infected progenitors in the early stage of Friend disease. Finally, our results clearly identify Pu.1 as a target gene for Stat3 in vivo following infection of mice with FV. The studies described herein support a model whereby the activation of Stat3 in the early stage of erythroleukemia induced by FV inhibits terminal differentiation of the infected erythroblasts, at least in part, through the upregulation of Pu.1 expression, thus promoting the polyclonal expansion of infected erythroblasts. The insertional activation of Pu.1 in the late stage of disease could, ultimately, circumvent the requirement for Stat3 in these cells.
Results
Stat3 is required for the development of Friend erythroleukemia in vivo
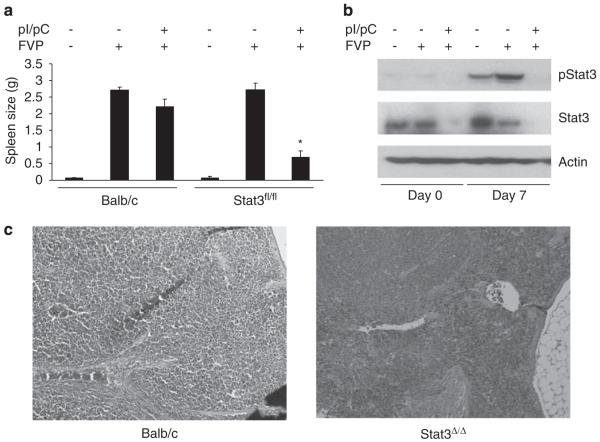
Infection of mice with FV drives a rapid polyclonal expansion of infected erythroblasts in the spleen of infected animals, resulting in severe splenomegaly. To determine whether Stat3 is required for the progression of Friend erythroleukemia in vivo, we utilized a conditional allele of Stat3 (Stat3fl/fl). Mice containing the Stat3fl/fl allele and the interferon (IFN)-inducible Mx1-Cre transgene were treated with pI/pC to induce deletion of Stat3 (Stat3Δ/Δ). These mice were infected with FV and spleen weights from these animals were assessed, as a measure of polyclonal erythroblast expansion. Although we observed enlarged spleens in response to FV infection in the control animals, the size of the spleens from Stat3Δ/Δ mice were significantly reduced (Figure 1a). Western blot analysis on spleens from Stat3Δ/Δ mice on days 0 and 7 following FV infection confirmed the absence of total or phosphorylated Stat3 in Stat3Δ/Δ mice (Figure 1b). Histological analysis revealed largely normal splenic architecture in the Stat3Δ/Δ mice on day 14 following infection with FVP, whereas the architecture of spleens from wild-type animals was entirely disrupted by infiltrating erythroblasts (Figure 1c). These results indicate that Stat3 plays an essential role in the expansion of primary erythroblasts in vivo in response to FV infection.
Figure 1.
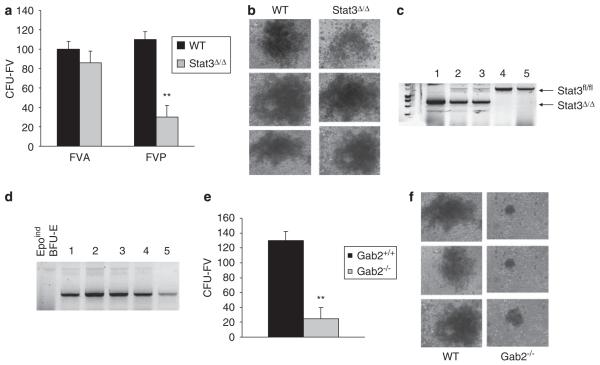
Deletion of Stat3 in vivo renders mice resistant to Friend disease. (a) Wild-type BALB/c and Balb/c-Stat3fl/fl;Mx1Cre mice were injected with phosphate-buffered saline (PBS) or pI/pC followed by infection with Friend virus. Two weeks following infection, spleens were harvested and splenic weights are shown. N=3 mice for each experimental group. (b) Analysis of Stat3 phosphorylation and protein expression. Spleens were harvested on days 0 and 7 following infection and expression of phosphorylated Stat3 and total Stat3 was assessed by western blot analysis. (c) Hematoxylin and eosin (H&E) staining of spleens from wild-type or Stat3Δ/Δ mice 14 days postinfection with FVP. *P<0.05.
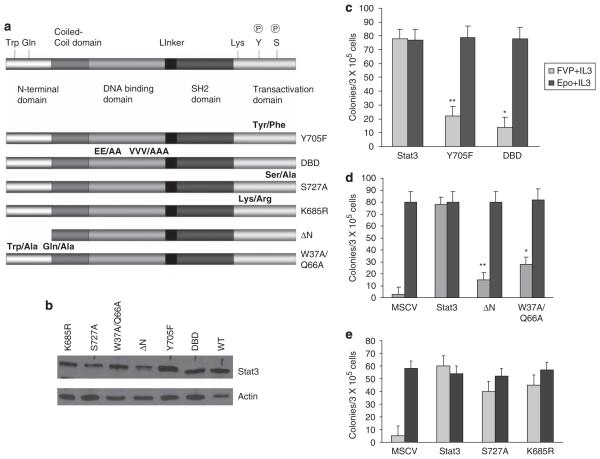
Infection of primary bone marrow with FVP in vitro results in the Epo-independent (Epoind) formation of BFU-E. Previously, we demonstrated that bone marrow from Stat3fl/fl mice transduced with Cre recombinase failed to form Epoind colonies in vitro in response to FVP infection (Ni et al., 2007). To confirm a role for Stat3 in FV induced Epoind colony formation, we transduced bone marrow from Stat3Δ/Δ mice with wild-type or mutant forms of Stat3. FVP-infected cells were plated in methylcellulose in the absence of Epo whereas mock-infected cells were plated in media containing Epo, and BFU-E colony formation was assessed. Using this approach, we demonstrated that bone marrow cells lacking Stat3 failed to form Epoind BFU-E colonies whereas cells transduced with Stat3 readily formed Epoind BFU-E colonies in response to FVP infection, the numbers of which were comparable to the number of colonies induced by Epo (Figures 2c–e). Recent studies have suggested that Stat3 mediates functions that are independent of tyrosine phosphorylation (Yang et al., 2005), however, mutants of Stat3 that blocked tyrosine phosphorylation (Y750F) or DNA binding (DBD) failed to support the Epoind growth of Stat3Δ/Δ bone marrow cells in response to FVP (Figure 2c). Tetramerization, mediated by the N-terminal domain of Stat3, has also been implicated in mediating transcriptional activation by Stat3 (Zhang and Darnell, 2001). Expression of mutants in which we deleted the N-terminal domain of Stat3 (Stat3ΔN) or mutated conserved N-terminal residues (W37A/Q66A) in bone marrow cells from Stat3Δ/Δ mice also failed to rescue FVP induced Epoind colony formation (Figure 2d). Serine phosphorylation and acetylation have been implicated in promoting Stat3 transactivation and stable dimer formation, respectively (Shen et al., 2004; Yuan et al., 2005). However, mutation of previously identified serine phosphorylation (S727) and acetylation (L685) sites in Stat3 did not impede the ability of Stat3 to support Epoind colony formation in response to FVP infection (Figure 2e). These results suggest that tyrosine phosphorylation, DNA binding and tetramerization of Stat3 are critical for the ability of Stat3 to promote Epoind growth of primary erythroblasts in response to FVP.
Figure 2.
Epo-independent colony formation in vitro in response to Friend virus infection requires tyrosine phosphorylation, DNA binding and tetramerization of Stat3. (a) Schematic representation of the Stat3 mutants used in this study. (b) Expression of the Stat3 mutants in the 293 packaging cells. (c–e) Bone marrow was extracted from Stat3Δ/Δ animals and transduced in vitro with wild-type or mutant forms of Stat3 followed by infection with Friend virus. The cells were plated in methylcellulose in the presence and absence of Epo, and BFU-E colonies were assessed on day 8 following staining with benzidine. DBD, DNA binding domain mutant; ΔN, deletion of the first 131 amino acids. **P<0.01; *P<0.05.
Stat3 regulates the differentiation of primary erythroblasts infected by FV
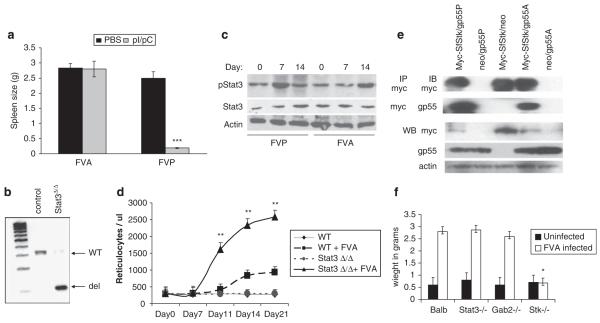
To determine whether Stat3 regulates the differentiation of FV-infected cells, we infected Stat3Δ/Δ mice or control Stat3fl/fl mice with FVP or FVA, which is a weak activator of EpoR signaling. Consistent with our previous results, Stat3Δ/Δ mice did not develop splenomegaly in response to infection with FVP. In contrast, FVA was able to induce splenomegaly in both Stat3Δ/Δ and control mice with equal efficiency (Figure 3a). PCR analysis confirmed the efficient deletion of the Stat3 locus in these mice (Figure 3b). To determine whether Stat3 is present and active in FVA-infected spleens, we performed western blot analysis on FVA- and FVP-infected splenocytes on days 0, 7 and 14 postinfection. As we have observed previously, phosphorylation of Stat3 is observed by 7 days postinfection. Although FVA also induces phosphorylation of Stat3, the phosphorylation was seen predominantly on day 14. To determine whether Sf-Stk interacts with both gp55P and gp55A, we mutated the transmembrane domain of gp55P to resemble that of gp55A. We found that both gp55A and gp55P immunoprecipitated with Sf-Stk (Figure 3e).
Figure 3.
Stat3Δ/Δ mice are resistant to disease induced by FVP, but not FVA. (a) Stat3Δ/Δ mice were infected with FVP or FVA. Two weeks following infection, spleens were isolated and splenic weights are shown. (b) Genomic DNA was isolated from the spleens of Stat3Δ/Δ or control mice followed by infection with FVA. Deletion of the Stat3 locus in the spleen 2 weeks following infection was verified by PCR analysis. (c) Spleens were harvested from Balb/c mice infected with FVA or FVP on the indicated days. Phosphorylated Stat3 and total Stat3 levels were assessed by western blot analysis. (d) Circulating reticulocyte counts from wild-type (WT) and Stat3Δ/Δ mice on the indicated days following infection with FVA. WT, uninfected Balb/c control; WT+FVA, BALB/c control infected with FVA; Stat3Δ/Δ uninfected Stat3Δ/Δ control; Stat3Δ/Δ FVP, Stat3Δ/Δ mice infected with FVA. (e) HEK 293 cells were transiently transfected with murine stem cell virus (MSCV)-based plasmids expressing myc-Sf-Stk, gp55P, gp55A and vector alone as indicated. Cell lysates were immunoprecipitated with anti-myc antibody and blotted with anti-myc or anti-gp55 antibodies. Expression of the individual proteins was confirmed by direct western blot with the indicated antibodies. (f) Wild-type BALB/c, Stat3Δ/Δ, Gab2−/− and Stk−/− mice were infected with FVA, and spleen size was assessed 2 weeks later. Uninfected spleen weights are shown for each genotype. ***P<0.001, **P<0.01, *P<0.05.
We have shown previously that Stat3 is recruited to the Sf-Stk signaling complex via a Stat3 docking site on Gab2 (Ni et al., 2007). To determine whether there is also a differential requirement for Gab2 in the presence or absence of EpoR signaling, we infected wild-type and Gab2−/− mice with FVA or FVP and examined spleen size 2 weeks following infection. As reported previously, we found that Gab2 plays a critical role in the development of splenomegaly following FVP infection (data not shown). However, like Stat3Δ/Δ mice, Gab2−/− mice exhibited splenomegaly following infection with FVA (Figure 3f). To determine whether susceptibility to FVA, like FVP, also requires Sf-Stk, we infected Stk−/− mice with FVA, and examined spleen size 2 weeks following infection. Although Stat3Δ/Δ and Gab2−/− mice were susceptible to the early stage of disease induced by FVA, Stk−/− mice remained resistant to disease induced by FVA (Figure 3f). Therefore, although Sf-Stk mediates susceptibility to both FVA and FVP, the downstream signals Gab2 and Stat3 are required for the polyclonal expansion of infected progenitors induced by FVP, but not FVA.
To determine whether Stat3 plays a role in the regulation of erythroblast differentiation following FV infection, we infected control and Stat3Δ/Δ mice with FVA. Blood samples were collected and analysed for the presence of reticulocytes. By days 10–21 postinfection, reticulocyte numbers in the Stat3Δ/Δ mice were significantly higher than those observed in control animals (Figure 3d). However, consistent with previous observations that the anemia that develops following infection with FVA is due primarily to hemodilution (Tambourin et al., 1973; Ney and D’Andrea, 2000), hematocrits dropped in both strains of FVA-infected mice and there was no significant difference in hematocrit between the two strains (data not shown). Taken together, our studies suggest that Stat3 plays a critical role in regulating the proliferation of primary erythroblasts infected with FVP, at least in part, by inhibiting differentiation thus allowing for the expansion infected erythroblasts in the presence of differentiation signals induced by the EpoR. This early regulation of differentiation signals would be dispensable for the development of erythroblastosis induced by FVA, which is a weak activator of EpoR signaling.
Stat3 regulates the expression of Pu.1 in FV-infected cells
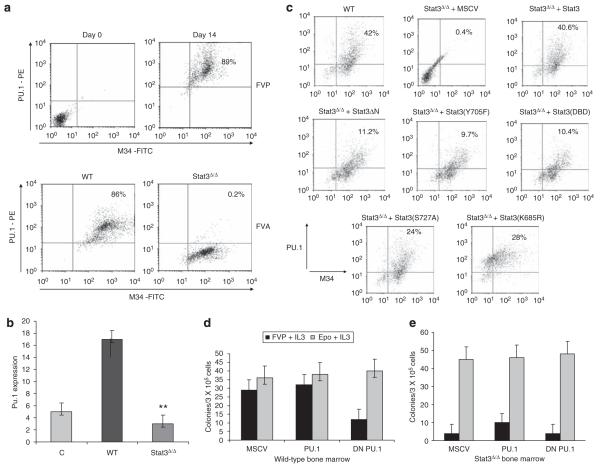
Expression of Pu.1 in myeloid cells is regulated by Stat3 (Panopoulos et al., 2002). Therefore, we set out to determine whether FV induces Pu.1 expression in a Stat3-dependent manner. Splenocytes from FVP-infected mice were harvested on days 0 and 14, followed by intracellular staining for Pu.1. The cells were cosorted for M34, a monoclonal antibody which recognizes gp55 from SFFV and gp70 from F-MuLV (Chesebro et al., 1981). We found that Pu.1 was highly expressed in FVP-infected splenocytes on day 14 postinfection (89%), compared with uninfected control spleens (Figure 4a, upper panel).
Figure 4.
Stat3 regulates Pu.1 expression following infection with Friend virus in vivo. (a) Wild-type BALB/c mice were injected with FVP (upper panel). Splenocytes were harvested on day 0 and day 14 and Pu.1 expression in infected cells (M34+) was assessed by intracellular staining and flow cytometry. Wild-type BALB/c and Stat3Δ/Δ mice were injected with FVA (lower panel) and Pu.1 expression in infected splenocytes on day 14 was assessed by flow cytometry. (b) Splenic RNA harvested from uninfected BALB/c mice (Control), FV-infected BALB/c mice (WT) and FV-infected Stat3Δ/Δ mice on day 14 was assessed for Pu.1 expression by real-time PCR. (c) Bone marrow from WT BALB/c or Stat3Δ/Δ mice transduced in vitro with vector alone (MSCV), WT Stat3 or the indicated mutants was infected with FVP overnight and expression of Pu.1 in infected cells was assessed by flow cytometry. (d) Bone marrow was harvested from wild-type BALB/c mice and transduced with empty murine stem cell virus (MSCV) or an MSCV-based retroviral vector expressing wild-type or dominant-negative Pu.1 followed by infection with FVP. The cells were plated in methylcellulose and BFU-E numbers were assessed on day 8 following staining with benzidine. (e) Bone marrow from Stat3Δ/Δ mice was transduced with wild-type or dominant-negative Pu.1 as described above followed by infection with FVP. Cells were plated in methylcellulose and BFU-E numbers were assessed on day 8 following benzidine staining. **P<0.01.
To determine whether Pu.1 expression is regulated by Stat3 in vivo, we infected mice with FVA, which promotes erythroblastosis in both control and Stat3Δ/Δ mice. Although Pu.1 was highly expressed in FVA-infected cells from control mice, little or no expression of Pu.1 was observed in Stat3Δ/Δ animals (Figure 4a, lower panel). To quantify the levels of Pu.1 expression, we examined Pu.1 expression in spleens from control and Stat3Δ/Δ mice infected with FVA by real-time PCR. We observed a significant increase in Pu.1 expression levels in spleens from FVA-infected control mice when compared with spleens from uninfected animals (Figure 4b). However, the levels of Pu.1 expression were greatly reduced in spleens from infected Stat3Δ/Δ mice, which also develop splenomegaly in response to FVA, indicating that the induction of Pu.1 expression in response to FV infection is Stat3 dependent.
To determine what domains of Stat3 are required for the induction of Pu.1 expression, we isolated bone marrow cells from Stat3Δ/Δ mice, transduced these cells with wild-type or mutant forms of Stat3, infected the cells overnight with FVP and assessed Pu.1 expression by flow cytometry. When wild-type Stat3 was expressed in Stat3Δ/Δ cells, the percentage of infected cells expressing Pu.1 (40.6%) was similar to that observed for wild-type bone marrow cells (Figure 4c). When the Stat3Δ/Δ cells were reconstituted with Stat3(Y705F), Stat3(DBD) or Stat3Δ/ΔN, the percentage of cells (11.2, 9.7 and 10.4%, respectively) expressing Pu.1 was significantly reduced compared with control. In contrast, a higher percentage of Stat3(S727A)- and Stat3(L685A)-transduced cells expressed Pu.1 (24 and 28%, respectively). These data support a role for tyrosine phosphorylation, DNA binding and tetramerization of Stat3 in the induction of Pu.1 expression.
Epoind colony formation in response to FVP requires the expansion of infected erythroblasts, followed by terminal differentiation, induced by EpoR activation. To determine whether Pu.1 plays a role in the Epoind colony formation induced by FVP in vitro, we transduced primary bone marrow cells from control mice with retroviral vectors expressing wild-type or dominant-negative Pu.1. These cells were infected with FVP, and BFU-E formation in the absence of Epo was assessed. Although exogenous expression of wild-type Pu.1 had no effect on colony formation, expression of the dominant-negative Pu.1 inhibited Epoind colony formation in response to FV infection (Figure 4d), indicating that Pu.1 expression may be critical in promoting the expansion of infected cells in vitro, before terminal differentiation. However, expression of exogenous Pu.1 alone in Stat3Δ/Δ bone marrow failed to rescue the response of these cells to FVP infection (Figure 4e), suggesting that there may be additional targets downstream of Stat3 required for disease development.
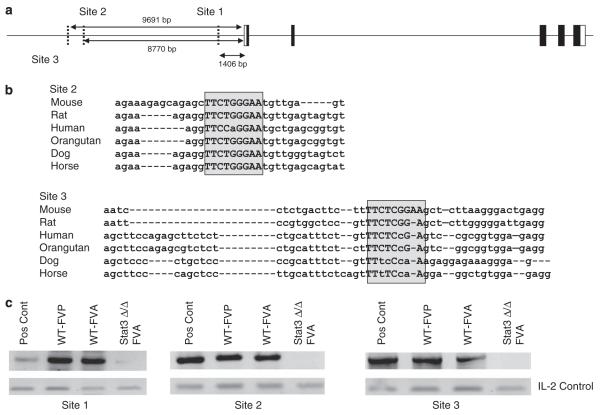
To determine whether Stat3 directly interacts with the Pu.1 promoter in infected cells, we analysed an 18 kb genomic sequence upstream of the murine Pu.1 start site. We identified three potential Stat3 binding sites which we termed sites 1, 2 and 3 (Figure 5a). Site 1 is not conserved in mammals, however, sites 2 and 3 are highly conserved among all species examined (Figure 5b). Using chromatin immunoprecipitation (ChIP) analysis on splenocytes from control mice infected with FVP or FVA, we observed Stat3 binding to all three sites in the Pu.1 promoter. However, little or no binding was observed in splenocytes from Stat3Δ/Δ animals infected with FVA, which also develop splenomegaly (Figure 5c). These results are consistent with a role for Stat3 in the direct regulation of Pu.1 expression in vivo.
Figure 5.
Stat3 binds to sites in the Pu.1 promoter region in vivo following infection with Friend virus. (a) Schematic representation of the putative Stat3 binding elements in the Pu.1 promoter region. Their positions are shown relative to the start of transcription. (b) The sequences of sites 2 and 3 are aligned from multiple species. The boxed region indicates the putative Stat3 binding sites. Within the boxed region, capital letters indicate conserved nucleotides and lower case letters indicate nonconserved nucleotides relative to the mouse sequence. (c) Wild-type BALB/c mice were infected with FVA or FVP and Stat3Δ/Δ mice were infected with FVA. Splenocytes were harvested from these animals on day 10 following infection, and binding of Stat3 to elements in the Pu.1 promoter was assessed by chromatin immunoprecipitation (ChIP) analysis. Pos Cont; input DNA. IL-2, nonspecific control for DNA content.
Stat3 is not required for the expansion of leukemic cells following FV infection
Following the initial stage of Friend disease, in which infected erythroblasts proliferate and differentiate in the spleen in a polyclonal manner, leukemic clones emerge which are capable of forming spleen colonies in FV resistant mice on transplantation. These cells, termed CFU-FV, form blast-like colonies in methylcellulose in the absence of exogenous growth factor stimulation (Mager et al., 1981). To determine whether Stat3 is required for the generation and/or growth of leukemic cells, we infected Stat3Δ/Δ or control mice with FVA or FVP, isolated splenocytes 4 weeks postinfection, and assessed CFU-FV colony formation. We found that there were reduced numbers of CFU-FV in response to FVP infection (Figure 6a). However, the size of the CFU-FV colonies derived from Stat3Δ/Δ mice following FVP infection are not significantly different than those that develop from control animals (Figure 6b). To determine whether these CFU-FV colonies contain Stat3, we performed PCR analysis to genotype individual colonies for deletion of the Stat3 locus. We found that, although some of the colonies contained the wild-type allele of Stat3, others developed from cells in which Stat3 was deleted (Figure 6c). To determine whether these colonies harbor retroviral insertions in the PU.1 locus, we performed PCR analysis of the PU.1 insertion site. Regardless of Stat3 genotype, all of the CFU-FV colonies contained proviral insertions at the PU.1 locus (Figure 6d). Insertion at this locus was not observed in Epoind colonies following FVP infection in vitro. These results indicate that Stat3 is not required for the cytokine-independent expansion of leukemic cells that develop in response to FVP infection, and that the requirement for Stat3 may be overcome by genetic alterations that occur at this stage, including insertional activation of PU.1.
Figure 6.
Gab2, but not Stat3, is required for the expansion of CFU-FV. (a) Stat3fl/fl mice (WT) or Stat3Δ/Δ mice were infected with FVP. Four weeks later, splenocytes were harvested and plated in methylcellulose in the absence of exogenous growth factors and numbers of CFU-FV were assessed. (b) Representative CFU-FV from WT and Stat3Δ/Δ mice infected with FVP. (c) Individual CFU-FV colonies from Stat3Δ/Δ mice infected with FVP were picked and genotyped for the Stat3 locus by PCR. (d) The same individual colonies were analysed by PCR for retroviral insertion at the Pu.1 locus. Epoind BFU-E; BFU-E formed in the absence of Epo following FVP infection of primary bone marrow cells in vitro. (e) Wild-type BALB/c and Gab2−/− mice were injected with FVP. Four weeks later, splenocytes were harvested and plated in methylcellulose in the absence of exogenous growth factors and numbers of CFU-FV were assessed. (f) Representative CFU-FV from wild-type and Gab2−/− mice. **P<0.01.
To determine whether Gab2 is required for the development or proliferation of leukemic cells in response to FV infection, we compared CFU-FV formation by splenocytes from wild-type or Gab2−/− mice 4 weeks postinfection with FVP. We also observed a significant reduction in CFU-FV development in the absence of Gab2 (Figure 6e). However, unlike the CFU-FV that arise from Stat3Δ/Δ cells, the colonies that develop from the spleens of Gab2−/− mice are much smaller in size than those derived from wild-type control animals, suggesting that Gab2 may provide signals, in addition to Stat3, that are required for the efficient proliferation of CFU-FV (Figure 6f). Taken together, these results suggest that, although Gab2 leads to the recruitment and activation of Stat3, which is required for the early stages of disease in response to FVP, Gab2, but not Stat3, plays a critical role in the proliferation of fully transformed leukemic cells.
Discussion
The erythroleukemia induced by FV is a multistep process, characterized by an early stage of erythroblast proliferation and differentiation followed by a late-stage differentiation block and leukemic transformation. The development of leukemia results from the cooperation of two classes of mutations, one conferring a proliferative advantage and one blocking differentiation. In Friend disease, the proliferative signal is a result of the activation of Sf-Stk by the viral glycoprotein, gp55, in the early stage of disease, whereas the block to differentiation is accomplished by insertional activation of the Pu.1 locus in the late stage of the disease. Here, we identify a novel role for Stat3 in the regulation of erythroblast differentiation in the early stage of the disease, thus promoting the expansion of infected erythroblasts induced by Sf-Stk. Furthermore, we have demonstrated, for the first time, a role for Stat3 in the regulation of Pu.1 expression in vivo, suggesting a mechanism by which Stat3 may achieve regulation of differentiation. We propose that Stat3-dependent activation of Pu.1 following infection with FVP, in the absence of proviral activation of Pu.1, tips the balance of signals toward an undifferentiated state in the early progenitor cells, thus promoting the efficient expansion of infected cells before terminal differentiation induced by activation of the EpoR. The result is the development of disease in which infected erythroblasts proliferate, and ultimately differentiate, resulting in the development of Epoind colonies in vitro and polycythemia in vivo.
Previous studies have demonstrated that transgenic expression of Pu.1 in mice results in the multistage development of erythroleukemia (Moreau-Gachelin et al., 1996). At the onset of disease, these mice become severely anemic, accompanied by a massive expansion of proerythroblasts that are partially blocked in differentiation, followed later by malignant transformation. Interestingly, the majority of tumors isolated from these mice during the late stage of the disease harbored acquired mutations in the Kit receptor (Kosmider et al., 2005). Conversely, knockdown of Gata-1 results in the development early-onset nonlymphoid leukemias, which are c-Kit-positive. Low-level GATA-1 expression in these animals is sufficient to support survival and proliferation but not differentiation (Shimizu et al., 2004). These studies support the prediction that two events are required for the development of leukemogenesis, one that promotes proliferation and one that blocks differentiation. Our studies indicate that Stat3 plays a critical role in promoting the block to differentiation in Friend disease, and that the requirement for Stat3 in the late stage of the disease may be negated by the insertional activation of Pu.1 expression by the SFFV provirus.
In conclusion, our data support a model, in which, during the early polyclonal expansion of FVP-infected cells, the viral protein, gp55, leads to the constitutive activation of Sf-Stk which, in turn, recruits a Grb2/Gab2 complex. The activation of Gab2 by Sf-Stk promotes the Epoind proliferation and/or survival of infected erythroblasts and, at the same time, inhibits the differentiation of these cells through the activation of Stat3, and the subsequent upregulation of Pu.1 expression. In the absence of EpoR activation following infection with FVA, however, this block to differentiation is not required to promote the expansion of infected cells. Alternatively, the clonal expansion of leukemic cells during the late stage of Friend disease requires Gab2, but not Stat3, resulting in signals that promote proliferation and survival, whereas retroviral insertion into the Pu.1 locus plays a central role in maintaining the undifferentiated state of the leukemic population. The insertional activation of Pu.1, and possibly other genetic alterations occurring at this stage, would ultimately override the requirement for Stat3 in maintaining leukemic transformation.
Materials and methods
Mice
STAT3fl/fl mice (B6. Cg-Stat3tm2Aki; Sano et al., 1999) were crossed four generations onto the BALB/c background. The genotype for the Fv2 locus was determined by PCR using the primers 5′-GGTGGGTTTAACGGTTAGGG-3′ and 5′-TCTGGGCTCTGCCTCCTTAT-3′. These mice were crossed to the IFN-inducible Cre recombinase transgenic mouse line, B6.Cg-Tg(Mx1-cre)1Cgn/J (Kuhn et al., 1995), also crossed onto the BALB/c background. To delete Stat3, mice were injected with 200 μl of PolyI/PolyC (Cat no. p-1958, Sigma St Louis, MO, USA) in phosphate-buffered saline (PBS) at a concentration of 2 mg/ml intraperitoneally daily for three injections and on alternate days for five injections. The Stat3 locus was genotyped using the primers 5′-CCTGAAGACCAAGTTCATCTGTGTGAC-3′, 5′-CACACAAGCCATCAAACTCTGGTCTCC-3′ and 5′-GATTTGAGTCAGGGATCCATAACTTCG-3′ to produce amplified products of the germline (2000 bp), floxed (270 bp) and deleted (75 bp) alleles. The Gab2−/− mice used in this study were described previously (Ni et al., 2007). All research involving the use of mice was performed in strict accordance with protocols approved by the Institutional Animal Care and Use committee of the Pennsylvania State University.
DNA constructs
Stat3(Y705)F and Stat3(EE/VVV) (DNA-binding domain mutant (DBD)) were obtained from C Horvath (Northwestern University) and MSCV-Stat3, MSCV-Stat3(Y705F) and MSCV-Stat3(DBD) were generated as described previously (Ni et al., 2007). MSCV-Stat3ΔN was generated by digesting MSCV-Stat3 with Pst1 and Xba1, resulting in deletion of the first 131 amino acids. MSCV-Stat3W37A/Q66A was generated from MSCV-Stat3 using the primers 5′-CGGCAGTTCCTGGCACCTGCGATTGAGAGTCAAGACTGG-3′ and 5′-CCAGTCTTGACTCTCAATCGCAGGTGCCAGGAACTGCCG-3′ for W37A and the primers 5′-CTCTTGGGTGAAATTGACGCGCAATATAGCCGATTCCTG-3′ and 5′-CAGGAATCGGCTATATTGCGCGTCAATTTCACCCAAGAG-3′ for Q66A as described previously (Zhang and Darnell, 2001). Serine 727 was mutated to alanine using the primers 5′-AGCCAAGGAGACTGCCGGATCTGA3′ and 5′-TCAGATCCGGCAGTCTCCTTGGCT-3′. Lysine 685 was mutated to arginine using the primers 5′-CGGAAGCGAGTGCAGCAGGATCTA-3′ and 5′-TAGATCCTGCTGCACTCGCTTCCG-3′.
To generate gp55A, a M390I mutation was introduced into gp55P using primers 5′-CGTTGATATCCGCCATCATCGGGTCTCTCATTATACTCC-3′ and 5′-GGAGTATAATGAGAGACCCGATGATGGCGGATATCAACG-3′. A double deletion of L399 and L400 was introduced using primers 5′-GTCTCTCATTATACTCCTACTCATTCTGCTTATTTGGACCCTG-3′ and 5′-CAGGGTCCAAATAAGCAGAATGAGTAGGAGTATAATGAGAGAC-3′. All point mutations and deletions were generated using the Stratagene (La Jolla, CA, USA) QuikChange mutagenesis kit.
Generation of viral supernatants
293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum, penicillin-strepto-mycin and L-glutamine. Cells were transiently transfected with 1–2 μg pEco and 2.5 μg murine stem cell virus (MSCV) using the Trans IT-293 transfection reagent (Mirus corporation, Madison, WI, USA) at 37 °C for 48–72 h before harvest of the viral supernatant.
In vitro bone marrow infections and colony assays
Primary bone marrow cells were infected with viral supernatants from the transient transfections for 20 h. For Epoindependent colony assays, the bone marrow cells were incubated with supernatants from cells expressing FVP or DMEM on ice for 1 h. The cells were then added to Methocult Medium M3234 (Stemcell Technologies, Vancouver, BC, USA), along with 2.5 ng interleukin (IL)-3 (Peprotech, Rocky Hill, NJ, USA), in triplicate with or without 1 U/ml Epo (R&D Systems, Minneapolis, MN, USA). Cultures were incubated for 8 days in 5% CO2 at 37 °C. Erythroid colonies were visualized by acid-benzidine staining. For the CFU-FV assay, mice were injected with FVP and 4 weeks later, splenoctyes were isolated and plated on Methocult M3231 (Stemcell Technologies). CFU-FV colonies were scored after 2 weeks. To detect the presence of the provirus in the Pu.1 locus we utilized a primer in the viral LTR, 5′-CACTAGAATACGAGCCACGATAAAT-3′ and a primer in the Pu.1 gene, 5′-CTTTCACTTGTGTAGTTGAAGATGG-3′. DNA from individual CFU-FV were subjected to 37 cycles of PCR of 1 min at 94C, 30 s at 60 °C and 3 min at 68 °C, in a final volume of 50 μl, using Takara Extaq (Code no. RR001).
Antibodies and western blot analysis
Anti-Stat3 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA; Cat no. SC-482). Anti-phospho-Stat3 (Cat no. 9131) and anti-myc were purchased from Cell Signaling (Danvers, MA, USA). Rabbit anti-β-actin was purchased from Sigma. Protein G magnetic beads were purchased from New England BioLabs. Cells were lysed in lysis buffer (1% NP-40, 150 mm Nac1, 1 mm EDTA, 0.25% deoxycholate, 2 mm Na3VO4, 10 mm NaF, 2 mm phenylmethylsulfonyl fluoride). After 15 min of centrifugation, the cleared lysates were immunoprecipitated and/or resolved by reduced SDS–polyacrylamide gel electrophoresis, the blots were incubated with the indicated antibodies and the bands were visualized with ECL (Amersham, Piscataway, NJ, USA).
Flow Cytometry
Mice were injected with FVP retroorbitally. Splenocytes were blocked with Fc block (BD Pharmingen) and stained with the M34 antibody (Chesebro et al., 1981) conjugated with FITC and Pu.1 antibody conjugated with PE (Santa Cruz Biotechnology) for analysis by flow cytometry.
RT–PCR
Quantitative RT–PCR was performed for Pu.1. using superscript II RNase H reverse transcriptase (Invitrogen Life Technologies), the 7300 Real-time PCR system (Applied Biosystems) and the Taqman genexpression assay for Pu.1 (Mm01719550) and GapDH (Mm99999915g1) from Applied Biosystems.
Chromatin immunoprecipitation
Mice were injected with FVP or FVA and splenocytes were isolated on day 10. Chromatin was prepared using the EZZyme chromatin kit. The samples were immunoprecipitated with anti-Stat3 (Cell Signaling; Cat no. 9132) and DNA was isolated using the Magna ChIP kit (Upstate Biotechnology, Lake Placid, NY, USA). The Primers used for PCR were: Site 1, 5′-ACTTTAAAGCCCAGCACTCG-3′ and 5′-ATCCATCCATCCATGCATCT-3′; Site 2, 5′-CCAGGAACCGGAATAGAACA-3′ and 5′-CATGCCAACCAGAAGACTCA-3′; Site 3, 5′-GAGAGCAGAGGGCTCAGAGA-3′ and 5′-AGTCCCGTCTCCAGAACTCA-3′. Nonspecific DNA in the immunoprecipitates was analysed using 5X excess template and the primers in the IL-2 gene: 5′-CTAGGCCACAGAATTGAAAGATCT-3′ and 5′-GTAGGTGGAAATTCTAGCATCATCC-3′.
Intracellular staining for Pu.1
Cells were fixed for 40 min at room temperature with 200 μl of 1× formaldehyde buffer (2.7 ml 37% formaldehyde solution in 1× PBS). The cells were centrifuged and the fixation solution was decanted. The cells were permeabilized with 200 μl 70% EtOH and incubated for 50 min at −20 °C. EtOH was decanted, the cells were washed with PBS and incubated with anti-Pu.1 antibody (Santa Cruz; Cat no. SC-352) for 45 min on ice. Cells were washed with PBS and incubated with anti-rabbit Alexa 660 (Invitrogen; A21074) for 45 min on ice. Cells were washed and Pu.1 staining was detected by flow cytometry.
Statistics
Statistical evaluation was performed using the Student’s t-test. P<0.05 was considered statistically significant.
Acknowledgements
This work was supported by grant R01 HL066571 from the National Institutes of Health and by a scholar award from the Leukemia and Lymphoma Society to P Hankey.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Afrikanova I, Yeh E, Bartos D, Watowich SS, Longmore GD. Oncogene cooperativity in Friend erythroleukemia: erythropoietin receptor activation by the env gene of SFFV leads to transcriptional upregulation of PU.1 independent of SFFV proviral insertion. Oncogene. 2002;21:1272–1284. doi: 10.1038/sj/onc/1205183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K, Cloyd M, Britt W, Portis J, Collins J, et al. Characterization of mouse monoclonal antibodies specific for Friend murine leukemia virus-induced erythroleukemia cells: friend-specific and FMR-specific antigens. Virology. 1981;112:131–144. doi: 10.1016/0042-6822(81)90619-x. [DOI] [PubMed] [Google Scholar]
- Choe KS, Radparvar F, Matushansky I, Rekhtman N, Han X, Skoultchi AI. Reversal of tumorigenicity and the block to differentiation in erythroleukemia cells by GATA-1. Cancer Res. 2003;63:6363–6369. [PubMed] [Google Scholar]
- Constantinescu SN, Keren T, Russ WP, Ubarretxena-Belandia I, Malka Y, Kubatzky KF, et al. The erythropoietin receptor transmembrane domain mediates complex formation with viral anemic and polycythemic gp55 proteins. J Biol Chem. 2003;278:43755–43763. doi: 10.1074/jbc.M302974200. [DOI] [PubMed] [Google Scholar]
- Fang C, Choi E, Nie L, Li JP. Role of the transmembrane sequence of spleen focus-forming virus gp55 in erythroleukemogenesis. Virology. 1998;252:46–53. doi: 10.1006/viro.1998.9453. [DOI] [PubMed] [Google Scholar]
- Finkelstein LD, Ney PA, Liu QP, Paulson RF, Correll PH. Sf-Stk kinase activity and the Grb2 binding site are required for Epo-independent growth of primary erythroblasts infected with Friend virus. Oncogene. 2002;21:3562–3570. doi: 10.1038/sj.onc.1205442. [DOI] [PubMed] [Google Scholar]
- Kosmider O, Denis N, Lacout C, Vainchenker W, Dubreuil P, Moreau-Gachelin F. Kit-activating mutations cooperate with Spi-1/PU.1 overexpression to promote tumorigenic progression during erythroleukemia in mice. Cancer Cell. 2005;8:467–478. doi: 10.1016/j.ccr.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Mager DL, Mak TW, Bernstein A. Quantitative colony method for tumorigenic cells transformed by two distinct strains of Friend leukemia virus. Proc Natl Acad Sci USA. 1981;78:1703–1707. doi: 10.1073/pnas.78.3.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau-Gachelin F, Wendling F, Molina T, Denis N, Titeux M, Grimber G, et al. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol. 1996;16:2453–2463. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ney PA, D’Andrea AD. Friend erythroleukemia revisited. Blood. 2000;96:3675–3680. [PubMed] [Google Scholar]
- Ni S, Zhao C, Feng GS, Paulson RF, Correll PH. A novel Stat3 binding motif in Gab2 mediates transformation of primary hematopoietic cells by the Stk/Ron receptor tyrosine kinase in response to Friend virus infection. Mol Cell Biol. 2007;27:3708–3715. doi: 10.1128/MCB.01838-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, Bartos D, Zhang L, Watowich SS. Control of myeloid-specific integrin alpha Mbeta 2 (CD11b/CD18) expression by cytokines is regulated by Stat3-dependent activation of PU.1. J Biol Chem. 2002;277:19001–19007. doi: 10.1074/jbc.M112271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papetti M, Skoultchi AI. Reprogramming leukemia cells to terminal differentiation and growth arrest by RNA interference of PU.1. Mol Cancer Res. 2007;5:1053–1062. doi: 10.1158/1541-7786.MCR-07-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persons DA, Paulson RF, Loyd MR, Herley MT, Bodner SM, Bernstein A, et al. Fv2 encodes a truncated form of the Stk receptor tyrosine kinase. Nat Genet. 1999;23:159–165. doi: 10.1038/13787. [DOI] [PubMed] [Google Scholar]
- Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Schlessinger K, Zhu X, Meffre E, Quimby F, Levy DE, et al. Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol. 2004;24:407–419. doi: 10.1128/MCB.24.1.407-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu R, Kuroha T, Ohneda O, Pan X, Ohneda K, Takahashi S, et al. Leukemogenesis caused by incapacitated GATA-1 function. Mol Cell Biol. 2004;24:10814–10825. doi: 10.1128/MCB.24.24.10814-10825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambourin PE, Gallien-Lartigue O, Wendling F, Huaulme D. Erythrocyte production in mice infected by the polycythaemia-inducing Friend virus or by the anaemia-inducing Friedn virus. Br J Haematol. 1973;24:511–524. doi: 10.1111/j.1365-2141.1973.tb01677.x. [DOI] [PubMed] [Google Scholar]
- Teal HE, Ni S, Xu J, Finkelstein LD, Cheng AM, Paulson RF, et al. GRB2-mediated recruitment of GAB2, but not GAB1, to SF-STK supports the expansion of Friend virus-infected erythroid progenitor cells. Oncogene. 2006;25:2433–2443. doi: 10.1038/sj.onc.1209288. [DOI] [PubMed] [Google Scholar]
- Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- Zhang J, Randall MS, Loyd MR, Li W, Schweers RL, Persons DA, et al. Role of erythropoietin receptor signaling in Friend virus-induced erythroblastosis and polycythemia. Blood. 2006;107:73–78. doi: 10.1182/blood-2005-05-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Darnell JE., Jr Functional importance of Stat3 tetramerization in activation of the alpha 2-macroglobulin gene. J Biol Chem. 2001;276:33576–33581. doi: 10.1074/jbc.M104978200. [DOI] [PubMed] [Google Scholar]