Abstract
Background
Vitamin D is crucial for maintaining musculoskeletal health. Recently, vitamin D insufficiency has been linked to a number of extraskeletal disorders, including diabetes, cancer, and cardiovascular disease. Determinants of circulating 25-hydroxyvitamin D (25-OH D) include sun exposure and dietary intake, but its high heritability suggests that genetic determinants may also play a role.
Methods
We performed a genome-wide association study of 25-OH D among ∼30,000 individuals of European descent from 15 cohorts. Five cohorts were designated as discovery cohorts (n=16,125), five as in silico replication cohorts (n=9,366), and five as de novo replication cohorts (n=8,378). Association results were combined using z-score-weighted meta-analysis. Vitamin D insufficiency was defined as 25-OH D <75 nmol/L or <50 nmol/L.
Findings
Variants at three loci reached genome-wide significance in the discovery cohorts, and were confirmed in the replication cohorts: 4p12 (overall P=1.9 × 10-109 for rs2282679, in GC); 11q12 (P=2.1 × 10-27 for rs12785878, near DHCR7); 11p15 (P=3.3 × 10-20 for rs10741657, near CYP2R1). Variants at an additional locus (20q13, CYP24A1) were genome-wide significant in the pooled sample (P=6.0 × 10-10 for rs6013897). A genotype score was constructed using the three confirmed variants. Those in the top quartile of genotype scores had 2- to 2.5-fold elevated odds of vitamin D insufficiency (P≤1 × 10-26).
Interpretation
Variants near genes involved in cholesterol synthesis (DHCR7), hydroxylation (CYP2R1, CYP24A1), and vitamin D transport (GC) influence vitamin D status. Genetic variation at these loci identifies individuals of European descent who have substantially elevated risk of vitamin D insufficiency.
Background
Vitamin D insufficiency affects as many as one-half of otherwise healthy adults in developed countries.1 The musculoskeletal consequences of inadequate vitamin D are well-established, and include childhood rickets, osteomalacia, and fractures.2 A growing number of other conditions have also been linked to vitamin D insufficiency, although causal associations have not yet been established in randomized trials. These extra-skeletal conditions include type 1 and 2 diabetes,2-4 cardiovascular disease,5,6 falls,7 and cancers of the breast, colon, and prostate.8-10 A recent meta-analysis suggested that vitamin D supplementation led to significant reductions in mortality.11
Personal, social, and cultural factors are important determinants of vitamin D status via their influence on sun exposure and diet. Maintaining vitamin D status requires sufficient exposure to ultraviolet light or adequate intake from diet or supplements. Levels of 25-OH D, the widely-accepted biomarker of vitamin D status, are highest in the summer and lowest in the winter in northern latitudes. However, only about a quarter of the inter-individual variability in 25-OH D is attributable to season of measurement, geographic latitude, or reported vitamin D intake.12,13 Previous twin and family studies suggest that genetic factors contribute substantially to variability in 25-OH D,13,14 with estimates of heritability as high as 53%. Although several rare Mendelian disorders cause functional vitamin D insufficiency, there are few data on the influence of common genetic variation on vitamin D status. Candidate gene studies have been performed to examine the effect of specific vitamin D-pathway genes, but these studies have been limited by modest samples sizes and the small numbers of variants examined.15-18
Thus, we conducted a large, multicentre genome-wide association study (GWAS) involving approximately 30,000 subjects from 15 cohorts in Europe, Canada, and the United States. Our aim was to identify common genetic variants influencing vitamin D levels and the risk of vitamin D insufficiency.
Methods
Study samples and genotyping
The discovery sample consisted of 16,125 individuals of European descent drawn from five epidemiological cohorts: the Framingham Heart Study, Twins UK, the Rotterdam Study, the 1958 British Birth Cohort (1958BC), and the Amish Family Osteoporosis Study (AFOS). There were five additional cohorts (n=9,366) with genome-wide association data used for in silico replication: the Cardiovascular Health Study, the North Finland Birth Cohort 1966 (NFBC1966), the Indiana cohort, the Dynamics of Health, Aging, Body, and Composition study (Health ABC), and the Gothenburg Osteoporosis and Obesity Determinants study (GOOD). We also performed genotyping of selected variants in 5,715 participants from four additional epidemiologic cohorts (Canadian Multicentre Osteoporosis Study [CaMos], Chingford, Hertfordshire, and the Aberdeen Prospective Osteoporosis Screening Study [APOSS]), and 2,715 additional participants from one of the discovery cohorts (1958BC). Full descriptions of all participating cohorts, as well as details of genotyping methods, quality control, and imputation procedures, are provided in the Supplemental Methods. Characteristics of the study samples are summarized in Supplemental Table 1.
25-OH D measurements
Concentrations of 25-OH D were measured by radioimmunoassay or chemiluminescent assay (DiaSorin Inc, Stillwater, MN) in the following cohorts: Framingham Heart Study, Twins UK, Rotterdam Study, Health ABC, AFOS, the GOOD cohort, and CaMoS. Detection limits ranged from 4 to 10 nmol/L. In the 1958BC samples, 25-OH D was measured using an automated application of the IDS OCTEIA ELISA on the Dade-Behring BEP2000 analyser (sensitivity of 5.0 nmol/L).19 In the Cardiovascular Health Study, NFBC1966, the Indiana cohort, Chingford, Hertsfordshire, and APOSS, total 25-OH D was measured using high performance liquid chromatography-tandem mass spectrometry. Serum vitamin D binding protein (DBP) was measured by an immunonephelometric assay in the Twins UK cohort.20 The detection limit was 50 mg/L.
Statistical analyses
Genome-wide analyses were conducted within each cohort. In the Framingham Heart Study, Twins UK, the Rotterdam Study, 1958BC, AFOS, NFBC1966, the Indiana cohort, Health ABC, and the GOOD study, linear regression models were used to generate cohort-specific residuals of naturally log transformed 25-OH D levels adjusted for age, sex, body mass index (BMI), and season. Log transformation was used to reduce skewness in the distribution of 25-OH D. Season was modeled using categorical variables for summer (July-September), fall (October-December), winter (January to March), and spring (April to June). A single set of definitions was used for season because the majority of the cohorts were at similar latitudes, and all of them were in the northern hemisphere. In cohorts that included related individuals (Framingham, Twins UK, AFOS, Indiana women), association between the additively-coded SNP genotypes and the standardized 25-OH D residuals was assessed using either linear mixed effect models or the score test implemented in MERLIN.21 For imputed SNPs, expected number of minor alleles (i.e. dosage) was used in assessments of association between genotype and 25-OH D residuals. In the Cardiovascular Health Study, analyses were adjusted for age, sex, and study site by including each as a covariate in the model. In all samples, the genomic control approach was used to adjust the P-values for potential effects of mild population stratification and to prevent inflation of type I error occurring from any departure from normality of the trait variable.
A priori, we designated the first five GWAS, all of which used immunoassays for measuring 25-OH D levels, as the “discovery samples.” The remaining five GWAS, three of which measured 25-OH D by mass spectrometry and two by immunoassay, were designated as in silico replication samples. We selected SNPs for replication if they had meta-analytic P-values < 5 × 10-8 in the discovery samples. Additionally, we considered SNPs at or near six pre-specified vitamin D pathway candidate genes: vitamin D receptor (VDR), 1-α-hydroxylase (CYP27B1), 25-hydroxylase (CYP2R1), 24-hydroxylase (CYP24A1), vitamin D binding protein (GC, DBP), and 27- and 25-hydroxlyase (CYP27A1). These SNPs were tested in the replication samples if they met a P-value threshold of 10-3 in the discovery samples. Lastly, selected SNPs were assessed for 25-OH D association in the de novo replication samples, using the same analytic approach. We then generated combined P-values across the 15 studies.22
Meta-analysis was conducted using a weighted z-score based approach, as implemented in the software METAL (www.sph.umich.edu/csg/abecasis/metal/). In this approach, association P-values are converted to signed z-statistics, where the sign reflects the direction of effect with respect to a reference allele. Each z-score is assigned a weight proportional to the square root of the sample size. Weighted z-statistics are summed across studies to obtain a global z-score and a corresponding two-sided P-value. A P-value < 5 × 10-8 was considered genome-wide significant.23
We also assessed whether selected genetic variants from the continuous trait analyses were associated with vitamin D insufficiency in the Framingham Heart Study, Twins UK, CaMoS, and 1958BC. Vitamin D insufficiency was defined using 2 cutpoints, 25-OH D < 75 nmol/L (30 ng/ml) and < 50 nmol/L (20 ng/ml).1 Covariates included age, sex, season, and BMI. Effect estimates from the logistic regression analysis were combined across cohorts by meta-analysis using inverse-variance weighting approach. We also performed analyses using a lower cutpoint, 25 nmol/L (10 ng/ml), to examine whether genetic variants were associated with severe vitamin D deficiency.
Additionally, a genotype score was constructed by taking a weighted average of the number of risk alleles for members of a cohort, with the weights determined by the beta coefficients from the meta-analysis. Logistic regression was performed to determine the odds of vitamin D insufficiency according to quartile of the genotype score. For this analysis, data from the Framingham Heart Study, Twins UK, and 1958BC were combined using a multivariate approach, with beta coefficients for each quartile of genotype score meta-analysed jointly, as previously described.24
Results
Results of genome-wide association analyses are summarized in Table 1. In the analysis of data from the five discovery samples, SNPs at three unique loci met the pre-specified threshold for genome-wide significance: 4p12 (lowest P=4.6 × 10-63 for rs2282679), 11q12 (P=1.6 × 10-13 for rs7944926), and 11p15 (P=3.9 × 10-8 for rs10741657). The 4p12 SNPs were within or near the GC gene, and the top results included a non-synonymous SNP in this gene, rs7041 (P=3.7 × 10-42 for association with 25-OH D). The 11q12 and 11p15 SNPs were near DHCR7/NADSYN1 (7-dehydrocholesterol reductase, NAD synthetase 1) and CYP2R1 (cytochrome P450, subfamily IIR), respectively.
Table 1. Results of genome-wide association analyses for 25-OH D concentrations.
| SNP | Chrom | Position | Nearest gene(s) | MAF | Combined P Discovery samples (up to N=16,124) | Combined P Replication samples (up to N=17,744) | Overall P |
|---|---|---|---|---|---|---|---|
| rs2282679 | 4 | 72827247 | GC | 0.29 | 4.57 × 10-63 | 2.88 × 10-48 | 1.9 × 10-109 |
| rs3755967 | 4 | 72828262 | GC | 0.29 | 7.41 × 10-53 | 3.00 × 10-24 | 2.42 × 10-75 |
| rs17467825 | 4 | 72824381 | GC | 0.29 | 3.85 × 10-52 | 1.61 × 10-23 | 6.75 × 10-74 |
| rs1155563 | 4 | 72862352 | GC | 0.30 | 4.70 × 10-55 | 4.26 × 10-20 | 2.37 × 10-73 |
| rs2298850 | 4 | 72833131 | GC | 0.28 | 8.94 × 10-49 | 2.12 × 10-24 | 2.03 × 10-71 |
| rs7041 | 4 | 72837198 | GC | 0.44 | 3.74 × 10-42 | 1.78 × 10-18 | 6.31 × 10-59 |
| rs12785878 | 11 | 70845097 | DHCR7/NADSYN1 | 0.23 | 1.27 × 10-12 | 2.39 × 10-16 | 2.12 × 10-27 |
| rs7944926 | 11 | 70843273 | DHCR7/NADSYN1 | 0.23 | 1.56 × 10-13 | 7.57 × 10-4 | 8.96 × 10-16 |
| rs12800438 | 11 | 70848651 | DHCR7/NADSYN1 | 0.23 | 5.98 × 10-13 | 6.39 × 10-4 | 2.54 × 10-15 |
| rs3794060 | 11 | 70865327 | DHCR7/NADSYN1 | 0.23 | 8.09 × 10-13 | 6.44 × 10-4 | 3.38 × 10-15 |
| rs4945008 | 11 | 70898896 | DHCR7/NADSYN1 | 0.24 | 8.98 × 10-13 | 6.11 × 10-4 | 4.55 × 10-15 |
| rs4944957 | 11 | 70845683 | DHCR7/NADSYN1 | 0.23 | 1.43 × 10-12 | 7.36 × 10-4 | 8.70 × 10-15 |
| rs10741657 | 11 | 14871454 | CYP2R1 | 0.40 | 3.91 × 10-8 | 2.09 × 10-14 | 3.27 × 10-20 |
| rs2060793 | 11 | 14871886 | CYP2R1 | 0.40 | 2.69× 10-6 | 2.36 × 10-7 | 1.73 × 10-11 |
| rs1993116 | 11 | 14866810 | CYP2R1 | 0.40 | 2.94 × 10-6 | 1.28 × 10-6 | 6.25 × 10-11 |
| rs12794714 | 11 | 14870151 | CYP2R1 | 0.43 | 6.24 × 10-5 | 8.71 × 10-7 | 1.84 × 10-9 |
| rs10500804 | 11 | 14866849 | CYP2R1 | 0.43 | 7.43 × 10-5 | 1.12 × 10-6 | 2.67 × 10-9 |
| rs7116978 | 11 | 14838347 | CYP2R1 | 0.36 | 1.17 × 10-5 | 7.59 × 10-5 | 4.99 × 10-9 |
The top 6 SNP results are shown at each chromosomal locus, ordered from smallest to largest overall P.
MAF: minor allele frequency.
The associations at all three loci were confirmed in the replication samples. The SNP at GC, rs2282679, had a combined P-value of 2.9 × 10-48 in the in silico replication samples, with a consistent direction of effect. Additional genotyping for this SNP was not performed. SNP rs10741657 at CYP2R1 had P=2.1 × 10-14 in the in silico and de novo replication samples, also with a consistent direction of effect. At the DHCR7/NADSYN1 locus, a perfect proxy for rs7944926 (rs12785878, r2=1.0) was genotyped in the de novo replication samples. The combined replication P-value (in silico and de novo replication samples) for rs12785878 was 2.4 × 10-16. Overall P-values (discovery and replication samples) for the three confirmed SNPs ranged from 3.3 × 10-20 to 1.9 × 10-109, as shown in Table 1. Regional plots for the results at each locus are shown in Figures 1A-C. In the discovery cohorts, SNPs at the three confirmed loci (GC, DHCR7/NADSYN1, and CYP2R1) explained up to 1-4% of the variation in 25-OH D concentrations.
Figure 1.
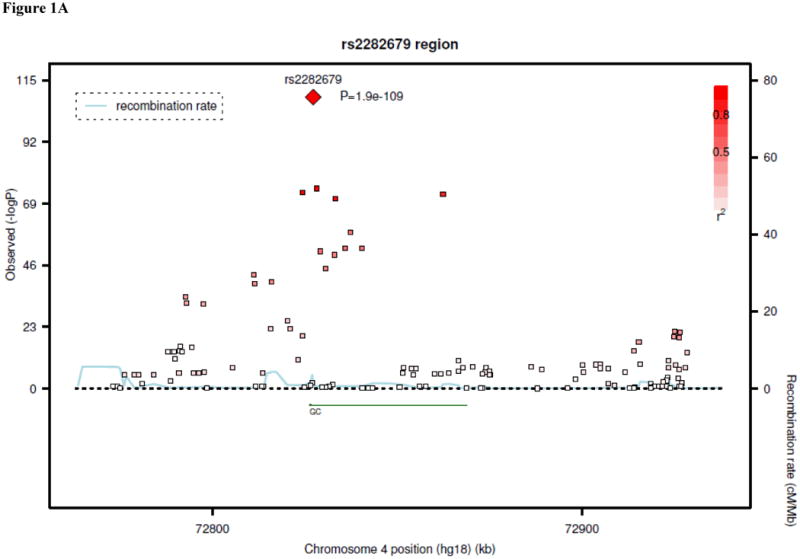
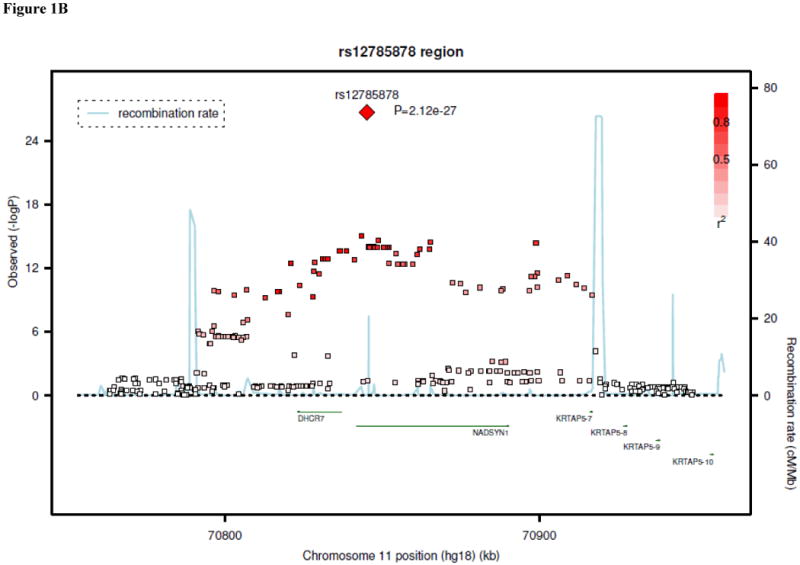
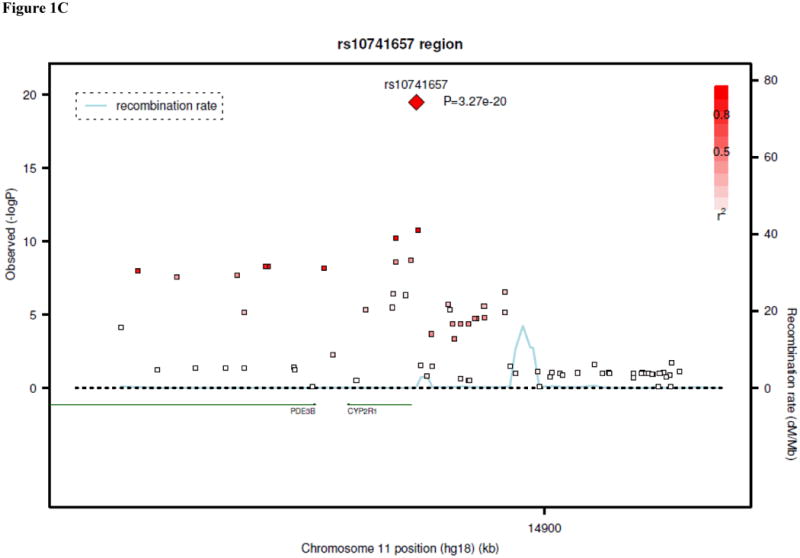
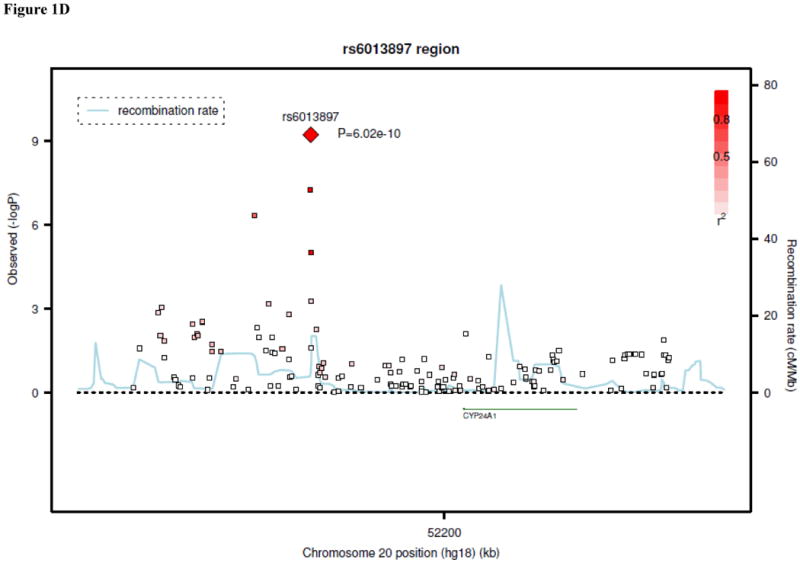
Regional linkage disequilibrium plots for SNPs at GC (panel 1A), DHCR7/NADSYN1 (panel 1B), CYP2R1 (panel 1C), and CYP24A1 (panel 1D).
Table 2 compares the influence of the genetic variants at the 3 validated loci with the observed effects of supplementation and season. Means levels of 25-OH D by genotype category at the 3 validated loci are shown from the 2 largest cohorts (N=12,208), along with mean levels by supplementation status and season. The relative differences in mean 25-OH D between minor and major homozygotes for the strongest genetic variants were comparable to those seen with supplementation in these cohorts, and nearly as large as those seen with a one season change.
Table 2. Mean 25-OH D levels by genotype, season, and supplementation status.
| Framingham Heart Study (n=5,656) | 1958 British Birth Cohort (n=6,552) | |
|---|---|---|
| GC* | ||
| Major homozygotes | 82.6 (0.73) | 61.9 (0.33) |
| Heterozygotes | 74.8 (0.81) | 57.0 (0.30) |
| Minor homozygotes | 64.6 (1.79) | 52.8 (0.27) |
| DHCR7† | ||
| Major homozygotes | 79.7 (0.71) | 59.6 (0.31) |
| Heterozygotes | 76.3 (0.86) | 56.3 (0.30) |
| Minor homozygotes | 71.7 (2.01) | 55.7 (0.32) |
| CYP2R1** | ||
| Major homozygotes | 75.4 (0.87) | 56.8 (0.34) |
| Heterozygotes | 78.6 (0.76) | 60.2 (0.32) |
| Minor homozygotes | 81.6 (1.26) | 61.1 (0.29) |
| Season | ||
| Winter | 61.6 (1.00) | 43.2 (0.26) |
| Spring/Fall | 77.4 (0.68) | 57.1 (0.30) |
| Summer | 95.8 (1.00) | 71.7 (0.30) |
| Supplementation | ||
| Yes | 83.4 (0.80) | 65.9 (0.30) |
| No | 74.7 (0.69) | 56.9 (0.31) |
Means (SE) are shown, in nmol/L. Sample from 1958 British Birth cohort is combined from the GWAS sample and the de novo genotyping sample (see Supplementary Methods).
rs2282679 in Framingham, rs4588 in 1958 Birth Cohort (r2 between SNPs >0.99).
rs7944926 in Framingham, rs12785878 in 1958 Birth Cohort (r2 between SNPs >0.99).
rs10741657 in Framingham and 1958 Birth Cohort.
In the candidate gene analysis, SNP rs6013897 near CYP24A1 (cytochrome P450, family 24, subfamily A) had a P-value of 7.2 × 10-4 in the discovery cohorts, and was tested for replication. The P-value was 8.4 × 10-8 in the replication cohorts, resulting in an overall P-value (discovery and replication) of 6.0 × 10-10. A regional plot for the results at the CYP24A1 locus is shown in Figure 1D. An additional candidate SNP, rs2544037 near VDR, had a P-value of 6.2 × 10-4 in the discovery cohorts, but failed to replicate in the replication samples. There were no SNPs near CYP27B1 or CYP27A1 with P-values less than 10-3 in the discovery cohorts.
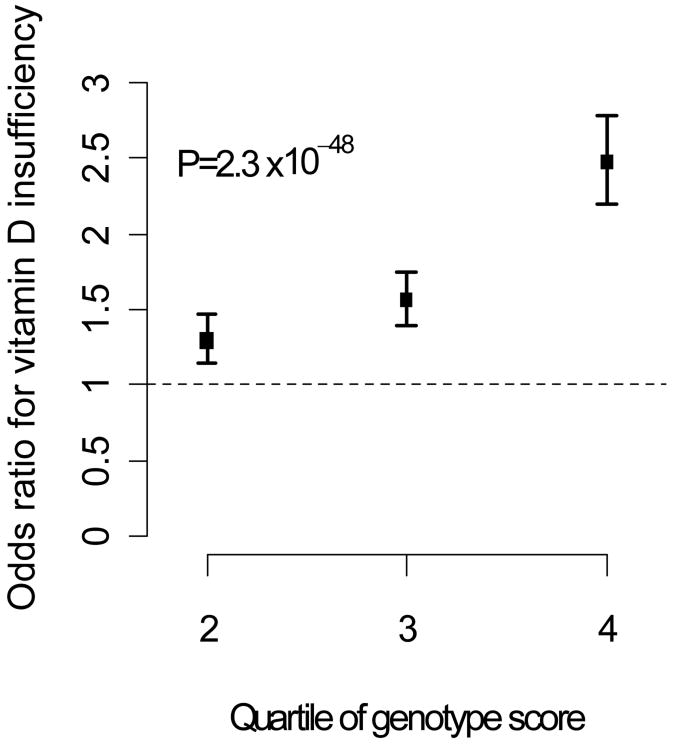
We performed additional analyses to assess the influence of the three variants identified by GWAS on the risk of clinical vitamin D insufficiency (25-OH D < 75 nmol/L or < 50 nmol/L). Results for the variants, individually and in combination, are shown in Table 3. Individuals with a “genotype score” (combining the 3 variants) in the top quartile had a 2- to 2.5-fold increased odds of vitamin D insufficiency (Figure 2). The genotype score was also associated with the risk of severe vitamin D deficiency (25-OH D < 20 nmol/L), with an adjusted odds ratio in the top quartile of 1.43 (95% confidence interval, 1.13-1.79; p=0.002).
Table 3. Genetic variants and risk of vitamin D insufficiency.
| < 75 nmol/L | < 50 nmol/L | |||
|---|---|---|---|---|
| Odds ratio* | P-value | Odds ratio* | P-value | |
| Individual variants | ||||
| GC (rs2282679) | 1.63 (1.53-1.73) | 3.5 × 10-50 | 1.49 (1.40-1.59) | 7.5 × 10-33 |
| DHCR7 (rs7944926) | 1.21 (1.14-1.29) | 4.1 × 10-10 | 1.21 (1.14-1.29) | 4.7 × 10-09 |
| CYP2R1 (rs10741657) | 1.21 (1.45-1.29) | 9.4 × 10-11 | 1.06 (1.00-1.13) | 0.06 |
| Genotype score | ||||
| Quartile 1 | 1.0 (Referent) | 1.0 (Referent) | ||
| Quartile 2 | 1.29 (1.15-1.46) | 1.10 (0.97-1.25) | ||
| Quartile 3 | 1.56 (1.39-1.75) | 1.38 (1.22-1.57) | ||
| Quartile 4 | 2.47 (2.20-2.78) | 1.92 (1.70-2.16) | ||
| P-for-trend | 2.3 × 10-48 | 1.0 × 10-26 | ||
For individual variants, odds ratios are per copy of the “risk allele” with 95% confidence intervals shown in parentheses. All logistic regressions were adjusted for age, sex, body mass index, and season.
Figure 2.
Risk of vitamin D insufficiency (using threshold of 75 nmol/L), according to quartile of genotype score. Bars indicate 95% confidence intervals.
Given the strong association of genetic variants at GC with 25-OH D concentrations, we also examined whether these variants were associated with serum DBP, which was measured in 1,674 individuals in the Twins UK cohort. SNP rs2282679 was strongly associated with DBP (P=4.0 × 10-42), with the minor allele related to lower DBP concentrations.
Discussion
Vitamin D insufficiency has been implicated in a wide range of musculoskeletal and extraskeletal diseases,1,2 which has led to substantial interest in the determinants of vitamin D status. Our findings establish a role for common genetic variants in the regulation of circulating 25-OH D levels. Indeed, the presence of deleterious alleles at the three confirmed loci more than doubled the odds of vitamin D insufficiency. These findings improve our understanding of vitamin D homeostasis and may assist in the identification of a subgroup of Caucasians at risk for vitamin D insufficiency.
DHCR7/NADSYN1 is a novel locus for association with vitamin D status, but one with compelling biological plausibility. DHCR7 encodes the enzyme 7-dehydrocholesterol reductase, which converts 7-dehydrocholesterol (7-DHC) to cholesterol, thereby removing the substrate from the synthetic pathway of vitamin D3, a precursor of 25-OH D3. Rare mutations in DHCR7 lead to Smith-Lemli-Opitz syndrome, which is characterized by reduced activity of 7-dehydrocholesterol reductase, accumulation of 7-DHC, low cholesterol, and multiple congenital abnormalities.25 It has been speculated that mutations in DHCR7 also confer a competitive advantage to heterozygous carriers, because high levels of 7-DHC could provide protection against rickets and osteomalacia from hypovitaminosis D.26 However, there are few data on vitamin D status in individuals with Smith-Lemli-Opitz syndrome or carriers of known mutations.27 The finding that common variants at DHCR7 are strongly associated with circulating 25-OH D suggests that this enzyme could play a larger role in the normal regulation of vitamin D status than previously recognized.
The gene at the second locus, CYP2R1, encodes a hepatic, microsomal enzyme first described in 2003. It has been suggested that CYP2R1 is the enzyme responsible for 25-hydroxylation of vitamin D in the liver, but this is uncertain because many other enzymes with 25-hydroxylase activity in vitro have been described.28 Prior clinical studies have been limited to a case report of a Nigerian man with a point mutation in CYP2R1 who had a history of rickets,28 and a previous candidate gene study in 133 individuals with type 1 diabetes.18 Because affected individuals with CYP2R1 polymorphisms have been difficult to identify, it has been proposed that there is redundancy in the enzymes responsible for the 25-hydroxylation step. Thus, our finding that common variants at the CYP2R1 locus are associated with circulating 25-OH D represents the strongest evidence to date that CYP2R1 is the enzyme responsible for the critical first step in vitamin D metabolism.
The third gene, GC, encodes DBP, a 52-59 kDA protein synthesized in the liver that binds and transports vitamin D and its metabolites (including 25-OH D and 1,25(OH)2D).29 A few recent studies have reported associations between nonsynonymous SNPs in this gene,15-17,30,31 and 25-OH D concentrations. However, these studies were relatively small (≤ 1500 subjects) and lacked replication. The most commonly studied GC variants are the nonsynonymous SNPs rs7041 (Asp→Glu) and rs4588 (Thr→Lys). The older nomenclature for GC haplotypes (GC1S, GC1F, and GC2) is based on specific combinations of alleles at these nonsynonymous SNPs.15 Our data strongly confirm the association of rs7041 with circulating 25-OH D (overall meta-analytic P=1.9 × 10-109). The other variant, rs4588, is not in the HapMap dataset and thus not part of our imputed results. However, rs4588 is only 11 bp away from rs7041, and direct genotyping of rs4588 in one of our samples (Twins UK) confirms that it is in linkage disequilibrium (r2>0.99) with multiple associated variants from our genome-wide association study.
We also showed that GC variants associated with lower 25-OH D concentrations were strongly related to lower levels of DBP. Whether variation in the amount of circulating DBP influences further metabolism and availability of vitamin D is not well established. It has been hypothesized that levels of DBP may affect the delivery of 25-OH D and activated vitamin D (1,25(OH)2D) to target organs, as well as clearance of vitamin D metabolites from the circulation.15,16 On the other hand, alterations in quantity or function of DBP could be accompanied by changes in the relative proportions of free and bound 25-OH D, with the free fraction being the potential rate-limiting factor for 1,25(OH)2D production. Further studies are needed to assess the effects of variation in serum DBP.
In a screen of candidate gene variants, we observed an additional association at the locus containing CYP24A1 that was genome-wide significant in pooled analyses of the discovery and replication samples (P=6 × 10-10). CYP24A1 encodes 24-hydroxylase, which initiates the degration of both 25-OH D and 1,25(OH)2D. Prior candidate gene and linkage studies have failed to show an association of variants at this locus with 25-OH D levels, but these studies were small and based on highly-selected cohorts.30,32
A high “genotype score” using the three confirmed GWAS variants conferred a 2- to 2.5-fold risk of having vitamin D insufficiency to individuals in the upper quartile of the score compared with those in the lower quartile, after accounting for the usual environmental factors. This observation indicates that variation at a relatively small number of genetic loci could have a clinically-important impact on the risk of vitamin D insufficiency. The genotype score was also associated with an elevated risk of severe vitamin D deficiency (25-OH D < 25 nmol/L), although the odds ratio was slightly lower (1.4 in the highest quartile). It is unclear whether the lower odds ratio reflects a greater contribution of environmental factors to the most severe forms of vitamin D deficiency, because the prevalence of severe deficiency was low in our community-based cohorts.
Whether this genetic predisposition modifies the response to sun exposure or dietary supplementation warrants further study, particularly given the large inter-individual differences that have been observed in response to treatment with identical doses of vitamin D.33 Furthermore, these variants might serve as useful genetic tools for studies in disease cohorts to determine whether vitamin D insufficiency plays a causal role in a variety of chronic diseases with which it has been epidemiologically linked.
The validity of our findings is supported by the large study sample (more than 30,000 subjects combined in discovery and validation samples), consistent results across several standard assays for 25-OH D, and the strong biological plausibility of genes at the principal loci. Several limitations of the study also deserve mention, however. The study was not designed to identify uncommon or rare variants. Resequencing at selected loci, based in part upon our results, could be used to identify less common variants of potentially larger effects.
We used a multi-stage design in order to maximize the homogeneity of the assays used in the discovery analyses. It is possible that more genome-wide significant associations would have been identified by combining all GWAS cohorts into a single stage, although we would have lacked a large replication sample. Other factors that may have contributed to reduced statistical power include second-order interactions (such as with age) and the use of a stringent p-value threshold in the discovery stage.34 Accordingly, the absence of certain candidate genes from our top hits, such as those affecting vitamin D action or skin pigmentation, doesn't exclude a role for genetic variation at these loci in influencing vitamin D levels, but their contribution may be small compared with the genes identified.
Some cohorts used different assays for measurement of 25-OH D levels. To minimize the potential variability introduced by cohort-specific measurement techniques, we performed analyses in which 25-OH D levels were standardized within cohort and analyzed as a continuous trait. Furthermore, primary results were meta-analysed using a z-score weighted approach, which is not scale-dependent. Specific information on dietary intake and sunlight exposure was not available from all of the cohorts. Such factors likely contribute to non-genetic variability in 25-OH D concentrations, which would tend to reduce the effect observed in our analyses.
The identified SNPs may not be the causal variants, but rather in linkage disequilibrium with them. We did not examine “downstream” markers of vitamin D status, because 25-OH D concentrations are considered the most reliable indicators of vitamin D status. Other molecules, such as 1,25(OH)2D or parathyroid hormone, have greater intra-individual variability and reflect the influence of multiple determinants in addition to vitamin D status. Lastly, we studied only white individuals of European descent. Whether the genetic variants identified in the present study influence vitamin D status in other racial/ethnic groups is unknown and warrants further study.
In conclusion, we report the results of the first large-scale, systematic investigation of genetic determinants of vitamin D insufficiency. Elucidating the genetic architecture of this trait provides a better understanding of the regulation of vitamin D metabolism. Additionally, the genetic variants described in this report identify individuals at substantially elevated risk for vitamin D insufficiency.
Supplementary Material
Acknowledgments
Framingham Heart Study: The Framingham Heart Study of the National Heart, Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine is supported by the NIH/NHLBI contract N01-HC-25195. The present study received support from the American Heart Association, the U.S. Department of Agriculture, Agricultural Research Service (under Cooperative Agreement No. 58-1950-7-707), and the National Institute of Aging (AG14759). Dr. Kiel was supported by a grant from the National Institute of Arthritis, Musculoskeletal, and Skin Diseases and the National Institute on Aging (R01 AR/AG 41398). The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by a contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
Twins UK and Chingford: The study was funded by the Wellcome Trust, Arthritis Research Campaign, European Community's Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F2-2008-201865-GEFOS and EC Framework 7programme grant 200800 Treat OA/(FP7/2007-2013), ENGAGE project grant agreement HEALTH-F4-2007-201413 and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). The study also receives support from the Dept of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London. TDS is an NIHR senior Investigator. The project also received support from a Biotechnology and Biological Sciences Research Council (BBSRC) project grant. (G20234). The authors acknowledge the funding and support of the National Eye Institute via an NIH/CIDR genotyping project (PI: Terri Young). We thank the staff from the Genotyping Facilities at the Wellcome Trust Sanger Institute for sample preparation, Quality Control and Genotyping led by Leena Peltonen and Panos Deloukas; Le Centre National de Génotypage, France, led by Mark Lathrop, for genotyping; Duke University, North Carolina, USA, led by David Goldstein, for genotyping; and the Finnish Institute of Molecular Medicine, Finnish Genome Center, University of Helsinki, led by Aarno Palotie. Genotyping was also performed by CIDR as part of an NEI/NIH project grant.
The Rotterdam Study: This study was funded by the Netherlands Organization of Scientific Research NWO Investments (175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2) the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) project 050-060-810 and the European Comission (HEALTH-F2-2008-201865-GEFOS, and HEALTH-F2-2008-00-TREAT-OA). We thank P. Arp, M. Jhamai, M. Moorhouse, M. Verkerk and S. Bervoets for their help in creating the GWAS database. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII) and the Municipality of Rotterdam. We thank the staff from the Rotterdam Study, particularly L. Buist and J.H. van den Boogert and also the participating general practitioners and pharmacists.
1958 British Birth Cohort: The project was funded by the MRC (project grant G0601653), and 25-hydroxyvitamin D assays by the BUPA foundation. EH is funded by Department of Health (UK) Public Health Career Scientist Award. The use of the DNA from the 1958 British Birth Cohort (1958BC) collection is funded by MRC Grant G0000934 and Wellcome Trust Grant 068545/Z/02. This research utilizes resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113. The MRC Centre of Epidemiology for Child Health is funded by the MRC. Great Ormond Street Hospital/University College London, Institute of Child Health receives a proportion of funding from the Department of Health's National Institute of Health Research (‘Biomedical Research Centres’ funding).
Health, Aging, Body and Composition Study: This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging and National Institute on Aging contracts N01-AG-6-2101, N01-AG-6-2103, and N01-AG-6-2106. Assessment of 25-hydroxyvitamin D levels was funded by a National Institute on Aging grant, R01-AG029364. The genome-wide association study was funded by a National Institute on Aging grant, R01-AG032098, and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number HHSN268200782096C).
The Amish Family Osteoporosis Study: The Amish Family Osteoporosis Study was funded by a grant from the National Institute of Arthritis, Musculoskeletal and Skin Diseases (R01 AR46838).
Gothenberg Osteoporosis and Obesity Determinants Study: Financial support was received from the Academy of Finland (project grants 104781, 120315 and Center of Excellence in Complex Disease Genetics), University Hospital Oulu, Biocenter, University of Oulu, Finland, NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), ENGAGE project and grant agreement HEALTH-F4-2007-201413, the Medical Research Council (studentship grant G0500539, centre grant G0600705), the Wellcome Trust (project grant GR069224), UK, and the Research Council UK fellowship. The DNA extractions, sample quality controls, biobank up-keeping and aliquotting was performed in the national Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki. Financial support was received from the Swedish Research Council, the Swedish Foundation for Strategic Research, The ALF/LUA research grant in Gothenburg, the Lundberg Foundation, the Emil and Vera Cornell Foundation, the Torsten and Ragnar Söderberg's Foundation, Petrus and Augusta Hedlunds Foundation, the Västra Götaland Foundation, the Göteborg Medical Society, and the Sahlgrenska Center for Cardiovascular and Metabolic Research (CMR, no. A305:188), which is supported by the Swedish Strategic Foundation.
Study of Indiana Women: This work was supported by National Institutes of Health grants P01 AG-18397 and M01 RR-00750. Genotyping services were provided by CIDR. CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, National Library of Medicine.
North Finland Birth Cohort 1966: We thank Professor Paula Rantakallio (launch of NFBC1966 and 1986), Ms Outi Tornwall and Ms Minttu Jussila (DNA biobanking). Financial support was received from the Academy of Finland (project grants 104781, 120315, 1114194 and Center of Excellence in Complex Disease Genetics), University Hospital Oulu, Biocenter, University of Oulu, Finland, NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), ENGAGE project and grant agreement HEALTH-F4-2007-201413, the Medical Research Council (studentship grant G0500539, centre grant G0600705), the Wellcome Trust (project grant GR069224), UK. The DNA extractions, sample quality controls, biobank up-keeping and aliquotting was performed in the national Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki.
Cardiovascular Health Study: The CHS research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01-HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295, R01 HL084443, R01 HL087652, and R01 AG027002 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01RR00069 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Hertfordshire: The Hertfordshire Cohort Study was funded by the Medical Research Council of Great Britain and the arthritis research campaign.
Aberdeen Prospective Osteoporosis Screening Study: Funding for APOSS cohort sample collection and analysis was supported in part by grants from the European Commission (QLRT-2001-02629) and the UK Food Standards Agency. We are also extremely grateful for the hard work of the clinical research staff in the Bone and Musculoskeletal Research Programme and to all the women who kindly participated in the study.
Canadian Multicentre Osteoporosis Study: CaMos is funded by the Canadian Institutes of Health Research. Dr. Richards is a Clinical Investigator of the Canadian Institutes of Health Research (CIHR) and an Osteoporosis Canada New Investigator. This work was supported by grants to Dr. Richards from the CIHR and Canadian Foundation for Innovation. We acknowledge the support and funding of the Public Health Agency of Canada to assay vitamin D levels and the input of Linda S. Greene-Finestone, M. de Groh, Nicholas Hiridoglou, Ross Duncan, Alaa Badawi and Suneil Malik.
Other: The Cambridge Institute for Medical Research (CIMR) is in receipt of a Wellcome Trust Strategic Award (079895). This work was also supported by the Juvenile Diabetes Research Foundation International, the Wellcome Trust, and the National Institute for Health Research Cambridge Biomedical Centre.
Footnotes
Author Disclosures: TJW has served on the scientific advisory board of Diasorin. None of the other authors report conflicts of interest relevant to this manuscript.
References
- 1.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357:266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 2.Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF, Lieben L, Mathieu C, Demay M. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008;29:726–776. doi: 10.1210/er.2008-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hypponen E, Laara E, Reunanen A, Jarvelin MR, Virtanen SM. Intake of vitamin D and risk of type 1 diabetes: a birth-cohort study. Lancet. 2001;358:1500–1503. doi: 10.1016/S0140-6736(01)06580-1. [DOI] [PubMed] [Google Scholar]
- 4.Zipitis CS, Akobeng AK. Vitamin D supplementation in early childhood and risk of type 1 diabetes: a systematic review and meta-analysis. Arch Dis Child. 2008;93:512–517. doi: 10.1136/adc.2007.128579. [DOI] [PubMed] [Google Scholar]
- 5.Wang TJ, Pencina MJ, booth sl, Jacques PF, Ingelsson E, Lanier K, Benjamin EJ, D'Agostino RB, Wolf M, Vasan RS. Vitamin D deficiency and risk of cardiovascular disease. Circulation. 2008;117:503–511. doi: 10.1161/CIRCULATIONAHA.107.706127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giovannucci E, Liu Y, Hollis BW, Rimm EB. 25-hydroxyvitamin D and risk of myocardial infarction in men: a prospective study. Arch Intern Med. 2008;168:1174–1180. doi: 10.1001/archinte.168.11.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bischoff-Ferrari HA, Dawson-Hughes B, Staehelin HB, Orav JE, Stuck AE, Theiler R, Wong JB, Egli A, Kiel DP, Henschkowski J. Fall prevention with supplemental and active forms of vitamin D: a meta-analysis of randomised controlled trials. BMJ. 2009;339:b3692. doi: 10.1136/bmj.b3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez ME, Giovannucci EL, Colditz GA, Stampfer MJ, Hunter DJ, Speizer FE, Wing A, Willett WC. Calcium, vitamin D, and the occurrence of colorectal cancer among women. J Natl Cancer Inst. 1996;88:1375–1382. doi: 10.1093/jnci/88.19.1375. [DOI] [PubMed] [Google Scholar]
- 9.Garland CF, Comstock GW, Garland FC, Helsing KJ, Shaw EK, Gorham ED. Serum 25-hydroxyvitamin D and colon cancer: eight-year prospective study. Lancet. 1989;2:1176–1178. doi: 10.1016/s0140-6736(89)91789-3. [DOI] [PubMed] [Google Scholar]
- 10.John EM, Schwartz GG, Dreon DM, Koo J. Vitamin D and breast cancer risk: the NHANES I Epidemiologic follow-up study, 1971-1975 to 1992. National Health and Nutrition Examination Survey. Cancer Epidemiol Biomarkers Prev. 1999;8:399–406. [PubMed] [Google Scholar]
- 11.Autier P, Gandini S. Vitamin D supplementation and total mortality: a meta-analysis of randomized controlled trials. Arch Intern Med. 2007;167:1730–1737. doi: 10.1001/archinte.167.16.1730. [DOI] [PubMed] [Google Scholar]
- 12.Livshits G, Karasik D, Seibel MJ. Statistical genetic analysis of plasma levels of vitamin D: familial study. Ann Hum Genet. 1999;63:429–439. doi: 10.1046/j.1469-1809.1999.6350429.x. [DOI] [PubMed] [Google Scholar]
- 13.Shea MK, Benjamin EJ, Dupuis J, Massaro JM, Jacques PF, D'Agostino RB, Sr, Ordovas JM, O'Donnell CJ, Dawson-Hughes B, Vasan RS, Booth SL. Genetic and non-genetic correlates of vitamins K and D. Eur J Clin Nutr. 2009;63:458–464. doi: 10.1038/sj.ejcn.1602959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter D, De Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, Spector TD. Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J Bone Miner Res. 2001;16:371–378. doi: 10.1359/jbmr.2001.16.2.371. [DOI] [PubMed] [Google Scholar]
- 15.Lauridsen AL, Vestergaard P, Hermann AP, Brot C, Heickendorff L, Mosekilde L, Nexo E. Plasma concentrations of 25-hydroxy-vitamin D and 1,25-dihydroxy-vitamin D are related to the phenotype of Gc (vitamin D-binding protein): a cross-sectional study on 595 early postmenopausal women. Calcif Tissue Int. 2005;77:15–22. doi: 10.1007/s00223-004-0227-5. [DOI] [PubMed] [Google Scholar]
- 16.Sinotte M, Diorio C, Berube S, Pollak M, Brisson J. Genetic polymorphisms of the vitamin D binding protein and plasma concentrations of 25-hydroxyvitamin D in premenopausal women. Am J Clin Nutr. 2009;89:634–640. doi: 10.3945/ajcn.2008.26445. [DOI] [PubMed] [Google Scholar]
- 17.Engelman CD, Fingerlin TE, Langefeld CD, Hicks PJ, Rich SS, Wagenknecht LE, Bowden DW, Norris JM. Genetic and environmental determinants of 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D levels in Hispanic and African Americans. J Clin Endocrinol Metab. 2008;93:3381–3388. doi: 10.1210/jc.2007-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos-Lopez E, Bruck P, Jansen T, Herwig J, Badenhoop K. CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes Metab Res Rev. 2007;23:631–636. doi: 10.1002/dmrr.719. [DOI] [PubMed] [Google Scholar]
- 19.Hypponen E, Turner S, Cumberland P, Power C, Gibb I. Serum 25-hydroxyvitamin D measurement in a large population survey with statistical harmonization of assay variation to an international standard. J Clin Endocrinol Metab. 2007;92:4615–4622. doi: 10.1210/jc.2007-1279. [DOI] [PubMed] [Google Scholar]
- 20.Haughton MA, Mason RS. Immunonephelometric assay of vitamin D-binding protein. Clin Chem. 1992;38:1796–1801. [PubMed] [Google Scholar]
- 21.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 23.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 24.Becker BJ, Wu M. The synthesis of regression slopes in meta-analysis. Statistical Science. 2007;3:414. [Google Scholar]
- 25.Tint GS, Irons M, Elias ER, Batta AK, Frieden R, Chen TS, Salen G. Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N Engl J Med. 1994;330:107–113. doi: 10.1056/NEJM199401133300205. [DOI] [PubMed] [Google Scholar]
- 26.Porter FD. Malformation syndromes due to inborn errors of cholesterol synthesis. J Clin Invest. 2002;110:715–724. doi: 10.1172/JCI16386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi M, Federico G, Corso G, Parenti G, Battagliese A, Frascogna AR, Della Casa R, Dello Russo A, Strisciuglio P, Saggese G, Andria G. Vitamin D status in patients affected by Smith-Lemli-Opitz syndrome. J Inherit Metab Dis. 2005;28:69–80. doi: 10.1007/s10545-005-3676-8. [DOI] [PubMed] [Google Scholar]
- 28.Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc Natl Acad Sci U S A. 2004;101:7711–7715. doi: 10.1073/pnas.0402490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Speeckaert M, Huang G, Delanghe JR, Taes YE. Biological and clinical aspects of the vitamin D binding protein (Gc-globulin) and its polymorphism. Clin Chim Acta. 2006;372:33–42. doi: 10.1016/j.cca.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Ahn J, Albanes D, Berndt SI, Peters U, Chatterjee N, Freedman ND, Abnet CC, Huang WY, Kibel AS, Crawford ED, Weinstein SJ, Chanock SJ, Schatzkin A, Hayes RB. Vitamin D-related genes, serum vitamin D concentrations and prostate cancer risk. Carcinogenesis. 2009;30:769–776. doi: 10.1093/carcin/bgp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janssens W, Bouillon R, Claes B, Carremans C, Lehouck A, Buysschaert I, Coolen J, Mathieu C, Decramer M, Lambrechts D. Vitamin D Deficiency is Highly Prevalent in COPD and Correlates with Variants in the Vitamin D Binding Gene. Thorax. 2009 doi: 10.1136/thx.2009.120659. [DOI] [PubMed] [Google Scholar]
- 32.Wjst M, Altmuller J, Braig C, Bahnweg M, Andre E. A genome-wide linkage scan for 25-OH-D(3) and 1,25-(OH)2-D3 serum levels in asthma families. J Steroid Biochem Mol Biol. 2007;103:799–802. doi: 10.1016/j.jsbmb.2006.12.053. [DOI] [PubMed] [Google Scholar]
- 33.Heaney RP. Vitamin D and calcium interactions: functional outcomes. Am J Clin Nutr. 2008;88:541S–544S. doi: 10.1093/ajcn/88.2.541S. [DOI] [PubMed] [Google Scholar]
- 34.Lasky-Su J, Lyon HN, Emilsson V, Heid IM, Molony C, Raby BA, Lazarus R, Klanderman B, Soto-Quiros ME, Avila L, Silverman EK, Thorleifsson G, Thorsteinsdottir U, Kronenberg F, Vollmert C, Illig T, Fox CS, Levy D, Laird N, Ding X, McQueen MB, Butler J, Ardlie K, Papoutsakis C, Dedoussis G, O'Donnell CJ, Wichmann HE, Celedon JC, Schadt E, Hirschhorn J, Weiss ST, Stefansson K, Lange C. On the replication of genetic associations: timing can be everything! Am J Hum Genet. 2008;82:849–858. doi: 10.1016/j.ajhg.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.