Abstract
The MUC1 oncoprotein is aberrantly expressed in human multiple myeloma cells by mechanisms that are not understood. Moreover, the functional role of MUC1 in multiple myeloma is not known. The present studies demonstrate that the MUC1 gene locus is amplified in multiple myeloma cell lines and in primary cells from patients. The KMS28PE multiple myeloma cell line, which was found to have MUC1 gene amplification, was stably silenced for MUC1 using different siRNAs. Silencing MUC1 was associated with a decrease in nuclear ß-catenin levels, consistent with the function of MUC1 in stabilizing ß-catenin. MUC1 is also known to activate the IKKß→NF-κB pathway and KMS28PE cells silenced for MUC1 were found to have downregulation of IKKß and IκBα phosphorylation, and decreased nuclear targeting of NF-κB p65. The results also demonstrate that MUC1: i) contributes to KMS28PE cell proliferation, and ii) protects against apoptosis and loss of self-renewal in the response to melphalan and dexamethasone. These findings indicate that MUC1 activates the ß-catenin and NF-κB pathways in multiple myeloma cells and contributes to their growth and survival.
Keywords: MUC1, multiple myeloma, ß-catenin, NF-κB, growth, self-renewal
Introduction
The canonical Wnt pathway regulates cytoplasmic levels of ß-catenin and is of importance to tumorigenesis (1). Glycogen synthase kinase 3ß (GSK3ß) in a complex with the adenomatous polyposis coli (APC) protein and axin targets ß-catenin for ubiquitination and degradation. Downregulation of GSK3ß by Wnt signaling increases levels of ß-catenin, promotes the formation of ß-catenin complexes with the Tcf/Lef family of transcription factors and thereby activates Wnt target genes (2,3). The Wnt pathway is constitutively activated in human multiple myeloma cells by an unclear mechanism that does not appear to involve: i) loss of APC or axin function, or ii) activating ß-catenin mutations (4,5). Aberrant expression of ß-catenin in multiple myeloma as compared to normal progenitor cells is associated with increased proliferation (4). Moreover, disruption of the ß-catenin/Tcf transcription complex reduces growth and survival of multiple myeloma cells (6). These findings and those obtained with agents that disrupt ß-catenin/Tcf-mediated transcription (7,8) have indicated that the Wnt/ß-catenin pathway is of importance to the pathogenesis of multiple myeloma.
The classical nuclear factor-κB (NF-κB) pathway is also constitutively activated in multiple myeloma cells by mechanisms that are not clearly understood (9,10). NF-κB p65 is sequestered in the cytosol by binding to IκBα (11,12). The IκB kinase ß (IKKß) in a complex with IKKγ is necessary and sufficient for phosphorylation and thereby degradation of IκBα. In turn, NF-κB p65 is released for nuclear targeting and activation of NF-κB-dependent genes. Importantly, inhibition of constitutive NF-κB activation in multiple myeloma cells with agents that block IKKß is associated with loss of proliferation and death (13–16). These findings have indicated that, as found for the Wnt/ß-catenin pathway, the multiple myeloma cell phenotype is dependent on NF-κB signaling.
The MUC1 heterodimeric transmembrane protein is aberrantly expressed by human multiple myeloma cells (17–23). The extracellular MUC1 N-terminal subunit (MUC1-N) contains variable numbers of 20 amino acid tandem repeats that are modified by O-glycosylation (24,25). The transmembrane MUC1 C-terminal subunit (MUC1-C) consists of a 58 amino acid extracellular region and a 72 amino acid cytoplasmic tail (26). MUC1-C accumulates in the cytoplasm of multiple myeloma cells and is targeted to the nucleus (27). Studies in multiple myeloma and other cell types have demonstrated that the MUC1 cytoplasmic domain (MUC1-CD) binds to ß-catenin (27–29) and stabilizes ß-catenin by blocking function of the destruction complex (30). Recent work has further shown that MUC1-C plays a role in constitutive activation of the IKKß-NF-κB p65 pathway (31). In addition, other studies have demonstrated that MUC1 induces transformation (30,32). However, it is not known whether MUC1 has similar functions in multiple myeloma cells.
The present studies demonstrate that the MUC1 locus is amplified in multiple myeloma cell lines and primary cells. The results also demonstrate that overexpression of MUC1 contributes to: i) constitutive activation of the ß-catenin and NF-κB pathways, and ii) growth and survival of multiple myeloma cells.
Materials and methods
Cell culture
Human multiple myeloma cell lines were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS; Mediatech, Herndon, VA), 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine (ATCC, Manasses, VA). Cells were treated with melphalan and dexamethasone (Sigma, St. Louis, MO).
Primary multiple myeloma cells
Bone marrow mononuclear cells from patients with multiple myeloma were CD138-enriched using anti-CD138 magnetic microbeads (Miltenyi Biotec, Auburn, CA). The resulting populations were >95% CD138+ cells.
Quantitative real-time PCR
Quantification of MUC1, TRIM46, THBS3 and GAPDH gene copy number was performed on genomic DNA using primer pairs with TaqMan probes or SYBR Green (Table I) and the ABI PRISM 7000 sequence detection system (Applied Biosystems, Foster City, CA). Relative gene copy number was determined using the comparative Ct method with GAPDH as the internal control.
Table I.
The primers used in this study.
| Gene | Forward primers | Reverse primers | Fragments | TaqMan probe or SYBR Green |
|---|---|---|---|---|
| MUC1 | 5'-GCCAGGATCTGTGGTGGTACA-3' | 5'-CTCCAGGTCGTGGACATTGA-3' | 70-bp | 6FAM-TTGACTCTGGCCTTCCCGAGAAGGTACC-3'TAMRA |
| TRIM46 | 5'-CACTGGACAAATCATTCAACAACTT-3' | 5'-AAACATTATCTCACACCATCCTCTCA-3' | 127-bp | 6FAM-TGCCCTGCAATGTCCATACCTAAAGGGT-3'TAMRA |
| THBS3 | 5'-GCTTAAATAGTGACCACCGGGT-3' | 5'-AACGCTGCTCATGGAAGCC-3' | 65-bp | SYBR Green |
| GAPDH | 5'-GGTTTACATGTTCCAATATGATTCCA-3' | 5'-ATGGGATTTCCATTGATGACAAG-3' | 93-bp | 6FAM-ATGGCACCGTCAAGGCTGAGAACG-3'TAMRA |
Immunoblot analysis
Whole cell lysates (WCL), and nuclear and cytoplasmic fractions were prepared as previously described (30,31,33). Soluble proteins were subjected to immunoblot analysis with anti-MUC1-C (LabVision, Fremont, CA), anti-ß-actin (Sigma), anti-ß-catenin (BD Biosciences, San Jose, CA), anti-lamin B (Calbiochem, San Diego, CA), anti-IκBα, anti-NF-κB p65 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-IκBα, anti-phospho-IKKß, anti-IKKß (Cell Signaling Technology, Danvers, MA). Reactivity was detected with horseradish peroxidase-conjugated second antibodies and chemiluminescence (ECL Plus; GE Healthcare Biosciences, Piscataway, NJ).
Silencing of MUC1 expression
The BLOCK-iT Target Screening System (Invitrogen, Carlsbad, CA) was used to generate siRNAs that target two different MUC1 sequences (#1: AAGGTACCATCAATGTCCACG; and #2: AAGTTC AGTGCCCAGCTCTAC) or a control sequence (CR: CGCT TACCGATTCAGAATGG). The siRNA cassettes were transferred to pLenti4/BLOCK-iT-DEST by LR recombination for generation of lentiviral vectors. Cells were infected with the lentiviruses at a multiplicity of infection of 5 in the presence of 8 μg/ml polybrene. Cell clones were selected in methylcellulose semi-solid medium containing 200 μg/ml zeocin and assayed for downregulation of MUC1 by immunoblotting.
Apoptosis assays
Cells were fixed with 70% ethanol and incubated in PBS containing 20 ng/ml RNase (Roche, Indianapolis, IN) for 60 min at 37°C. Cells were then stained with 2.5 μg/ml propidium iodide for 30 min at room temperature in the dark. DNA content was then analyzed by flow cytometry. The percentage of cells with sub-G1 DNA was determined by the Modfit LT Program (Verity Software, Topsham, ME).
Colony formation assays
Cells were seeded at 5,000 per well of a 6-well plate containing 0.4% methylcellulose in RPMI-1640 medium supplemented with 10% heat-inactivated FBS. Colonies were counted at 28 days under a stereomicroscope.
Results and Discussion
Amplification of the MUC1 locus in multiple myeloma cells
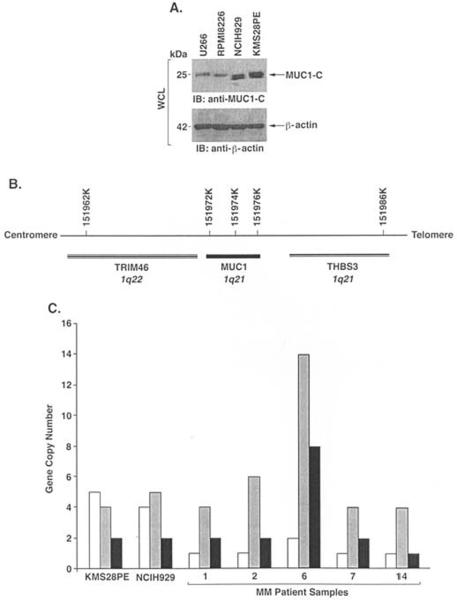
MUC1 is detectable in multiple myeloma cell lines and primary cells from patients, but not in CD34+ hematopoietic progenitor cells (17–23). Little, however, is known about the mechanisms responsible for MUC1 expression in multiple myeloma cells. Although subsets of B cell lymphomas have amplification of the MUC1 gene (34), there are no reports of alterations of the MUC1 locus in multiple myeloma cells. MUC1 gene copy number in multiple myeloma cells was assessed by quantitative real-time PCR amplification of a 70-bp fragment located outside the tandem repeat region. GAPDH was used as a reference gene with amplification of a 93-bp fragment. Copy number ratios for MUC1 as compared to GAPDH were determined for 21 multiple myeloma cell lines (Table II). Eleven of the multiple myeloma cell lines had ≤2 and were scored as not having significant amplification of the MUC1 locus (Table II). By contrast, 10 multiple myeloma cell lines had ratios ranging from 3 to 5 (Table II). Thus, 48% of the multiple myeloma cell lines exhibited a >2-fold increase in MUC1 copy number. Amplification of MUC1 as found in NCIH929 and KMS28PE cells was associated with an increase in MUC1-C expression as compared to that in U266 and RPMI8226 cells with a single MUC1 copy number (Fig. 1A). The MUC1 locus is flanked by the TRIM46 and THBS3 genes (Fig. 1B). To determine if increases in MUC1 copy number are associated with amplification of this region, we studied TRIM46 and THBS3 in NCIH929 and KMS28PE cells. The results show that amplification of MUC1 is associated with increases in TRIM46 copy number and, to a lesser extent, THBS3, indicating that the regions flanking MUC1 are also subject to amplification (Fig. 1C). Amplification of the MUC1 locus (>2-fold) was also identified in 14 of 21 (66%) samples obtained from multiple myeloma patients (Table II) and was associated with varying increases in TRIM46 and THBS3 gene copy number (Fig. 1C). These findings indicate that aberrant expression of MUC1 in multiple myeloma cells can be attributed, at least in part, to amplification of the MUC1 locus.
Table II.
MUC1 gene copy number in cell lines and primary cells.
| Cell lines |
Primary cells |
||
|---|---|---|---|
| Multiple myeloma cell line | MUC1 copy no. | Multiple myeloma Bone marrow | MUC1 copy no. |
| U266 | 1 | 1 | 4 |
| RPMI-8226 | 1 | 2 | 6 |
| INA6 | 1 | 3 | 2 |
| KMS-38 | 1 | 4 | 3 |
| KMS-12-PE | 1 | 5 | 3 |
| KMS-12-BM | 1 | 6 | 14 |
| OCIMY-5 | 1 | 7 | 4 |
| L363 | 2 | 8 | 3 |
| Korpas 620 | 2 | 9 | 1 |
| KMM-1 | 2 | 10 | 2 |
| ARK | 2 | 11 | 3 |
| S6B45 | 3 | 12 | 3 |
| MM1.S | 3 | 13 | 3 |
| SK-MM-1 | 3 | 14 | 4 |
| H11112 | 3 | 15 | 2 |
| LP1 | 3 | 16 | 3 |
| OCIMY-7 | 3 | 17 | 3 |
| KMS-18 | 3 | 18 | 3 |
| XG1 | 3 | 19 | 2 |
| KMS28PE | 4 | 20 | 2 |
| NCIH929 | 5 | 21 | 2 |
MUC1 gene copy number per haploid genome was determined using GAPDH as the internal control.
Figure 1.
Amplification of the MUC1 locus in multiple myeloma cells. A, Whole cell lysates from the indicated multiple myeloma cell lines were immunoblotted with anti-MUC1-C and anti-ß-actin. B, Location of the TRIM46, MUC1 and THBS3 genes in chromosome 1. C, Copy number for TRIM46 (open bars), MUC1 (shaded bars) and THBS3 (solid bars) in the KMS28PE and NCIH929 cell lines and primary multiple myeloma cells.
Silencing MUC1 in KMS28PE cells attenuates activation of the ß-catenin pathway and decreases cell growth
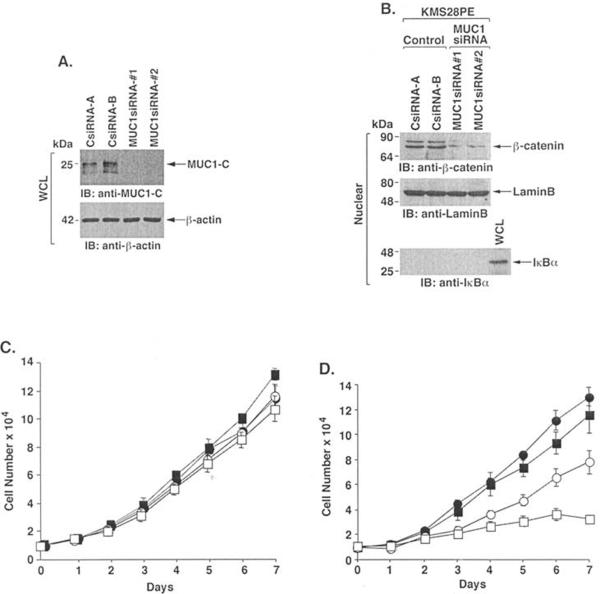
To assess the functional contribution of MUC1 amplification and over-expression to the multiple myeloma cell phenotype, we infected the KMS28PE cells with a lentivirus expressing a siRNA that targets the MUC1 sequence 5'-AAGGTACCATC AATGTCCACG-3' (MUC1siRNA#1) in MUC1-C. Compared to KMS28PE cells infected with a lentivirus expressing a control siRNA (CsiRNA), MUC1-C levels were down-regulated by MUC1siRNA#1 (Fig. 2A). To avoid potential off-target effects, we also infected KMS28PE cells with a lentivirus expressing a siRNA that targets a different MUC1 sequence (5'-AAGTTCAGTGCCCAGCTCTAC-3'; MUC1siRNA#2) in MUC1-N. Downregulation of MUC1-C was similar with the two MUC1siRNAs (Fig. 2A), consistent with a single MUC1 transcript encoding both MUC1-N and MUC1-C. The MUC1-C subunit stabilizes ß-catenin and contributes to activation of the Wnt pathway (28–30). In concert with this function, silencing MUC1-C in KMS28PE cells was associated with downregulation of nuclear ß-catenin levels (Fig. 2B). Silencing MUC1 had no apparent effect on growth of KMS28PE cells in 10% FBS (Fig. 2C). However, proliferation of the KMS28PE/MUC1siRNA cells was significantly decreased in the presence of 2% FBS (Fig. 2D). These findings indicate that silencing MUC1-C is associated with decreased: i) activation of ß-catenin and: ii) growth in the presence of low serum.
Figure 2.
Effects of silencing MUC1 on proliferation of KMS28PE cells. A, Lysates from KMS28PE cells stably infected with lentiviruses expressing the control siRNA (A and B clones), MUC1siRNA#1 or MUC1siRNA#2 were immunoblotted with anti-MUC1-C and anti-ß-actin. B, Nuclear lysates from the indicated KMS28PE cells were immunoblotted with anti-ß-catenin and, as controls for loading and purity, with antibodies against nuclear lamin B and cytosolic IκBα. Whole cell lysate (WCL) was used as a control for IκBα. C and D, Proliferation of the KMS28PE cell clones CsiRNA-A (solid circles), CsiRNA-B (solid squares), MUC1siRNA#1 (open circles) and MUC1siRNA#2 (open squares) in 10% (C) and 2% (D) FBS.
Silencing MUC1 in multiple myeloma cells attenuates NF-κB activation
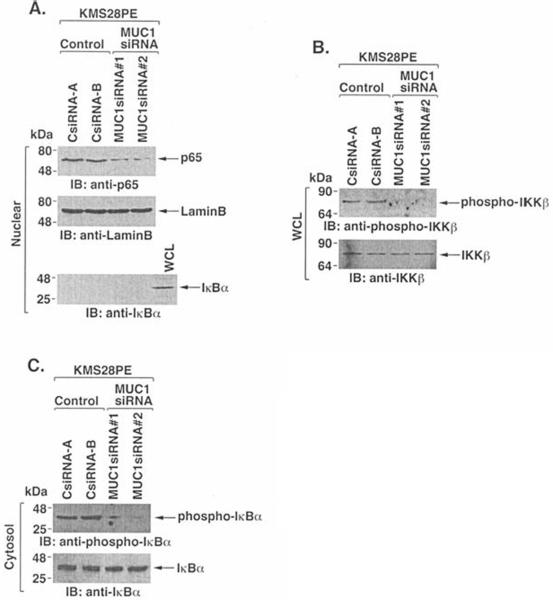
Recent work has demonstrated that MUC1-C binds directly to IKKß and confers constitutive activation of the NF-κB pathway in carcinoma cells (31). The NF-κB pathway is also activated in multiple myeloma cells (13–15,35). Consistent with these findings, NF-κB p65 was detectable in nuclear lysates of KMS28PE cells (Fig. 3A). Notably, however, silencing MUC1 decreased nuclear levels of NF-κB p65 (Fig. 3A). The results also demonstrate that silencing MUC1 decreases phospho-IKKß levels (Fig. 3B), supporting a role for MUC1 in IKKß activation. Moreover, phosphorylation of IκBα was decreased in KMS28PE cells silenced for MUC1 (Fig. 3C). These findings indicate that, as found in carcinoma cells (31), MUC1 contributes to activation of the IKKß→ NF-κB pathway in KMS28PE cells.
Figure 3.
Constitutive activation of the IKKß→NF-κB pathway in KMS28PE cells is attenuated by silencing MUC1. A, Nuclear fractions from the indicated cells were immunoblotted with anti-NF-κB p65 and antibodies against nuclear lamin B and cytosolic IκBα. B, Whole cell lysates were immunoblotted with anti-phospho-IKKß and anti-IKKß. C, Cytosolic fractions were blotted with anti-phospho-IκBα and anti-IκBα.
Silencing MUC1 sensitizes KMS28PE cells to melphalan and dexamethasone
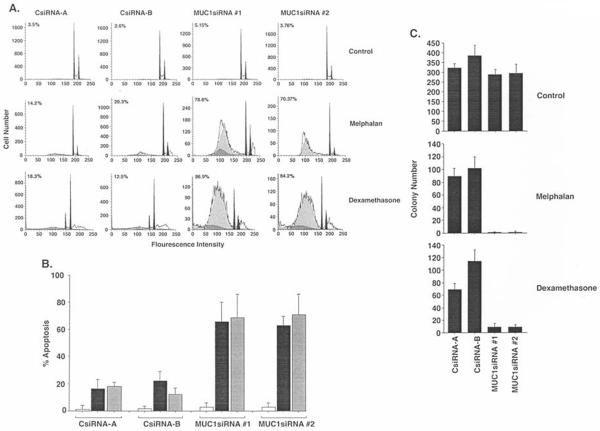
To determine if silencing MUC1 affects death drug-induced apoptosis, KMS28PE/CsiRNA and KMS28PE/MUC1siRNA cells were treated with melphalan and then monitored for sub-G1 DNA content. In concert with the effects of MUC1-C on NF-κB activation, the results show that silencing MUC1 sensitizes cells to melphalan-induced apoptosis (Fig. 4A). Silencing MUC1 also increased the apoptotic response of KMS28PE cells to dexamethasone (Fig. 4A). These results were confirmed in repetitive experiments with cells expressing MUC1siRNA#1 and MUC1siRNA#2 (Fig. 4B). To further assess the effects of silencing MUC1, the KMS28PE cells were treated with melphalan and then assayed for colony formation. The results demonstrate that silencing MUC1 increases melphalan-induced loss of self-renewal (Fig. 4C). Dexamethasone-induced decreases in self-renewal were also potentiated by silencing MUC1 (Fig. 4C). These findings thus demonstrate that MUC1 blocks induction of apoptosis and protects against loss of self-renewal capacity in the response to melphalan and dexamethasone.
Figure 4.
Silencing MUC1 sensitizes to melphalan- and dexamethasone-induced death. A, The indicated KMS28PE cells were treated with 4 μM melphalan or 50 μM dexamethasone for 72 h, stained with propidium iodide and assayed for sub-G1 DNA content by flow cytometry. The percentage of cells with sub-G1 DNA is indicated in each panel. B, The percentage of apoptotic cells with sub-G1 DNA is expressed as the mean ± SD obtained from three experiments for the indicated control (open bars), melphalan-treated (solid-bars) and dexamethasone-treated (shaded bars) cells. C, The indicated KMS28PE cells were treated with 4 μM melphalan or 50 μM dexamethasone for 24 h and then seeded in 0.4% methycellulose/RPMI-1640 medium with 10% FBS. Colonies were counted at 28 days. The results are expressed as the colony number (mean ± SD for three experiments).
Role of MUC1 expression in multiple myeloma cells
MUC1-C interacts with diverse signaling molecules that include growth factor receptors (36,37), kinases (c-Src, c-Abl, Bcr, Lyn, PKCδ, IKKß) (27,38–41) and transcription factors (ß-catenin, p53) (30,42). The available evidence indicates that MUC1-C functions as a chaperone that holds client proteins in certain configurations that affect stability or activity (30,42,43). In multiple myeloma cells, constitutive activation of the Wnt/ß-catenin and NF-κB pathways has been identified as being of importance to the malignant phenotype. The present results demonstrate that expression of MUC1 in multiple myeloma cells contributes to nuclear targeting of ß-catenin. MUC1 expression was also associated with activation of IKKß, phosphorylation of IκBα and targeting of NF-κB p65 to the nucleus. Notably, recent work has demonstrated that mutations in the NF-κB pathway are frequently associated with its constitutive activation in multiple myeloma cells (16,44). Thus, further studies will be needed to assess involvement of MUC1 in multiple myeloma cells with activating mutations of the NF-κB pathway. In concert with findings that blocking the Wnt/ß-catenin (4,6–8) or NF-κB (13–16) pathways affects growth and survival of multiple myeloma cells, our results using KMS28PE cells indicate that silencing MUC1 has similar effects. In this context, MUC1 expression was associated with increased proliferation in low serum concentrations, indicating that MUC1 can protect against growth factor deprivation. Moreover, MUC1 protected against the induction of cell death in response to melphalan and dexamethasone. These findings thus indicate that MUC1 activates the ß-catenin and NF-κB pathways in multiple myeloma cells and contributes to their growth and survival.
Acknowledgements
This work was supported by the National Cancer Institute Grant CA100707.
References
- 1.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 2.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a ß-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 3.Morin PJ, Sparks AB, Korinek V, et al. Activation of ß-catenin-Tcf signaling in colon cancer by mutations in ß-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 4.Derksen PW, Tjin E, Meijer HP, et al. Illegitimate WNT signaling promotes proliferation of multiple myeloma cells. Proc Natl Acad Sci USA. 2004;101:6122–6127. doi: 10.1073/pnas.0305855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiang YW, Endo Y, Rubin JS, Rudikoff S. Wnt signaling in B-cell neoplasia. Oncogene. 2003;22:1536–1545. doi: 10.1038/sj.onc.1206239. [DOI] [PubMed] [Google Scholar]
- 6.Sukhdeo K, Mani M, Zhang Y, et al. Targeting the beta-catenin/TCF transcriptional complex in the treatment of multiple myeloma. Proc Natl Acad Sci USA. 2007;104:7516–7521. doi: 10.1073/pnas.0610299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasui H, Hideshima T, Hamasaki M, et al. SDX-101, the R-enantiomer of etodolac, induces cytotoxicity, overcomes drug resistance, and enhances the activity of dexamethasone in multiple myeloma. Blood. 2005;106:706–712. doi: 10.1182/blood-2005-02-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasui H, Hideshima T, Ikeda H, et al. Novel etodolac analog SDX-308 (CEP-18082) induces cytotoxicity in multiple myeloma cells associated with inhibition of beta-catenin/TCF pathway. Leukemia. 2007;21:535–540. doi: 10.1038/sj.leu.2404561. [DOI] [PubMed] [Google Scholar]
- 9.Chauhan D, Uchiyama H, Akbarali Y, et al. Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-kappa B. Blood. 1996;87:1104–1112. [PubMed] [Google Scholar]
- 10.Hideshima T, Chauhan D, Schlossman R, Richardson P, Anderson KC. The role of tumor necrosis factor alpha in the pathophysiology of human multiple myeloma: therapeutic applications. Oncogene. 2001;20:4519–4527. doi: 10.1038/sj.onc.1204623. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto Y, Gaynor R. IκB kinases: key regulators of the NF-κB pathway. Trends Biochem Sci. 2003;29:72–79. doi: 10.1016/j.tibs.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 13.Hideshima T, Chauhan D, Richardson P, et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277:16639–16647. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 14.Hideshima T, Neri P, Tassone P, et al. MLN120B, a novel IkappaB kinase beta inhibitor, blocks multiple myeloma cell growth in vitro and in vivo. Clin Cancer Res. 2006;12:5887–5894. doi: 10.1158/1078-0432.CCR-05-2501. [DOI] [PubMed] [Google Scholar]
- 15.Jourdan M, Moreaux J, Vos JD, et al. Targeting NF-kappaB pathway with an IKK2 inhibitor induces inhibition of multiple myeloma cell growth. Br J Haematol. 2007;138:160–168. doi: 10.1111/j.1365-2141.2007.06629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi T, Makiguchi Y, Hinoda Y, et al. Expression of MUC-1 on myeloma cells and induction of HLA-unrestricted CTL against MUC1 from a multiple myeloma patient. J Immunol. 1994;153:2102–2109. [PubMed] [Google Scholar]
- 18.Treon SP, Mollick JA, Urashima M, et al. Muc-1 core protein is expressed on multiple myeloma cells and is induced by dexamethasone. Blood. 1999;93:1287–1298. [PubMed] [Google Scholar]
- 19.Burton J, Mishina D, Cardillo T, et al. Epithelial mucin-1 (MUC1) expression and MA5 anti-MUC1 monoclonal antibody targeting in multiple myeloma. Clin Cancer Res. 1999;5:S3065–S3072. [PubMed] [Google Scholar]
- 20.Paydas S, Sahin B, Gonlusen G, Hazar B, Zorludemir S. MUC1 expression in plasmacytoma. Leuk Res. 2001;25:221–225. doi: 10.1016/s0145-2126(00)00111-9. [DOI] [PubMed] [Google Scholar]
- 21.Choi C, Witzens M, Bucur M, et al. Enrichment of functional CD8 memory T cells specific for MUC1 in bone marrow of patients with multiple myeloma. Blood. 2005;105:2132–2134. doi: 10.1182/blood-2004-01-0366. [DOI] [PubMed] [Google Scholar]
- 22.Cloosen S, Gratama J, van Leeuwen EB, et al. Cancer specific Mucin-1 glycoforms are expressed on multiple myeloma. Br J Haematol. 2006;135:513–516. doi: 10.1111/j.1365-2141.2006.06331.x. [DOI] [PubMed] [Google Scholar]
- 23.Baldus SE, Palmen C, Thiele J. MUC1 (EMA) expressing plasma cells in bone marrow infiltrated by plasma cell myeloma. Histol Histopathol. 2007;22:889–893. doi: 10.14670/HH-22.889. [DOI] [PubMed] [Google Scholar]
- 24.Gendler S, Taylor-Papadimitriou J, Duhig T, Rothbard J, Burchell JA. A highly immunogenic region of a human polymorphic epithelial mucin expressed by carcinomas is made up of tandem repeats. J Biol Chem. 1988;263:12820–12823. [PubMed] [Google Scholar]
- 25.Siddiqui J, Abe M, Hayes D, Shani E, Yunis E, Kufe D. Isolation and sequencing of a cDNA coding for the human DF3 breast carcinoma-associated antigen. Proc Natl Acad Sci USA. 1988;85:2320–2323. doi: 10.1073/pnas.85.7.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merlo G, Siddiqui J, Cropp C, et al. DF3 tumor-associated antigen gene is located in a region on chromosome 1q frequently altered in primary human breast cancer. Cancer Res. 1989;49:6966–6971. [PubMed] [Google Scholar]
- 27.Li Y, Chen W, Ren J, et al. DF3/MUC1 signaling in multiple myeloma cells is regulated by interleukin-7. Cancer Biol Ther. 2003;2:187–193. doi: 10.4161/cbt.2.2.282. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and ß-catenin in cell adhesion. J Biol Chem. 1997;272:12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3ß with the DF3/MUC1 carcinoma-associated antigen and ß-catenin. Mol Cell Biol. 1998;18:7216–7224. doi: 10.1128/mcb.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3ß-mediated phosphorylation and degradation of ß-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 31.Ahmad R, Raina D, Trivedi V, et al. MUC1 oncoprotein activates the IκB kinase ß complex and constitutive NF-κB signaling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren J, Agata N, Chen D, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anti-cancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dyomin VG, Palanisamy N, Lloyd KO, et al. MUC1 is activated in a B-cell lymphoma by the t(1;14)(q21;q32) translocation and is rearranged and amplified in B-cell lymphoma subsets. Blood. 2000;95:2666–2671. [PubMed] [Google Scholar]
- 35.Ni H, Ergin M, Huang Q, et al. Analysis of expression of nuclear factor kappa B (NF-kappa B) in multiple myeloma: downregulation of NF-kappa B induces apoptosis. Br J Haematol. 2001;115:279–286. doi: 10.1046/j.1365-2141.2001.03102.x. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Ren J, Yu W-H, et al. The EGF receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and ß-catenin. J Biol Chem. 2001;276:35239–35242. doi: 10.1074/jbc.C100359200. [DOI] [PubMed] [Google Scholar]
- 37.Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Kuwahara H, Ren J, Wen G, Kufe D. The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3ß and ß-catenin. J Biol Chem. 2001;276:6061–6064. doi: 10.1074/jbc.C000754200. [DOI] [PubMed] [Google Scholar]
- 39.Raina D, Ahmad R, Kumar S, et al. MUC1 oncoprotein blocks nuclear targeting of c-Abl in the apoptotic response to DNA damage. EMBO J. 2006;25:3774–3783. doi: 10.1038/sj.emboj.7601263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren J, Li Y, Kufe D. Protein kinase C δ regulates function of the DF3/MUC1 carcinoma antigen in ß-catenin signaling. J Biol Chem. 2002;277:17616–17622. doi: 10.1074/jbc.M200436200. [DOI] [PubMed] [Google Scholar]
- 41.Kawano T, Ito M, Wu Z, et al. MUC1 oncoprotein regulates Bcr-Abl stability and pathogenesis in chronic myelogenous leukemia cells. Cancer Res. 2007;67:11576–11584. doi: 10.1158/0008-5472.CAN-07-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 44.Keats JJ, Fonseca R, Chesi M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]