Abstract
The spliceosome is the complex macromolecular machine responsible for removing introns from precursors to mRNAs (pre-mRNAs). We combined yeast genetic engineering, chemical biology, and multi-wavelength fluoresence microscopy to follow assembly of single spliceosomes in real time in whole cell extracts. We find that individual spliceosomal subcomplexes associate with pre-mRNA sequentially via an ordered pathway to yield functional spliceosomes, and that association of every subcomplex is reversible. Further, early subcomplex binding events do not fully commit a pre-mRNA to splicing; rather commitment increases as assembly proceeds. These findings have important implications for the regulation of alternative splicing. This experimental strategy should prove widely useful for mechanistic analysis of other macromolecular machines in environments approaching the complexity of living cells.
Complex macromolecular machines are involved in every aspect of cellular information processing, from DNA synthesis to protein destruction. These machines can involve dozens, or hundreds, of components and are often too complex to fully reconstitute from purified parts. One such machine is the spliceosome, responsible for removing introns from nascent transcripts via pre-mRNA (precursor to mRNA) splicing. Arguably the most complex macromolecular machine in the cell (1), the spliceosome is composed of 5 small nuclear RNAs (snRNAs) and ~100 core proteins minimally required for activity in vitro (2). The snRNAs and many core proteins are arranged into stable subcomplexes constituting small nuclear ribonucleoprotein particles (U1 and U2 snRNPs and the U4/U6.U5 tri-snRNP) and the multi-protein Prp19-complex (NTC). While association of U1 with pre-mRNA can occur in the absence of ATP, stable association of all other subcomplexes requires ATP hydrolysis. Intron excision occurs after the spliceosome has been fully assembled and activated via additional structural rearrangements (3).
Current models of spliceosome assembly, activation, and catalysis generally depict it as an ordered (U1→U2→tri-snRNP→NTC→activation→catalysis), one-way process (3). Yet deviations from the ordered assembly model have been reported (4–6), with some studies suggesting that both spliceosome assembly and catalysis are dynamic and reversible (7–9). None of these studies, however, directly examined the kinetics of subcomplex association with pre-mRNA. Here, we monitored subcomplex dynamics during spliceosome assembly in real time by combining yeast genetic engineering, chemical biology, and a multi-wavelength fluorescence technique: Co-localization Single Molecule Spectroscopy (CoSMoS) (10).
Labeling Spliceosome Subcomplexes by Combining Chemical Tags with Genetic Engineering
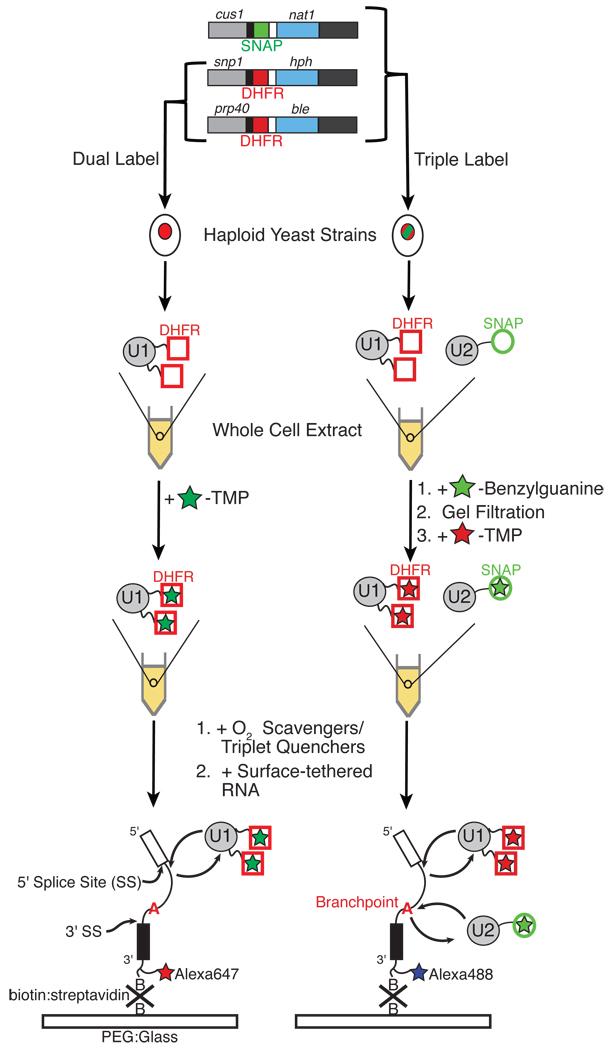
We previously established that splicing of single pre-mRNA molecules can be monitored by multi-wavelength total internal reflection fluorescence (TIRF) microscopy in the complex environment of S. cerevisiae whole cell extract (yeast WCE) (11). To enable kinetic analysis of spliceosome assembly, we have now developed methods to introduce fluorophores into individual spliceosomal subcomplexes in WCE. Protein labeling was accomplished using homologous recombination to fuse either a SNAP (an alkyl-guanine S-transferase) (12) or an E. coli DHFR (dihydrofolate reductase) tag (13) onto the C-terminus of numerous spliceosomal proteins. These tags enabled us to incorporate bright, photostable organic dyes into the subcomplexes and to avoid the poor photon output and blinking behavior of single fluorescent proteins (14). Integration of two orthogonal tags allows for simultaneous monitoring of two different subcomplexes by CoSMoS (Figure 1). To ensure functionality of the tagged species, we tagged only essential proteins and verified that the resultant strains (Table S1) had growth rates and in vitro splicing activities comparable to the parental strain (Figures S1–S3). By using several selectable markers, we were able to incorporate up to three tags into a single strain. Multiple tags present in the same subcomplex minimized artifacts due to incomplete labeling, photobleaching, and/or long-lived dark state formation of single fluorophores (15).
Figure 1.
Preparation and analysis of fluorescently-labeled spliceosome subcomplexes by CoSMoS.
DHFR tags were labeled by adding excess (20 nM) fluorophore-trimethoprim (TMP) conjugates (e.g., Cy3-TMP) to WCE. TMP binding to DHFR is non-covalent, but the ternary complex formed between DHFR, TMP, and endogenous NADPH (20–30 µM in WCE) is extremely long-lived [(koff = ~0.0005 s−1; data not shown) and (16)]. SNAP tags were covalently labeled by incubating WCE with benzylguanine dye conjugates (e.g. SNAP-DY549) and then removing excess dye by gel filtration. Gel electrophoresis (SDS-PAGE) confirmed labeling specificity (Figure S4A) and efficiency (70–90% labeling of functional SNAP tags; Figure S4B). None of the dye adducts or labeling procedures employed here significantly inhibit splicing in vitro (Figures S2,3).
Single molecule experiments were carried out in WCE containing fluorescently tagged proteins, an O2 scavenging system, and triplet-state quenchers (17). Data were acquired using a TIRF microscope with laser excitation at 488, 532, and 633 nm. Such TIRF experiments detect surface-bound molecules as discrete spots, while fluorescent components in solution remain as diffuse background. To monitor spliceosome assembly, a model pre-mRNA derived from the rp51a transcript (11, 18) containing a single fluorophore and 3' biotin was tethered to a streptavidin-derivatized glass surface at densities of 100–250 pre-mRNA molecules per 314 µm2 field of view (FOV) (Figure 1). Arrivals and departures of individual spliceosomal subcomplexes were visualized as the appearance and disappearance of fluorescent spots that co-localized with surface-tethered pre-mRNAs.
Subcomplexes Accumulate on Surface-tethered pre-mRNAs and form Functional Spliceosomes
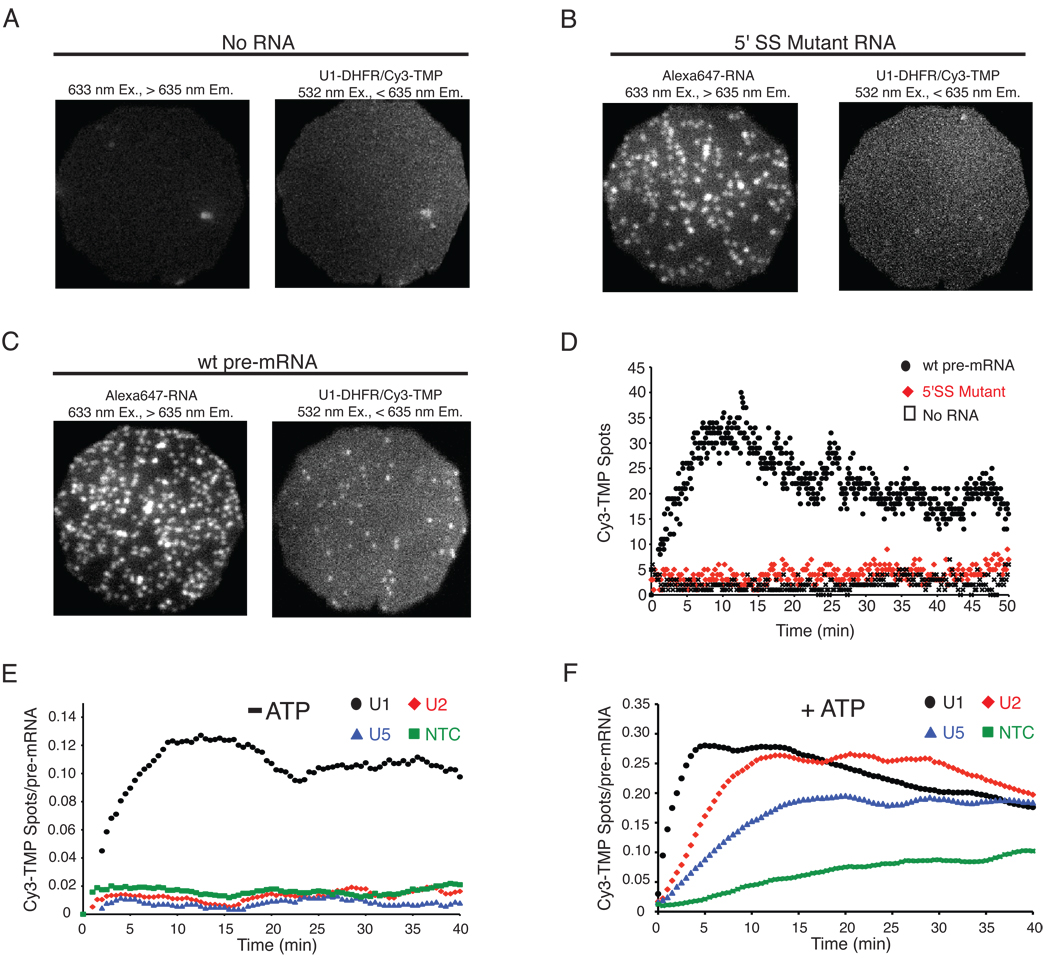
The first subcomplex to bind during spliceosome assembly is thought to be U1 snRNP, which interacts with the 5' splice site (SS). To validate our approach, we monitored U1 association (via DHFR/Cy3-TMP tags on U1 components Snp1 and Prp40) in the presence of ATP with either wild type (wt) pre-mRNA or a mutant version in which the 5' SS had been mutated (G/GUAUGU → c/aUAccU). No stable association was observed in the absence of tethered RNA or with the 5' SS mutant pre-mRNA (Figure 2A,B). As expected, U1 spots were present on a surface containing wt pre-mRNA (Figure 2C). Monitoring of U1 association with wt pre-mRNA over time revealed rapid surface accumulation of U1 signals during the first 5 minutes (Figure 2D, Movies S1–S2). In contrast, no time-dependent signal accumulation was observed in the absence of pre-mRNA or with the 5' SS mutant. Thus, the long-lived signals are dependent both on the presence of pre-mRNA and an intact 5' SS.
Figure 2.
Individual DHFR-labeled subcomplexes binding to surface-tethered pre-mRNAs. Ex. = excitation wavelength; Em. = emission wavelength. (A–C) Images of three fields of view (FOV, 20 × 20 µm), each at two different emission wavelengths, showing that U1 DHFR Cy3-TMP fluorescence signals (spots) are only observed when wt pre-mRNA is present (C). (D) U1 spots versus time in individual FOVs containing RNAs indicated. Note: For wt pre-mRNA, the decrease in spot number after 10 minutes is due to Cy3-TMP photobleaching in this experiment. Experiments in (A–D) contained ATP. (E–F) Smoothed (9-point moving block averaged) curves of indicated subcomplex spots per pre-mRNA versus time, minus (E) or plus (F) ATP. Each subcomplex was monitored in a different WCE. Data in (F) are the average of N = 4 replicates.
We next compared the kinetics of U1 association with those of U2, tri-snRNP, and the NTC. Binding events for individual subcomplexes were monitored in separate experiments using WCEs containing two DHFR/Cy3-TMP tags on a given subcomplex (Table S1). U2 was labeled via the U2-SF3b components Cus1 and Hsh155. Both U1 and U2-SF3b are thought to stably associate with pre-mRNA during assembly and then be expelled prior to catalytic activation (2, 19). The tri-snRNP and NTC were individually labeled via Brr2 and Snu114 (core U5 components) and Cef1 and Ntc90 (core NTC components). Both U5 and NTC are thought to remain spliceosome-associated throughout activation and catalysis, departing only upon mRNA product release.
As expected, only U1 spots accumulated on wt pre-mRNA in the absence of ATP (Figure 2E, Movie S1). In contrast, all subcomplexes accumulated in the presence of ATP, albeit at different rates (Figure 2F, Movie S2). These rates were consistent with an apparent order of assembly: U1 → U2 → tri-snRNP → NTC. Like U1, accumulation of U2, U5, and NTC was also dependent on an intact 5' SS (Figure S5A), confirming the specificity of the interactions for a splicing-competent pre-mRNA. Similar results were obtained with the analogous SNAP-tagged strains (Figure S5B–E).
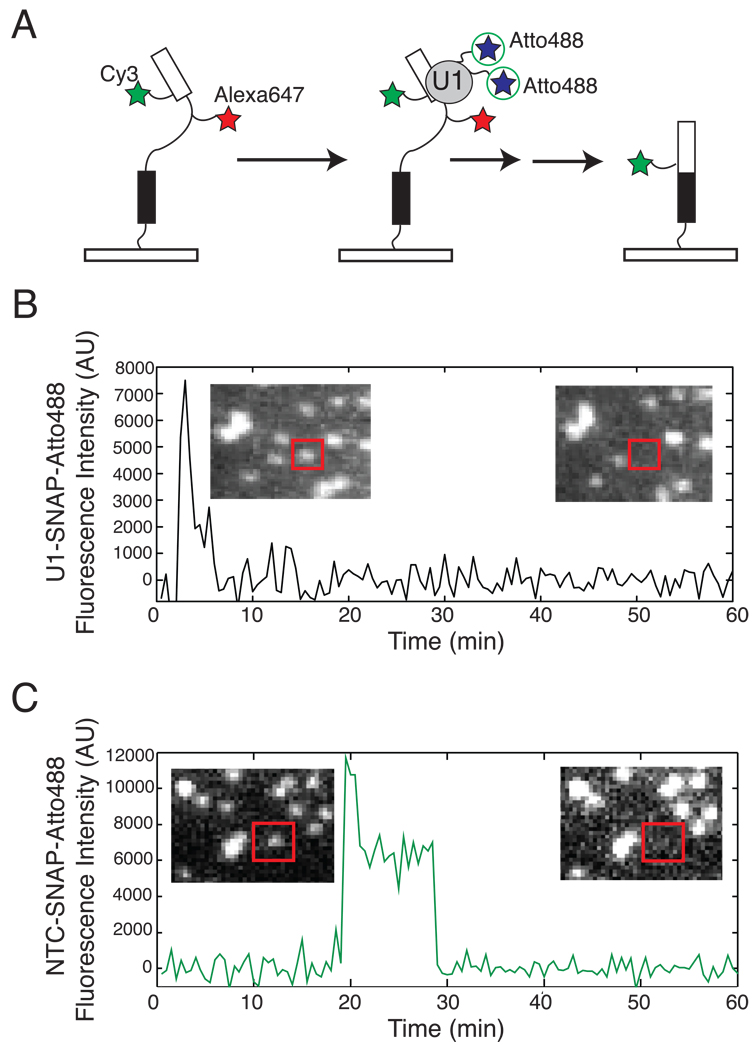
While the above results indicated that we could observe subcomplex association with surface-tethered pre-mRNA molecules, they did not reveal what fraction of those pre-mRNAs ultimately spliced. To address this, we combined our previously described Cy3/Alexa647 splicing reporter pre-mRNA (11) with extracts in which either U1 or NTC was labeled with SNAP-Atto488 (Figure 3). In these experiments, disappearance of fluorescence from the Alexa647-labeled intron without loss of the Cy3-labeled exon demonstrates that either the pre-mRNA was spliced and the spliceosome/lariat intron complex departed from the surface-tethered mRNA, or that the Alexa647 was photobleached (11).
Figure 3.
(A) Scheme for detecting subcomplex binding to and splicing of the same pre-mRNA molecule. (B and C) Example single molecule traces of U1-SNAP (B) and NTC-SNAP (C) binding to pre-mRNAs (red boxes in insets) that had Alexa647 fluorescence at t = 0 (left insets), but not at t = 60 (right insets).
Similar to our previous observations (11), the extent of intron fluorescence loss was 15 ± 2% (SE) and 18 ± 2% for ATP-containing U1-SNAP and NTC-SNAP extracts, respectively, compared to 4 ± 1% for an inactive no ATP control (where loss measures photobleaching). These results indicate that active spliceosomes are formed on the surface-linked pre-mRNAs in our labeled extracts. For both U1 and NTC, we could observe numerous pre-mRNAs that both gained the labeled subcomplex and lost intron fluorescence (Table S2). Interestingly, only 21 ± 3% of pre-mRNAs that had at least one U1 binding event also lost intron fluorescence. This indicates that interaction with U1 does not absolutely commit a pre-mRNA to splicing. In contrast, roughly half (53 ± 5%) of pre-mRNAs that acquired NTC lost intron fluorescence, suggesting that commitment increases as assembly proceeds. Analysis of individual U1 and NTC binding event lifetimes indicated that pre-mRNAs that ultimately lost their intron signals tended to have ~2-fold longer U1 lifetimes and ~2-fold shorter NTC lifetimes (Figure S6). One possible explanation is that productive U1 association is stabilized by binding of additional spliceosome assembly factors. Conversely, the shorter NTC lifetime may indicate that properly assembled spliceosomes proceed rapidly through activation, catalysis and mRNA product release soon after NTC binding.
Order and Kinetics of Spliceosome Assembly
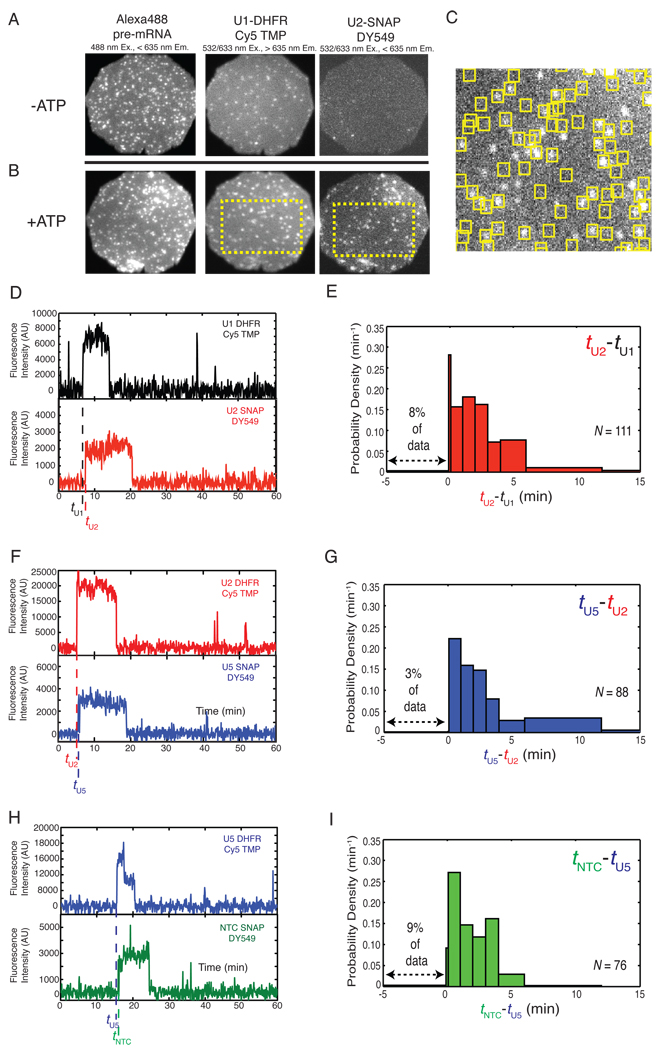
While the experiments in Figure 2 can define the population-averaged timing with which different subcomplexes arrive at the pre-mRNA, they do not directly assess the order of subcomplex addition on individual pre-mRNA molecules. Further, the data in Figures 2 and S5 are composites of subcomplex association and dissociation events, photobleaching, and TMP dye exchange, and are additionally complicated by variations in WCE splicing activity. These issues can be resolved by using CoSMoS to simultaneously follow the pre-mRNA association of two spliceosomal subcomplexes in the same WCE. To do so, we used two DHFR/Cy5-TMP tags and a single SNAP DY549 tag to label two subcomplexes (e.g., U1-DHFR and U2-SNAP) with different fluorophores in the same extract (triple label extracts, Figure 1), and visualized them binding to individual Alexa488-labeled pre-mRNA molecules (Figure 4A–B).
Figure 4.
(A and B) Images of two FOVs taken at three different wavelengths with triple label extract to monitor U1-DHFR/Cy5-TMP and U2-SNAP-DY549 association with Alexa488-labeled pre-mRNA, without (A) or with (B) ATP. (C) Magnification of dashed area in (B) showing co-localization of U1 (yellow boxes) with U2 (white spots). (D) Fluorescence intensity traces showing association of U1 and U2 with an individual pre-mRNA molecule (not shown) in the presence of ATP. Arrival times for each subcomplex (tU1 and tU2) are marked. (E) Histogram of the delay between subcomplex arrival times (tU2-tU1). (F, H) Single molecule fluorescence intensity traces for U2/U5 (F) and U5/NTC (H) bound to single pre-mRNA molecules (not shown). (G, I) Histograms of the delays between U2 and U5 binding (G) and U5 and NTC binding (I).
As was observed with the individually labeled extracts, when both U1 and U2 were labeled in the same WCE only U1 to co-localized with pre-mRNAs in the absence of ATP, while both U1 and U2 co-localized with pre-mRNAs in the presence of ATP (Figure 4B–C and Movie S3). When individual pre-mRNA molecules were followed over time, the largest class (49%, Table S3) exhibited at least one discrete onset of U1 fluorescence and at least one discrete onset of U2 fluorescence (Figures 4D and S7). Other classes exhibited only U1 binding (18%), only U2 binding (6%), or no binding events (27%). These latter subpopulations may arise from the presence of some nonfluorescent subcomplexes in the extract and/or from alternative conformations of the pre-mRNA (15) that prevent spliceosome assembly. U1 and U2 spots persisted for seconds to minutes before disappearing due to either dye photobleaching or subcomplex dissociation. For U1, which was labeled with two DHFR tags, fluorescence typically vanished in one or two discrete steps (96% of events, Table S4). Analogously, for U2, which was labeled with one SNAP tag, fluorescence most often vanished in a single step (88% of events). Thus only one copy each of U1 and U2 is present at any given time on the majority of pre-mRNAs.
To quantitatively evaluate the U1 and U2 binding order on individual pre-mRNA molecules (Figure 4D), we calculated tU2-tU1, the difference between the arrival times of the two subcomplexes (20). A histogram (Figure 4E) shows that the overwhelming majority (90%) of these delay times were positive, indicating that U2 binding nearly always followed U1 binding. This conclusion was confirmed by correlation analysis of the absolute binding times (Figure S8) which revealed that even U1 binding events occurring late in the experiment were soon followed by U2 binding. While U1 and U2 appeared to arrive simultaneously on a small minority (9 out of 223 events) of pre-mRNAs, some of these are likely cases of U1 and U2 arriving in rapid succession separated by a delay that the experimental time resolution (5–6s) was insufficient to resolve ((20) and Table S5). Thus, assembly is highly ordered, with U1 always or almost always binding before U2. Further, >95% of pre-mRNAs that acquire both U1 and U2 acquire them separately rather than as a preformed U1/U2 complex. Consequently, formation of a U1/U2 complex prior to association with pre-mRNA cannot be a requirement for splicing since the fraction of pre-mRNAs that splice is greater than 5% (Table S2).
To examine the ordering of later assembly steps, we used the same methodologies with other triply labeled yeast strains. U2 fluorescence almost always preceded onset of U5 fluorescence (Figures 4F and S9, Table S6); 97% of the tU5-tU2 values were positive (Figure 4G). Similarly, U5 fluorescence almost always preceded onset of NTC fluorescence (Figure 4H and S10, Table S7); 91% of the tNTC-tU5 delay values were positive (Figure 4I). In both the U2/U5 and U5/NTC data sets, very few traces (Table S5) exhibited apparent simultaneous binding of the subcomplexes, and analysis of all traces suggested that at most one copy each of U5 and NTC were present on the majority of pre-mRNAs (Table S4). In sum, our data indicate that when spliceosome assembly is followed on individual RP51A pre-mRNA molecules, the predominant reaction pathway is highly ordered (U1 → U2 → tri-snRNP → NTC). Further, the experiments indicate little or no preassociation for any pair of subcomplexes studied (Table S5). As with U1/U2, these data demonstrate that no preassociation of these subcomplexes is required for splicing.
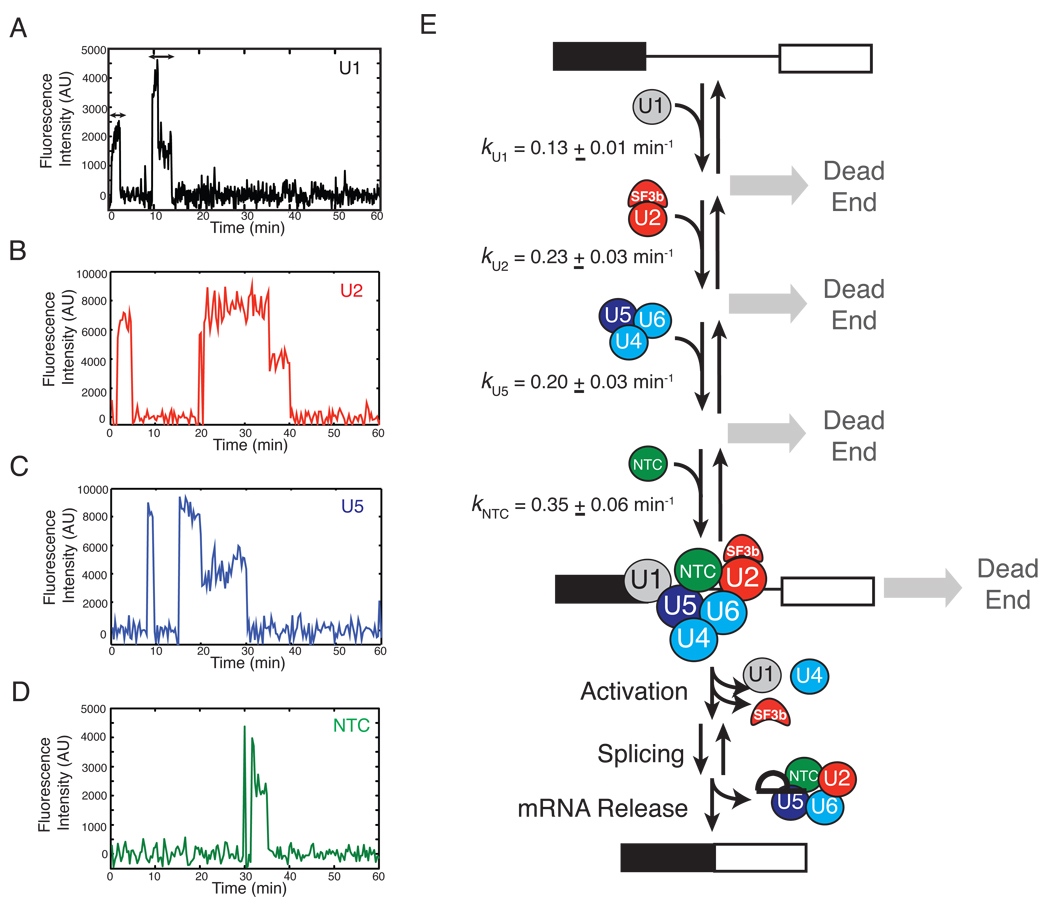
On top of providing information about binding order, the CoSMoS methodology permits measurement of defined kinetic parameters. The arrival times of the first U1 subcomplex on each pre-mRNA and all three time-delay data sets (tU2-tU1, tU5-tU2 and tNTC-tU5) are well fit by single exponential distributions (Figure S11), allowing determination of apparent first-order rate constants (Figure 5). All four rate constants fall in a narrow range (0.1–0.4 min−1), suggesting that no single subcomplex association step predominantly limits the rate of spliceosome assembly on RP51A pre-mRNA.
Figure 5.
(A–D) Single molecule traces of SNAP-DY549 labeled subcomplexes binding and dissociating multiple times from individual pre-mRNA molecules (not shown) in the presence of ATP. Arrows indicate durations of two U1 binding events (dwell times) used to analyze subcomplex lifetimes. (E) Kinetic scheme for spliceosome assembly and splicing of RP51A pre-mRNA. Our results provide evidence for reversible binding of all of the major subcomplexes (backward arrows), while others have provided evidence for reversibility of splicing chemistry (8). There is as yet no evidence for reversibility of the activation step prior to splicing or mRNA release.
In addition to arrival times, the triple label experiments also allowed us to examine the order of subcomplex loss from pre-mRNA. Preliminary analysis revealed that U1 fluorescence tended to be lost before U2 fluorescence, and U2 fluorescence tended to be lost before U5 fluorescence. Only with U5 and NTC did a significant number of pre-mRNAs lose fluorescence from both subcomplexes simultaneously (Table S8). These results are consistent with known post-assembly events, including ordered loss of U1 and the SF3b component of U2 during spliceosome activation and subsequent simultaneous loss of U5 and NTC coincident with spliced mRNA release (2, 19). While additional analyses of photobleaching and Cy5-TMP dye exchange rates will be required to fully interpret these results, they do indicate that subcomplex dissociation coupled to activation and spliceosome disassembly is detectable using this methodology. Definitive analysis of subcomplex dissociation relative to catalysis and intron release awaits future development of more photostable splicing reporters.
We also examined dissociation kinetics of each subcomplex (20). In all cases, good fits of dwell time distributions required a function containing more than one exponential term (Figure S12; Table S9). This presence of both short-(τ1< 1 min) and long-lived (τ2 > 1 min) characteristic dwell times indicates that there is more than one species from which each subcomplex can dissociate. Thus, subcomplex dissociation is more complex than some current models suggest, and there are multiple mechanisms consistent with our data (Figure S13). Elucidation of these mechanisms may be possible by combining CoSMoS with appropriate mutants and inhibitors of assembly.
Pre-mRNAs Can Engage Subcomplexes Multiple Times
Subsequent to dissociation of a particular subcomplex, many pre-mRNA molecules reacquired a copy of the same subcomplex. On individual pre-mRNAs, U1 often appeared to bind and dissociate repeatedly (Figures 5A and S14). Use of two covalent SNAP labels on U1 allowed us to verify by photobleaching that the majority of reoccurring U1-SNAP signals resulted from association and dissociation of different U1 molecules ((20), Figure S15, and Table S10) rather than blinking of a single molecule (15). Further, using the splicing reporter pre-mRNA (Figure 3), we could observe multiple U1 binding events on pre-mRNAs that spliced (20 ± 7% of pre-mRNAs that lost intron fluorescence acquired multiple U1 signals, Figure S16). Thus pre-mRNAs that have multiple encounters with U1 are not irreversibly trapped in an inactive state. In the absence of ATP, U1 had a dwell time distribution nearly identical to that observed in the presence of ATP (Figure S17, Table S9). This suggests that ATP-hydrolysis by RNA helicases or RNPases in WCE is not required for U1 dissociation.
A previous study using native PAGE reported two different ATP-independent U1:pre-mRNA complexes: δun and δcommit (21). The more abundant δun (unstable and uncommitted) did not survive challenge from competitor RNAs, while the minor δcommit represented a more stable, challenge-resistant species. Since it could be chased into subsequent steps of the splicing pathway, δcommit is likely the same species as U1-containing commitment complexes (CC1 and/or CC2) (22). Our analysis of U1 snRNP dwell times (Figure S12 and Table S9) and our observation of U1 dynamics (Figure 5) provide evidence for at least two types of U1:pre-mRNA complexes with dwell times differing by more than an order of magnitude –- an abundant short-lived component likely representing δun, and a longer lived component likely including CC1 and/or CC2. Consistent with this hypothesis, elimination of the branchsite (which is necessary to form CC2 but not CC1; (18)) from our transcript (UACUAAC → GUUAGUA) decreased abundance of the longer-lived component, but did not abolish it (Figure S18, Table S9). Thus the long-lived component must contain species in addition to CC2
We have also observed multiple arrivals and departures of U2, U5, and the NTC on individual pre-mRNAs (Figures 5B–D, S14, and S19). As seen with U1, multiple NTC binding events could be detected on the splicing reporter pre-mRNA (4 ± 2% of pre-mRNAs both lost their intron signal and acquired NTC more than once, Figure S16). The number of binding events observed per pre-mRNA molecule was dependent on the subcomplex being studied. U1 exhibited by far the largest number of binding events, with the number of events systematically decreasing for each successive subcomplex in the pathway (Figure S20). This suggests that at each step of subcomplex addition some fraction of the pre-mRNA molecules are lost to side pathways that do not lead to productive splicing (Figure 5E).
Summary and Perspective
Taken together the data from this first real-time kinetic analysis of spliceosome assembly are consistent with existing models and lead to new insights. Spliceosome assembly on the RP51A substrate is highly ordered (U1 → U2 → tri-snRNP → NTC), and pre-association of the subcomplexes is not required for splicing. While no single step appears to irreversibly commit this pre-mRNA to splicing, commitment increases as spliceosome assembly proceeds.
Further, spliceosome assembly on this pre-mRNA is kinetically efficient with no single subcomplex binding step predominantly restricting the overall rate. Finally, we have directly observed multiple binding events for all subcomplexes, demonstrating that subcomplex binding is reversible. Together, these findings have important implications for the regulation of alternative splicing. If spliceosome assembly is reversible and no single assembly step irreversibly commits a particular pair of splice sites to splicing, then alternative splice site choice can potentially be regulated at any stage of assembly. This hypothesis is bolstered by observations that some regulation of alternative splicing apparently occurs at late stages of assembly (23, 24).
By making possible kinetic analysis of spliceosome assembly in whole cell extracts, this work opens the door to answering fundamental questions concerning the mechanisms of pre-mRNA splicing. The combination of CoSMoS with chemical and genetic tools is a powerful approach for elucidating the mechanisms of complex biological processes, even when those processes can only be studied in cell extracts. These methods should prove broadly useful for analyzing many other complex macromolecular machines.
Supplementary Material
Footnotes
Competing Interests
V.W.C. is a co-inventor of the TMP-tag technology licensed and commercialized by Active Motif.
Supporting Online Information
Materials and Methods
Figures S1–S21
Tables S1–S12
Movies S1–S3
Scheme S1
REFERENCES
- 1.Nilsen TW. Bioessays. 2003;25:1147. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 2.Fabrizio P, et al. Mol. Cell. 2009;36:593. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 3.Wahl MC, Will CL, Luhrmann R. Cell. 2009;136:701. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Stevens SW, et al. Mol. Cell. 2002;9:31. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 5.Xu YZ, et al. EMBO J. 2004;23:376. doi: 10.1038/sj.emboj.7600050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider M, et al. Mol. Cell. 2010;38:223. doi: 10.1016/j.molcel.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 7.Abelson J, et al. Nat. Struct. Mol. Biol. 2010;17:504. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tseng CK, Cheng SC. Science. 2008;320:1782. doi: 10.1126/science.1158993. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Query CC, Konarska MM. Nat. Struct. Mol. Biol. 2007;14:519. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 10.Friedman LJ, Chung J, Gelles J. Biophys. J. 2006;91:1023. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. RNA. 2008;14:170. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juillerat A, et al. Chem. Bio.l. 2003;10:313. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 13.Miller LW, Cai Y, Sheetz MP, Cornish VW. Nat. Methods. 2005;2:255. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 14.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. Nature. 1997;388:355. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 15.Rasnik I, McKinney SA, Ha T. Nat. Methods. 2006;3:891. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 16.Dunn SM, King RW. Biochemistry. 1980;19:766. doi: 10.1021/bi00545a024. [DOI] [PubMed] [Google Scholar]
- 17.Dave R, Terry DS, Munro JB, Blanchard SC. Biophys. J. 2009;96:2371. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seraphin B, Rosbash M. EMBO J. 1991;10:1209. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lardelli RM, Thompson JX, Yates JR, 3rd, Stevens SW. RNA. 2010;16:516. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.See the supporting material available on Science Online.
- 21.Ruby SW. J. Biol. Chem. 1997;272:17333. doi: 10.1074/jbc.272.28.17333. [DOI] [PubMed] [Google Scholar]
- 22.Legrain P, Seraphin B, Rosbash M. Mol. Cell. Biol. 1988;8:3755. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M, Manley JL. Nat. Rev. Mol. Cell. Biol. 2009;10:741. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnal S, et al. Mol. Cell. 2008;32:81. doi: 10.1016/j.molcel.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 25.We thank J. Chung, A. Okonechnikov, J. Yan, J. Haber, S. Lovett, I. Correa, M.-Q. Xu, Z. Chen, and B. Smith for helpful discussions and assistance. This work was supported by NIH RO1's GM043369 (J.G.), GM81648 (J.G.), GM053007 (M.J.M), GM54469 (V.W.C.), RC1 GM091804 (V.W.C), NRSA fellowship GM079971 (A.A.H.) and K99/R00 GM086471(A.A.H.). D.J.C., S.S.G., and R.W were supported by NIH training grant GM759628, a National Defense Science and Engineering Graduate fellowship, and a Deutscher Akademischer Austausch Dienst fellowship, respectively. M.J.M. is a HHMI investigator.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.