Abstract
Fragile X-associated tremor/ataxia syndrome (FXTAS) is an adult-onset neurodegenerative disorder caused by CGG trinucleotide repeat expansions in the fragile X mental retardation 1 (FMR1) gene. The neuropathological hallmark of the disease is the presence of ubiquitin-positive intranuclear inclusions in neurons and in astrocytes. Ubiquitin-positive intranuclear inclusions have also been found in the neurons of transgenic mice model carrying an expanded CGG(98) trinucleotide repeat of human origin, but have not previously been described in glial cells. Therefore, we used immunocytochemical methods to determine the pathological features of nuclear and/or cytoplasmic inclusions in astrocytes, Bergmann glia and neurons, as well as relationships between inclusion patterns, age, and repeat length in CGG knock-in (KI) mice in comparison with wild type mice. In CGG KI mice, ubiquitin-positive intranuclear inclusions were found in neurons (e.g., pyramidal cells, GABAergic neurons) throughout the brain in cortical and subcortical brain regions; these inclusions increased in number and size with advanced age. Ubiquitin-positive intranuclear inclusions were also present in protoplasmic astrocytes, including Bergmann glia in the cerebellum. The morphology of intranuclear inclusions in CGG KI mice was compared to that of typical inclusions in human neurons and astrocytes in postmortem FXTAS brain tissue. This new finding of previously unreported pathology in astrocytes of CGG KI mice now provides an important mouse model to study astrocyte pathology in human FXTAS.
Keywords: FXTAS, mouse model, neurodegenerative disorder, cellular pathology, astrocytes, microglia, oligodendrocytes
1. Introduction
The Fragile X mental retardation gene (FMR1) is polymorphic for the length of CGG tandem trinucleotide repeat in the 5’-untranslated region (5’-UTR). Repeat lengths in the general population range from 5–55, while full mutations with repeat lengths above 200 result in hypermethylation of FMR1, transcriptional silencing and the clinical syndrome Fragile X mental retardation (FXS). Intermediate length CGG expansions between 55–200, referred to as the fragile X premutation, have now been identified in patient populations and are associated with a unique, progressive, late onset, neurodegenerative disorder called Fragile X-associated tremor/ataxia syndrome (FXTAS). The neurological symptoms of FXTAS include intention tremor and ataxia, peripheral neuropathy, neuropsychological problems (anxiety, depression), and cognitive impairments including dementia at late stages of the disorder (Berry-Kravis et al., 2007; Hagerman and Hagerman, 2004; Hagerman et al., 2007). Radiologic changes seen on MRI include patchy T2 signal hyperintensities in cerebral white matter and in the middle cerebellar peduncle (“the MCP sign”), and overall brain atrophy (Brunberg et al., 2002). The neuropathological hallmark of FXTAS is the presence of eosinophilic intranuclear inclusions in neurons and astrocytes throughout the brain that stain positive for ubiquitin (Greco et al., 2006; Greco et al., 2002; Tassone et al., 2004). In contrast to FXS where gene silencing occurs, FXTAS is associated with increased transcription resulting in 3–8 fold elevations in levels of FMR1 mRNA in leukocytes. However, translation of the FMR1 mRNA with expanded CGG repeats in the premutation range is inefficient so that levels of the gene product, Fragile X mental retardation protein (FMRP) are paradoxically low (Tassone et al., 2000). The disorder appears to be due to an “RNA toxic gain of function”, although the mechanisms for disease progression, including formation of intranuclear ubiquitin-positive inclusions in neurons and astrocytes are not well understood (Oostra and Willemsen, 2009).
Human carriers with the fragile X premutation underlying FXTAS have intranuclear inclusions in neurons and astrocytes throughout the brain. In humans, it is still unknown at what age inclusions form due to the progressive nature of the disorder and the advanced age at which FXTAS is typically diagnosed, as well as that the tissue diagnosis is made at autopsy. It has been shown that inclusions can form after as few as eight days in vitro after an expanded CGG repeat in the premutation range is introduced into human primary neural progenitor cells and into established cell lines (Arocena et al., 2005). In order to better characterize FXTAS and to study its molecular mechanisms, knock-in (KI) mice bearing expanded CGG trinucleotide repeats in the 5’-UTR have been created. The CGG KI mouse model of the fragile X premutation was generated by replacing the endogenous CGG8 trinucleotide repeat with a CGG98 trinucleotide repeat of human origin via homologous recombination (Bontekoe et al., 1997; Willemsen et al., 2003). Similar to the human cases of FXTAS, neurons of these CGG KI mice show intranuclear inclusions that stain for ubiquitin in neurons in a number of brain regions (Bontekoe et al., 1997; Brouwer et al., 2008; Willemsen et al., 2003). It is not yet known if the inclusions contribute directly to the neuropathology seen in FXTAS. It has been suggested that intranuclear inclusions may not be pathological of themselves, but may reflect pathology such as mRNA toxicity due to the increased gene transcription resulting from the premutation or perhaps due to the presence of the mutant FMR1 mRNA itself (Brouwer et al., 2008; Oostra and Willemsen, 2009; Willemsen et al., 2003).
Intranuclear inclusions in these CGG KI mice are common in neurons at 50–100 weeks of age, and have been observed as early as 20 weeks of age (Bontekoe et al., 1997; Brouwer et al., 2008; Willemsen et al., 2003). However, unlike FXTAS, intranuclear inclusions have not been previously reported in astrocytes in the CGG KI mice. The present study replicates the previous findings of intranuclear inclusions in neurons in the CGG KI mouse, and expands upon these findings by describing the presence and distribution of such inclusions in glial cells (i.e., astrocytes).
2. Results
Nuclear pathology in neurons of CGG KI mice and FXTAS patients
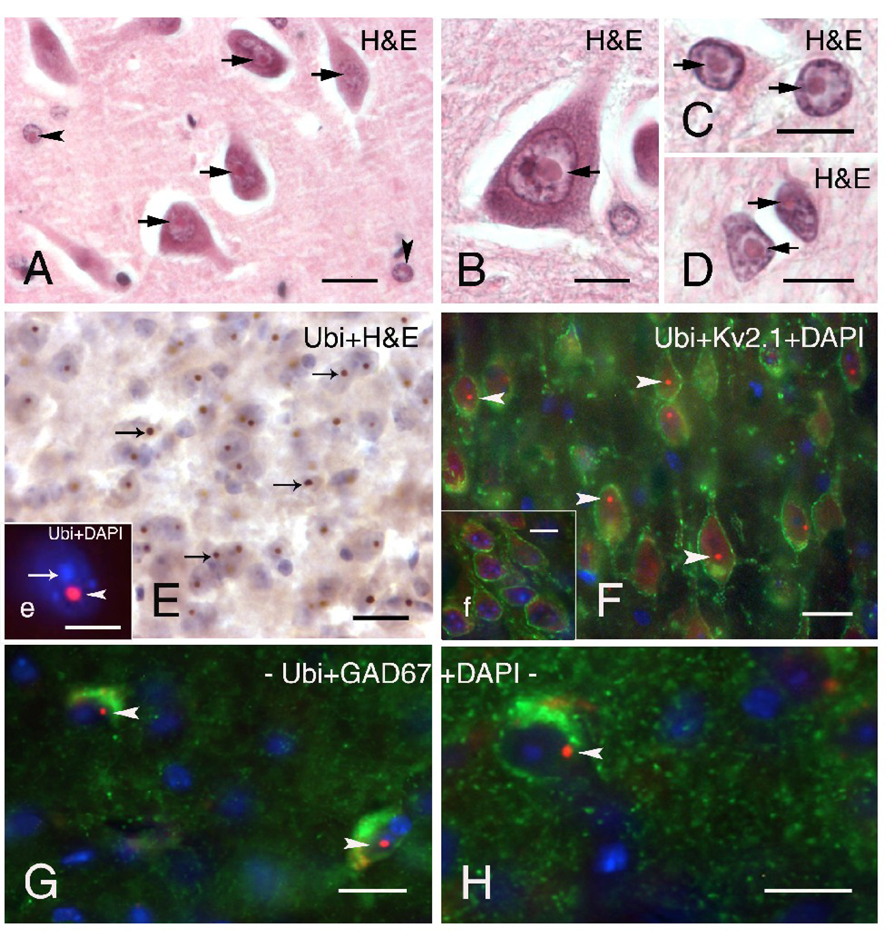
The presence of intranuclear inclusions in neurons and astrocytes is a pathological hallmark of FXTAS. Thus far, similar intranuclear inclusions have not been reported in oligodendrocytes or microglia in human FXTAS or CGG KI mice. Figures 1A, B show typical intranuclear inclusions in neurons and astrocytes in the hippocampus of a patient who was diagnosed with FXTAS at 65 years of age and later died at 78 (case 2 from (Greco et al., 2002)). These inclusions in a human patient are shown for comparison with intranuclear inclusions found in the brains of CGG KI mice, and both appeared as well-delineated spherical bodies, 2–3 µm diameter, that were intensely eosinophilic with hematoxylin and eosin staining (H&E). Identification of cell types in human tissue were based on methods reported by (Greco et al., 2006; Greco et al., 2002).
Figure 1.
Neuronal intranuclear inclusions in brain tissue from a patient with FXTAS and in CGG KI mice. A–B. Typical eosinophilic intranuclear inclusions in neurons (A, B; arrows) and astrocyte-like cells (A; arrowheads) in the dentate hilus of a FXTAS patient. (H&E; hematoxylin and eosin). C–D. Large eosinophilic intranuclear inclusions in protoplasmic astrocytes in gray matter (C; arrows) and in fibrous astrocytes (D; arrows) located in white matter of the entorhinal cortex of a FXTAS patient. E. Intranuclear inclusions (arrows) in neurons in the entorhinal cortex of CGG KI mice stained with DAB (brown reaction product) and counterstained with H&E. Inset e shows higher magnification fluorescent image of a DAPI-stained neuronal nucleus (blue, arrow) with a prominent ubiquitin-positive intranuclear inclusion (red, arrowhead). F. Pyramidal neurons in motor cortex of CGG KI mice immunolabeled for Kv2.1 potassium channels in the membrane (green), ubiquitin-positive intranuclear inclusions (red, arrowheads), and DAPI staining of the nucleus (blue). Inset f shows the absence of intranuclear inclusions in neurons in the cortex of a wildtype mouse. G–H. Ubiquitin-positive intranuclear inclusions (red, arrowhead) in GAD67-labeled interneurons (green) in the dentate gyrus molecular layer (H) and in the hippocampal CA1 stratum lacunosum moleculare (G). Scale bars: A, 20 µm; B–H, 10 µm.
Topographical Distribution of Intranuclear Neuronal Inclusions
Figure 1E shows intranuclear inclusions in neurons in the entorhinal cortex of a CGG KI mouse that immunostained stained for ubiquitin and were similar in appearance to inclusions found in human brain tissue. Immunofluorescent staining of ubiquitin (red) in Figure 1e clearly shows the localization of an intranuclear inclusion within a DAPI-stained nucleus (blue) in a CGG KI mouse brain. Using triple immunofluorescent staining for the potassium channel subunit Kv2.1 (green) to visualize neurons in the motor cortex, and DAPI to label the nucleus (blue), ubiquitin positive intranuclear inclusions (red) are readily observed in CGG KI mice (Fig. 1F), but not in wildtype control mice (Fig. 1f, inset). Inclusions in the entorhinal cortex and neocortex of older KI mice (e.g., 52–70 weeks of age) ranged in size from 1.6–2.7 µm diameter. Microscopic examination of both wildtype and male CGG KI mice revealed no additional evidence of gross-anatomical abnormalities and/or neuropathological features (e.g., no obvious neuronal cell loss, astrogliosis) as compared with wildtype mice.
In CGG KI mice older than 52 weeks of age, ubiquitin-positive intranuclear inclusions were found in neurons in most brain regions, but the frequency and size of individual inclusions differed depending on the brain region under examination and length of the CGG expanded repeats. In the neocortex intranuclear inclusions were observed across all layers, but were most frequent in layers II/III and V, and were more frequently observed in neurons of the rostral neocortex (i.e., sensory and motor areas) and caudal/ventral neocortex (i.e., retrosplenial, visual, and entorhinal cortices), the olfactory bulb and hippocampus (Figures 1E, F). Granule cells within the olfactory bulb and dentate gyrus, and neurons in the hippocampal-amygdala area presented the highest incidence of neuronal intranuclear inclusions, where >50% of neurons exhibited inclusions. Prominent intranuclear inclusions were also present in a high percentage of neurons in hypothalamic nuclei, thalamic subnuclei, inferior colliculus, and specific nuclei of the brain stem (e.g., vestibularis nuclei, and nuclei of the reticular formation). In the cerebellum, neuronal intranuclear inclusions were present in neurons within the cerebellar nuclei, and in the cerebellar cortex inclusions were present in granular cells and interneurons (likely stellate cells, Golgi cells, and/or basket cells based on their location). However, inclusions were rarely observed in Purkinje cells, and were also rarely seen in some brain regions such as the striatum.
Cell Type Distribution of Intranuclear Neuronal Inclusions
To determine whether GABAergic neurons exhibit ubiquitin-positive intranuclear inclusions, brain sections of CGG KI mice were immunoreacted for GAD67, one of the two GABA-synthetising isoenzymes used to identify GABAergic neurons. In neocortex and hippocampus, a high percentage of the GAD67-labeled interneurons showed the presence of intranuclear inclusions identified by their positive immunoreactivity for ubiquitin (Figure 1 G–H).
Age and Gender Dependence of Intranuclear Neuronal Inclusions Localization
The presence and size of intranuclear inclusions in neurons were clearly age-dependent as previously reported (Hunsaker et al., 2009; Willemsen et al., 2003). CGG KI mice at 12–25 weeks of age showed only a small number of neuronal intranuclear inclusions, regardless of brain region, and were more frequent in smaller neurons such as granule cells within the olfactory bulb and dentate gyrus. At this age inclusions were also seen in neurons of rostral neocortical areas, amygdala and hypothalamus, albeit rarely. In addition, intranuclear inclusions at these younger ages appeared to be smaller in size, becoming larger with increasing age (i.e., > than 40 weeks), suggesting a developmental progression in their number and size (data not shown). Wildtype mice ranging from 20–76 weeks of age were examined in parallel with the CGG KI mice, but no ubiquitin-positive inclusions were observed in any of the wildtype mice at any of the ages examined (e.g., Figure 1f, inset).
Additionally, two female CGG KI mice aged 70 and 75 weeks of age that were heterozygous for the CGG repeat expansion were examined for inclusions in the present study. These two female KI mice had CGG repeat expansions of 150 and 152 on one X allele, respectively, with the other allele having the wildtype length of 8–10 CGG trinucleotide repeats. The brains of these female mice showed the presence of ubiquitin-positive intranuclear inclusions that were similar in appearance to male CGG KI mice (data not shown). Female wildtype mice examined in parallel never showed the presence of intranuclear inclusions.
It is noteworthy that in both WT and CGG KI mice the majority of large neurons (e.g., pyramidal neurons in the neocortex and neurons in brainstem nuclei) showed diffuse ubiquitin-positive cytoplasmic staining (e.g., Figure 1F). This staining pattern was present in addition to lipofuscin-like accumulations of autofluorescent material within the cytoplasm of neurons in aging animals, but did not obscure the prominent staining pattern of inclusions in CGG KI mice. Control procedures using CuSO4 to reduce autofluorescence confirmed that the ubiquitin staining of intranuclear inclusions, as well as cytoplasmic staining of ubiquitin, were not due to lipofuscin-like autofluorescence.
Nuclear pathology in astrocytes of CGG KI mice
Previous studies of CGG KI mice did not find intranuclear inclusions in astrocytes of CGG KI mice (Willemsen et al., 2003), while such inclusions in astrocytes are consistently observed in the human brain from FXTAS patients (Figures 1C, D) (Greco et al., 2006; Greco et al., 2002). Because the CGG KI mouse models much of the other pathology of FXTAS, a major goal of the present study was to determine whether intranuclear inclusions could be demonstrated in the astrocytes of CGG KI mice.
General Topographic Distribution and Localization
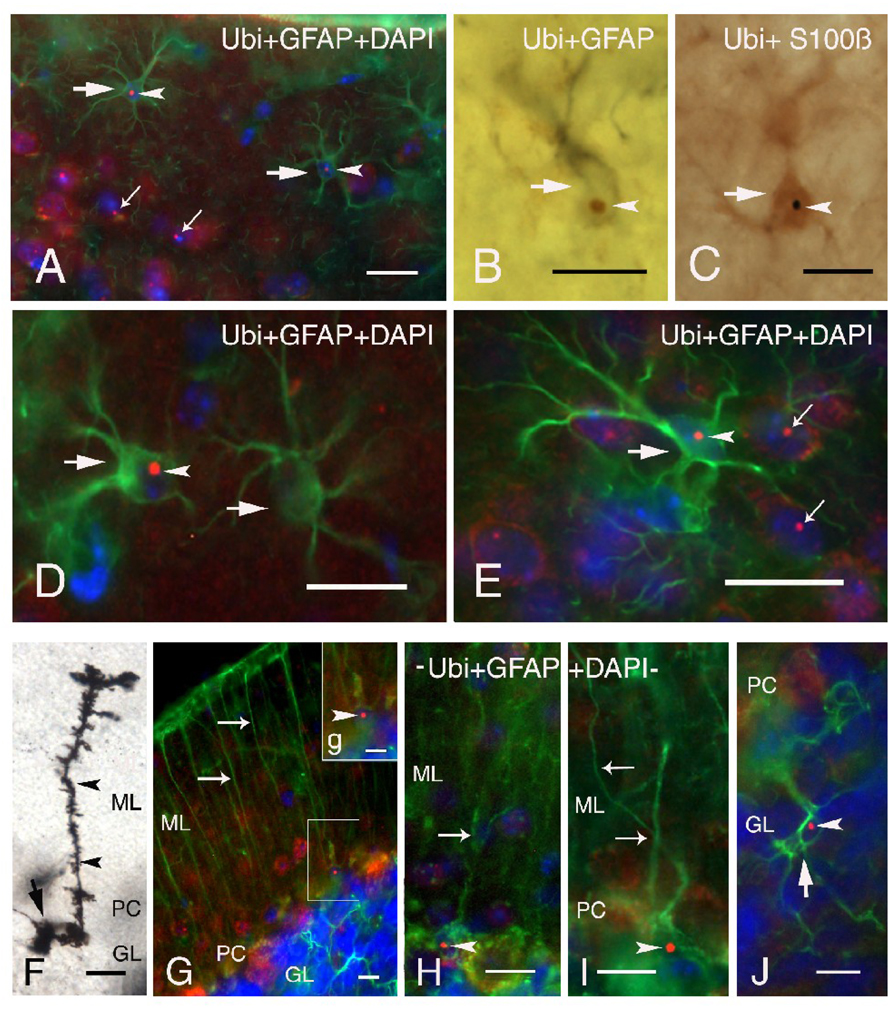
Protoplasmic astrocytes, predominantly located in grey matter, and fibrous astrocytes within the white matter were selectively immunolabeled with GFAP and/or S100β antisera and visualized using immunofluorescence (Figures 2A, D, E) or DAB peroxidase staining (Figures 2B, C). Both labeling techniques revealed the presence of ubiquitin-positive intranuclear inclusions in astrocytes. Inclusions were seen more frequently in protoplasmic astrocytes and only rarely in fibrous astrocytes in the white matter (e.g., fiber tracts). Intranuclear inclusions in astrocytes ranged in size from 1.1 to 2.1 µm diameter and were clearly identified in astrocytic nuclei as ubiquitin-positive spherical bodies (arrowheads; Figures 2A, D, E), that were easily distinguished from surrounding DAPI blue-stained nuclei. Protoplasmic astrocytes with intranuclear inclusions were more diffusely distributed in cortical layer I (large arrows; Figures 2A, D), and clearly distinguished from neuronal intranuclear inclusions as shown in Figures 2A and 2E (neuronal inclusions indicated by small arrows). It is notable that ubiquitin-positive intranuclear inclusions were rarely observed in protoplasmic astrocytes in other brain regions (e.g. hypothalamus, caudal brainstem nuclei). Astrocytes bearing ubiquitin-positive intranuclear inclusions were quantitatively assessed in lamina I of the neocortex. Only 10.2 ± 1.2% of total astrocytes in lamina I of the neocortex exhibited intranuclear inclusions. Quantification of the numbers of astrocytes with inclusions is problematic because of the high degree of variability in the numbers of astrocytes with inclusions found from one brain region to another, and even within a specific brain region. For example, in some lamina I areas of the neocortex as many as 30–40% of astrocytes show inclusions, while there may few or no astrocytes with inclusions in the immediately adjacent neocortical area. The reason for this pattern of distribution of astrocytes with inclusions is unknown. We are in the process of obtaining more quantitative data on the relationship between age, CGG repeat length, and distribution of neurons and astrocytes with inclusions to be reported as a later date. In the current study there was no evidence for the presence of reactive astrocytes and/or prominent astrogliosis in the brain of CGG KI mice as compared to wildtype mice.
Figure 2.
Immunofluorescent labeling of ubiquitin-positive intranuclear inclusions in astrocytes and Bergmann glia of CGG KI mice. A. GFAP immunofluorescent labeling (green) of two protoplasmic astrocytes (large arrows) in neurocortical lamina I showing ubiquitin-positive intranuclear inclusions (red, arrowheads) within the DAPI stained nucleus (blue). Note the ubiquitin-positive intranuclear inclusions in several neurons (small arrows) located in lamina II/III. B. Immunocytochemistry with DAB showing a GFAP-labeled astrocyte (gray, arrow) containing a ubiquitin-positive intranuclear inclusion (brown, arrowhead). C. Astrocyte (arrow) with an intranuclear inclusion (black, arrowhead) immunostained with S100β using a DAB chromogen with a blue-gray reaction product. D–E. Higher magnification of protoplasmic astrocytes (arrows) in neocortical lamina I (D) and II/III (E) labeled for GFAP (green), ubiquitin (red, arrowhead) and with nuclear DAPI staining (blue). Note ubiquitin-positive intranuclear inclusions in several neurons in panel E (small arrows). F. Golgi-Cox impregnated Bergmann glia in the cerebellar cortex of a CGG KI mouse with characteristic small cell body (arrow), and radial process (arrowheads) extending through the molecular layer (ML) to the pial membrane. G. Fluorescent image of GFAP-positive Bergman glia cells (green) with radial processes (arrows), some of which show ubiquitin-positive intranuclear inclusions (g inset, arrowhead). H–I. Higher magnification of Bergmann glia with radial processes (arrows) and intranuclear inclusions (arrowheads). J. Vellate astrocyte (arrow) in the granule layer of the cerebellar cortex labeled with GFAP (green) exhibiting extended processes and a ubiquitin-stained intranuclear inclusions (red, arrowhead) within the DAPI stained nucleus (blue). Abbreviations: GL, granule layer; PC, Purkinje cell layer; ML, molecular layer)
In addition, ubiquitin-positive intranuclear inclusions were also frequently observed in other non-neuronal cells in the brain, including the ependymal/subependymal cells and epithelial cells of the choroid plexus (data not shown) cells that may have the same precursor cell type as astrocytes (cf., (Greco et al., 2006)).
Localization of Intranuclear Inclusions in Bergmann Glia
In the cerebellum ubiquitin-positive intranuclear inclusions were found diffusely in Bergmann glia and astrocytes, and were widely present in neurons of CGG KI mice. Figure 2F shows an example of a Golgi impregnated Bergmann glia, with its soma located in the Purkinje cell layer (PC) and a long radial process ascending to the pial surface. These radial processes bear irregular, leaf-like appendages that end in bulbous tips or endfeet near the pial surface. Bergmann glia immunostained for GFAP showed a similar morphology, as well as ubiquitin-positive intranuclear inclusions in their soma (Figures 2G–I). These inclusions were always clearly defined, round and localized to the nucleus (Figure 2G, inset g, H and I). In addition to Bergmann glia, protoplasmic astrocytes were commonly seen within the molecular layer, but within the granular layer they represented a morphologically distinct neuroglia cell type, the velate protoplasmic astrocyte. These astrocytes are characterized by veil-like processes ramifying within the granular layer (Palay and Chan-Palay, 1974). A GFAP-labeled velate protoplasmic astrocyte bearing a ubiquitin-positive inclusion is shown in Figure 2J. There was no evidence of reactive astrocytes /or specifically Bergmann gliosis in the cerebellum of CGG KI mice as shown by GFAP staining and/or Golgi impregnation.
Cytoplasmic inclusions in oligodendrocytes of CGG KI mice
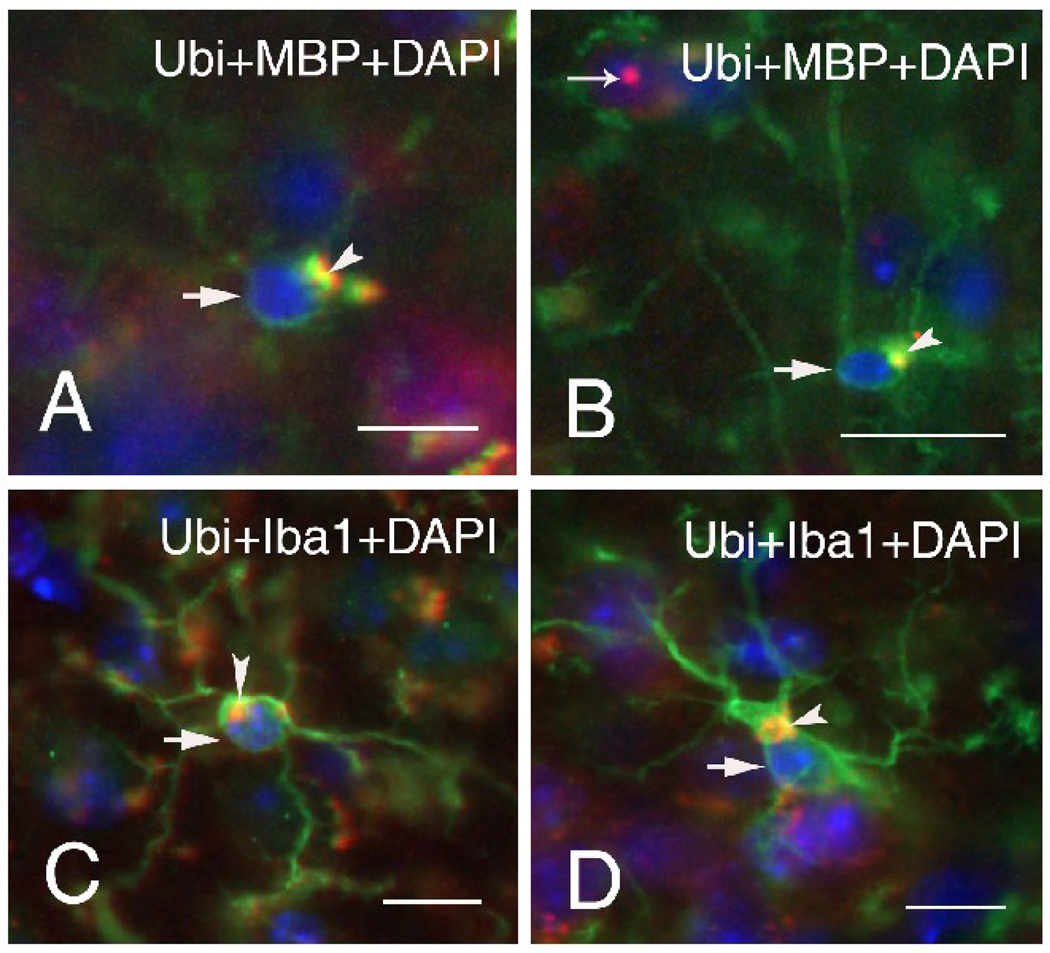
Many oligodendrocytes across cortical and subcortical regions stained positive for myelin basic protein (MBP), but none of these oligodendrocytes showed ubiquitin-positive intranuclear inclusions (Figure 3A, B). However, many oligodendrocytes in both wildtype (Figure 3A) and CGG KI (Figure 3B) mice contained irregular and amorphic inclusions within the cytoplasm which were immuno-positive for both MBP and ubiquitin (arrowheads; Figure 3A, B). These inclusions were clearly localized within the cytoplasm, were not in the nucleus (stained blue with DAPI), and measured between 1.1 to 3.3 µm in diameter.
Figure 3.
Representative immunofluorescent staining (arrows) of oligodendrocytes of wildtype (A) and CGG KI (B) mice, and microglia of wildtype (C) and CGG KI mice (D). Large, irregularly shaped cytoplasmic inclusion bodies were found in oligodendrocytes and microglia of both wildtype and CGG KI mice (arrow heads). Oligodendrocytes were labeled for myelin basic protein (MBP, green) and microglia for Iba1 (green), so that co-colocalization of ubiquitin in the inclusions with either MBP or Iba1 appears yellow. Nuclei were stained with DAPI (blue). Both wildtype mice were 60 weeks of age (A, C), and the two CGG KI mice were 70 (B) and 59 (D) weeks of age. Scale bars are 10 µm.
Cytoplasmic inclusions in microglia of CGG KI mice
Microglia identified by Iba1 staining were distributed throughout the brain, with the majority of cells displaying a ramified morphology and territorial distribution suggesting a “resting microglia phenotype”. However, in the neocortex and brain stem of CGG KI mice some of the microglial cells presented morphological features of activated, but not “phagocytic”, microglia (e.g., bushy morphology, retraction of the processes). Brain macrophages (“phagocytic” microglia) were occasionally observed. The major observation was that none of the microglia subtypes exhibited ubiquitin-positive intranuclear inclusions. However, in both wildtype (Figure 3C) and CGG KI (Figure 3D) mice the majority of microglial cells had ubiquitin-positive inclusions within the cytoplasm that had an amorphic and/or vacuolar appearance and ranged in size from 3–10 µm in diameter. The majority of those cytoplasmic inclusions also demonstrated autofluorescence in control sections in which the antibodies were omitted. In order to determine if this autofluorescence was due to lipofuscin, sections were treated with CuSO4 (Schnell et al., 1999). This procedure did not affect autofluorescence, suggesting that it was not lipofuscin-related. The source of autofluoresence of these cytoplasmic inclusions remains to be determined.
3. Discussion
Intranuclear inclusions in neurons and astrocytes that stain for ubiquitin are the neuropathological hallmark of FXTAS. Similar appearing ubiquitin-positive inclusions are also found in neurons in the brains of CGG KI mice, but previous studies have not demonstrated such inclusions in mouse astrocytes. In this study we extend the description of the cellular and brain regional distribution and appearance of intranuclear inclusions in a CGG KI mouse model of FXTAS to include astrocytes. New evidence is also presented for previously unreported neuropathology in the cytoplasm of oligodendroglia and microglia in both CGG KI as well as wildtype mice that may be related to aging.
Ubiquitin-positive intranuclear inclusions were found in astroglia in neocortex and cerebellum, including Bergmann glia of the cerebellum. Inclusions were preferentially found in protoplasmic astrocytes, and were only rarely observed in fibrous astrocytes. Overall, the occurrence of intranuclear inclusions in glia was markedly less common than in neurons. As expected, intranuclear inclusions were not observed in neurons or glia of wildtype mice. These results are an important extension of previous findings because intranuclear inclusions in astrocytes and Bergmann glia of CGG KI mice were not reported in previous studies (Brouwer et al., 2008; Entezam et al., 2007; Willemsen et al., 2003). This difference may be due to the use of different background strains or to differences in histological methodology (e.g., frozen sections versus paraffin embedding, H & E versus immunofluoresence; difference in section thickness). Since inclusions in astrocytes are a common feature of clinical FXTAS (Greco et al., 2006; Greco et al., 2002), these findings further validate the mouse model and enhance its value as a model of FXTAS. Although no remarkable histopathologies (e.g., Purkinje cell loss, Bergmann gliosis) were observed in the cerebellum of CGG KI mice in the present study, intranuclear ubiquitin-positive inclusions were found in Bergmann glia and other astroglial cells. These findings suggest involvement of cerebellar glia from CGG mice in the pathogenesis of the disease. The relatively small number of glia showing inclusions may explain, in part, the “milder” motor deficits found in the CGG KI mouse model of FXTAS than that seen in clinically advanced cases of FXTAS (Van Dam et al., 2005). The fact that murine astrocytes appear to be less prone to generate intranuclear inclusions in CGG KI mice when compared to astrocytes in human FXTAS raises important questions concerning differences between murine and human astrocytes in the cellular machinery and molecular mechanisms underlying inclusion formation.
Intranuclear inclusions in neurons of CGG KI mice were found preferentially in cortical/neocortical areas (e.g., rostral neocortex, olfactory bulb/nuclei, hippocampus, entorhinal cortex), subcortical regions (e.g., amygdala, hypothalamus, inferior colliculus, brain stem nuclei) and in selected neuronal cell types (e.g., neocortical pyramidal cells, GABAergic neurons/interneurons), similar to previous findings in KI mice (Brouwer et al., 2008; Entezam et al., 2007; Willemsen et al., 2003).
Inclusion formation, CGG repeat length, Fmr1 mRNA and Fmrp
The precise relationships between CGG repeat number and inclusion formation, neuropathology, and/or clinical involvement are still unclear. In humans, CGG length is highly correlated with the number of intranuclear inclusions in astrocytes and neurons, and appears to be an important predictor of brain pathology (Greco et al., 2006). Similarly, CGG KI mice with larger CGG repeat sizes typically have the greatest numbers and most widespread distributions of intranuclear inclusions in the brain (Brouwer et al., 2008; Entezam et al., 2007; Willemsen et al., 2003). However, it has also been reported that CGG KI mice with 70 CGG repeats, and CGG KI mice with CGG expansions above 200 show few intranuclear inclusions, suggesting that an upper and lower threshold for CGG repeat length and for inclusion formation may exist (Brouwer et al., 2009).
It is also possible that intranuclear inclusions in FXTAS may not be causally involved in pathology. They may even serve a protective function, possibly sequestering abnormal mRNAs and proteins, thereby protecting cells from pathology as has been suggested for polyglutamine diseases (Saudou et al., 1998). Other studies have shown that inhibition of ubiquination of huntingtin fragments and/or reduction of mutant huntingtin results in reduced nuclear inclusion formation which, in turn, significantly decreases the risk of cell death (Arrasate et al., 2004). Clarification of the involvement and role of intranuclear inclusions in FXTAS and other neurodegenerative diseases is central to elucidation of disease pathogenesis (Woulfe, 2007).
Similar to FXTAS patients (Hagerman and Hagerman, 2004), CGG KI mice exhibit elevated Fmr1 mRNA expression and reduced Fmrp levels (Brouwer et al., 2008). However, increased Fmr1 mRNA levels do not appear to correlate in any simple way with the length of the CGG repeat in these mice (Brouwer et al., 2008; Entezam et al., 2007), and the formation of inclusions may not require elevated Fmr1 mRNA since inclusions occur in Purkinje cells of mice with ectopic expression of just an expanded CGG trinucleotide repeat (Hashem, et al., 2009). These mice also showed a loss of Purkinje cells, other Purkinje cell pathology (swollen axons), and motor deficits on the rotarod test. These reports are in contrast to the positive correlation between FMR1 mRNA and CGG repeat length reported in human premutation carriers (Brouwer et al., 2008; Brouwer et al., 2007; Entezam et al., 2007; Greco et al., 2006; Hessl et al., 2005; Kenneson et al., 2001; Primerano et al., 2002).
Intranuclear inclusions and neuropathology
The absence of FXTAS in patients with FXS, where the FMR1 gene is silent and FMR1 mRNA is absent, has lead to the proposal that FXTAS may be due to an RNA gain-of-function toxicity resulting from increased FMRP mRNA levels (Greco et al., 2002; Hagerman and Hagerman, 2004; Hagerman et al., 2001; Jacquemont et al., 2003; Tassone et al., 2004; Tassone et al., 2000). Toxic RNA gain-of-function mechanisms have been proposed for other neurodegenerative diseases, including myotonic dystrophy and Huntington’s disease-like 2 (reviewed in (Ranum and Cooper, 2006; Woulfe, 2007)). More than 20 inclusion-associated proteins have been identified, including components of the proteasome, HSP27 and 70, αβ-crystallin, DNA-repair-ubiquitin-associated HR23B, MBP, members of the histone 2A family, and the RNA binding protein hnRNP (Bergink et al., 2006; Iwahashi et al., 2006). The presence of RNA binding proteins suggests that depletion of such proteins when bound to RNA could mediate the proposed toxic RNA gain-of-function mechanism for disease pathogenesis in FXTAS (Hagerman et al., 2001). This is supported by the demonstration that intranuclear inclusions can be formed in both primary neural progenitor cells and established neural cell lines engineered to contain an expanded CGG88 trinucleotide repeat in the FMR1 5’ untranslated region (UTR) (Arocena et al., 2005). The inclusions were αβ-crystallin-positive, although not associated with ubiquitin, suggesting that the incorporation of ubiquinated proteins into inclusions may be a later event in inclusion formation. Furthermore, the lamin A/C in the nuclear envelope appeared to become irregular and accumulate in the inclusions as well, demonstrating additional nuclear pathology.
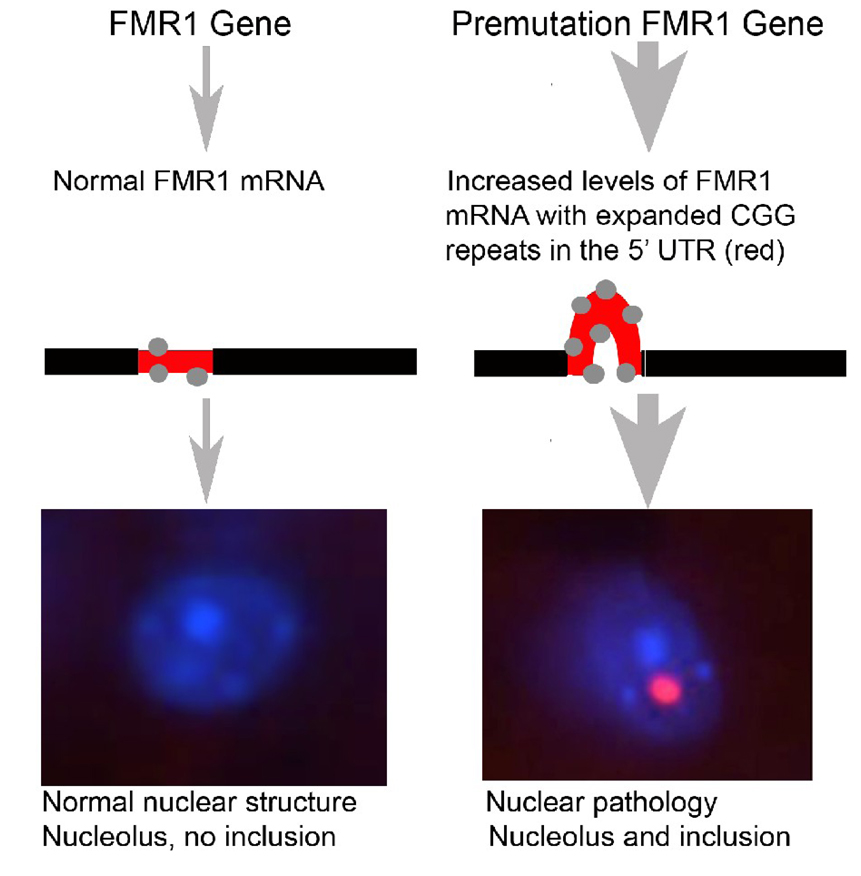
One model for inclusion formation in FXTAS is shown in Figure 4. In this model, transcription of normal FMR1 results in normal levels of FMR1 mRNA to which RNA binding proteins bind in the 5’ UTR (region shown in red), and no intranuclear inclusions are formed. In contrast, the presence of an expanded CGG repeat in the premutation FMR1 gene (e.g., carrying an expanded CGG repeat in the range of 55–200) leads to 3–8 fold higher FMR1 transcription levels, a corresponding increase in binding of RNA binding proteins to the expanded 5’ UTR region, and the development of intranuclear inclusions that contain FMR1 mRNA and several RNA binding proteins. However, the role of any specific binding protein in FXTAS pathology remains unclear as no single protein found in inclusions has yet to be shown to be dominant among the more than 20 proteins found (Iwahashi et al., 2006).
Figure 4.
Proposed model of inclusion formation in the fragile X permutation. Left: Normal FMR1 gene and transcribed FMR1 mRNA have between 5–40 CGG repeats in the 5’ untranslated region (UTR—short red segment on the mRNA). Proteins that bind to RNA, including CGG binding proteins, bind normally to the mRNA (gray dots) and no inclusions are formed (bottom left). Right: In the premutation condition FMR1 and mRNA have between 55–200 CGG repeats in the 5’UTR (long red segment on the mRNA). This results in increased levels of mutant FMR1 mRNA as well as excessive binding of RNA binding proteins to the mRNA (gray dots). Although the precise mechanism remains unclear, increased mutant FMR1 mRNA results in intranuclear inclusions that contain CGG binding proteins, ubiquitin, as well as the FMR1 mRNA itself (bottom right—pink inclusion body adjacent to the pale blue nucleolus).
In the current study intranuclear inclusions in neurons far outnumbered inclusions found in GFAP-labeled astrocytes. Currently, there is no clear explanation for this difference in the inclusion numbers between neurons and glial cells. There could be differences between neurons and glia in expression levels of Fmr1 mRNA, Fmrp, or specific RNA-associated proteins that may explain these differences, but such differences have not been demonstrated. Other neurological disorders, such as Huntington’s disease, also include the formation of prominent inclusions in neuronal nuclei that are associated with misfolded proteins. Aggregates of mutant huntingtin protein are found in neurons and in glia, although more prominently in neurons (Tydlacka et al., 2008). These huntingtin aggregates co-localize with components of the ubiquitin-proteasome system (UPS) which is involved in clearing misfolded proteins (Lehman, 2009). The observation in Huntington’s disease that UPS activity is lower in neurons than in astroglial cells, and also lower in the nucleus than the cytoplasm, suggests that lower neuronal UPS activity may be responsible for preferential accumulation of misfolded proteins in neurons, as well as for their selective vulnerability to subsequent cell death. In contrast, higher UPS activity in astroglial cells resulted in less severe glia pathology (i.e., less/or no nuclear inclusions and reduced huntingtin aggregates (Tydlacka et al., 2008)). Based on these findings it is tempting to speculate that there may be a similar relationship between UPS activity and inclusion formation in neurons and glial cells of CGG KI mice resulting in fewer inclusions in glial cells than in neurons. However, further research is needed to establish any relationship between UPS activity and inclusion formation in neuronal and or glial cells in the CCG KI mouse model of FXTAS.
Conclusions
The CGG KI mouse model exhibits many of the important clinical and neuropathological features of FXTAS in humans (Berman and Willemsen, 2009). In particular, the CGG KI mouse model develops ubiquitin-positive intranuclear inclusions in both astrocytes and neurons. The role of intranuclear inclusions in the pathophysiology of FXTAS remains unclear, and continued studies on the CGG KI mouse as a model of FXTAS should contribute to finding answers to these questions. Such studies will not only contribute important information to the study and treatment options for FXTAS and other trinucleotide repeat disorders, but also to other neurodegenerative disorders such as Alzheimer's and Parkinson's disease that are increasingly prevalent in an aging population.
4. Experimental Procedure
Animals
The generation of an expanded CGG trinucleotide repeat knock-in (CGG KI) mouse model of the fragile X premutation has been described previously in detail (Bontekoe et al., 1997; Willemsen et al., 2003). Briefly, the native CGG8 trinucleotide repeat of human origin in the 5’ UTR of the mouse Fmr1 gene was replaced by a human CGG98 trinucleotide repeat via homologous recombination. Across breedings, the CGG repeats showed mild instability, both expanding and contracting in length within the fragile X premutation range, which is defined as ~55–200 CGG repeats (Brouwer et al., 2009; Willemsen et al., 2003). The CGG KI mice were originally developed on a mixed FVB/N×C57BL/6J background, and were backcrossed with C57BL/6J mice from Jackson Laboratories (Bar Harbor, ME) until greater than 98% C57BL/6J by microsatellite analysis. Five male wildtype mice between 22–62 weeks old and 8 male CGG KI mice 16–76 weeks of age were used in the current study. The CGG KI mice used in the present study had between 128 and 198 CGG repeats. Brains of two female heterozygous CGG KI mice (70 and 75 weeks old and with 10/150 and 10/162 CGG repeats, respectively) as well as two wildtype female mice were examined for presence of intranuclear inclusions in neurons and astrocytes for comparison with males. Wildtype and CGG KI mice were housed under the same conditions of constant temperature and a 12 / 12 h light-dark cycle, and with food and water ad libitum. All experiments were conducted in compliance with the NIH Guide for Care and Use of Laboratory Animals, and were approved by the Institutional Animal Care and Use Committee of the University of California at Davis.
Genotyping
DNA was extracted from mouse tails by incubating with 10 mg/ml Proteinase K (Roche Diagnostics) in 300 µl lysis buffer containing 50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 150 mM NaCl, 1% SDS overnight at 55°C. One hundred µl saturated NaCl was then added and the suspension was centrifuged. One volume of 100% ethanol was added, gently mixed, and the DNA was pelleted by centrifugation and the supernatant discarded. The DNA was washed and centrifuged in 500 µl 70% ethanol. The DNA was then dissolved in 100 µl milliQ-H20. CGG repeat lengths were determined by PCR using the Expanded High Fidelity Plus PCR System (Roche Diagnostics). Briefly, approximately 500–700 ng of DNA was added to 50 µl of PCR mixture containing 2.0 µM of each primer, 250 µM of each dNTP (Invitrogen), 2% DMSO (Sigma), 2.5 M Betaine (Sigma), 5 U Expand HF buffer with Mg (7.5 µM). The forward primer was 5’-GCTCAGCTCCGTTTCGGTTTCACTTCCGGT-3’ and the reverse primer was 5’-AGCCCCGCACTTCCACCACCAGCTCCTCCA-3’. PCR steps were 10 min denaturation at 95°C, followed by 34 cycles of 1 min denaturation at 95°C, annealing for 1 min at 65°C, and elongation for 5 min at 75°C to end each cycle. PCR ends with a final elongation step of 10 min at 75°C. DNA CGG band sizes were determined by running DNA samples on a 2.5% agarose gel and staining DNA with ethidium bromide.
Case Report: Autopsy, Clinical History and Neuropathology
FXTAS neuropathology from a previously described human case is described in this study for comparison with that found in the CGG KI mice (case 2 from (Greco et al., 2002)). The FXTAS patient was a carrier of an FMR1 premutation allele of 80 CGG repeats, developed clinical features of FXTAS at 65 years of age, and died at 78 years of age. Description of the clinical history of this case and detailed neuropathology (e.g., standard techniques, including fixation, hematoxylin and eosin (H&E) staining, immunocytochemistry and neurohistological analysis) have been already reported and as described by (Greco et al., 2002) (case 2). Brain autopsy of this case was performed in a standard fashion and in accordance with University of California, Davis, IRB-approved protocols.
Tissue Preparation for Light Microscopy/Immunocytochemistry
General tissue preparation
Mice were anesthetized with sodium pentobarbital (100 mg/kg, i.p.), then perfused with isotonic saline with heparin (1000 units/ml saline), followed by a solution of 4% paraformaldehyde (PFA) in 0.1 M sodium phosphate buffer (PB; pH 7.4). The brains were immediately removed from skull and placed in the same fixative for 1 hour at 4°C. After post-fixation, the brains were rinsed in PB, cryoprotected in 10% sucrose in 0.1 M PB for 1 hour, followed by 30% sucrose in 0.1 M PB for 24 hours at 4°C, then rapidly frozen on dry ice. Thirty µm parasagittal serial sections were cut on a sliding microtome equipped with a freezing stage, and collected into series of every fifth section directly into 30% sucrose. Single sets of sections were selected for further processing which included: cresyl violet and/or H&E staining for general histological evaluation; immunocytochemistry for neuronal and glial cell markers and for ubiquitin to visualize intranuclear inclusions, the hallmark pathology of FXTAS patients that are also features of CGG KI mice (Greco et al., 2006; Greco et al., 2002; Willemsen et al., 2003).
Immunocytochemistry
Immunocytochemical and immunofluorescence techniques were used to visualize the occurrence and distribution of intranuclear and cytoplasmic inclusions, with a focus on their presence or absence in glial cells (i.e., astrocytes, including cerebellar Bergmann glia, oligodendrocytes, and microglial cells), as well as in GABA-ergic interneurons neurons of wild-type and CGG KI mice. Subsets of alternate sections were processed for immunocytochemistry using a modification of the avidin-biotin complex (ABC)-peroxidase technique (Hsu et al., 1981) as previously described (Wenzel et al., 2004). Briefly, free-floating sections were rinsed in PB (pH 7.4), and pretreated with 0.1% sodium borohydride for antigen retrieval for 15 min followed by treatment with 0.5–2% H2O2 in PB for 90 min to inactivate endogenous peroxidases. Sections were then treated with 3% goat, horse, or swine serum (Sigma, St.Louis, MO; DAKO, Inc., Carpinteria, CA) and 0.3% Triton X (TX) in 0.01 M PB, 0.15M NaCl, pH 7.4 (PBS) for 1 hour to reduce nonspecific staining. Sections were rinsed in PBS for 30 minutes and incubated for 48–72 hours at 4° C in the various antibodies and dilutions: mouse monoclonal anti-glutamic acid decarboxylase (GAD67), (Chemicon, Temecula, CA), 1:1000; mouse monoclonal anti-glial fibrillary acidic protein (GFAP), (DAKO, Inc.), 1:2000 (1:750 for immunofluorescence (IF)); rabbit polyclonal anti-S100β (Abcam, Inc., Cambridge, MA), 1:1000; mouse monoclonal anti-myelin basic protein (MBP), (Chemicon), 1:500; rabbit polyclonal anti-Iba1 (ionized calcium binding adaptor molecule 1; Wako Chemicals USA, Inc., Richmond, VA), 1:2000 (1:1000 for IF); mouse-monoclonal anti-Kv2.1 (provided by Dr. J.S. Trimmer; UC Davis), 1:500 for IF; and rabbit poly- and mouse monoclonal antibodies against ubiqutin (DAKO, Inc.; Abcam, Inc.) 1:2000 and 1:1000, respectively, (1:1000 for IF) in PBS containing 1% goat, horse or swine serum, 2% BSA and 0.3% TX. Following rinses for 2 hours in PBS, sections were incubated in biotinylated goat or swine anti-rabbit IgG or horse anti-mouse IgG (DAKO, Inc.; Vector Laboratories, Burlingame, CA), diluted 1:500 for 24 hours at 4°C, rinsed 2 hours in PBS and then incubated in ABC (Elite ABC Kit, Vector Laboratories), diluted 1:500 in 1% goat or horse serum, 2% BSA, 0.3% TX and PBS for 24 hours at 4°C. Sections were rinsed thoroughly in PB (pH 7.4), then transferred to Tris-HCl buffers (pH 7.4; 7.6) and then incubated for 15 minutes in 0.025% 3,3’-diaminobenzidine (DAB, Sigma) in TB (pH7.6). After reacting for 5–10 minutes in fresh DAB with 0.003% H2O2, sections were rinsed in TB, followed by PB. Iba1-immunostaining was visualized using 0.05% DAB with a blue/grey chromogen (DAB; Vector SG Substrate Kit, Vector Laboratories). Double-immunostaining was performed for GFAP/ubiquitin, as well as for MBP/ubiquitin co-localization using combined incubation of poly- and monoclonal antisera and differently-colored chromogens (Vector SG Substrate Kits) resulting in a blue staining for GFAP-positive cells and brown reaction product for ubiquitin-positivity of inclusions. Specificity of the immunostaining was evaluated by omitting primary antibodies from the regular staining. Sections were mounted on slides, dehydrated, cleared, and coverslipped with Permount.
Immunofluorescence staining
For single and double-immunofluorescent labeling of ubiquitin colocalized with neuronal/glial cell markers, sections were transferred into 10% sucrose in 0.1 M PB, then rinsed in 0.1 M PB and treated with 0.1% sodium borohydride for 15 min. Thereafter, sections were rinsed again with 0.1 M PB and then permeabilized with 0.5% H2O2 in 0.1 M PB for 15 min followed by rinses in 0.1 M PB and 0.01 M PBS. Free-floating sections were treated with 10% goat or horse serum in 0.01M PBS containing 0.3% TX-100 (vehicle) for 1 hr, and then incubated overnight at 4°C in vehicle containing different combinations of mouse monoclonal/rabbit polyclonal antibodies of different IgG isotypes (see above). After rinses in 0.01 M PBS and 10% goat or horse serum (vehicle), sections were incubated in isotype-specific Alexa-conjugated secondary antibodies (1:2000): Alexa 568-labeled goat anti-rabbit IgG and/or Alexa 488-labeled goat anti-mouse IgG (Invitrogen, Carlsbad, CA) for 1–2 hrs as described previously (Wenzel et al., 2007). Following rinses in vehicle, sections were mounted on gelatin-coated slides and coverslipped with mounting medium containing DAPI (4’, 6-diamidino-2-phenyindole di-lactate) for nuclear staining (Vectashield “Hard Set”, Vector Laboratories).
To differentiate between specific immunofluorescent labeling and nonspecific autofluorescence resulting from accumulations of lipofuscin in aging brains, sections were first treated with 10 mM CuSO4 in 50 mM ammonium acetate buffer (pH 5.0) for 20 min (Schnell et al., 1999). This treatment reduced autofluorescence but did not significantly affect the intensity of specific immunofluorescent labeling; although immunofluorescent staining for ubiquitin within the neuronal cytoplasm was slightly enhanced.
Golgi-Cox staining
The Bergmann glia in the CGG KI mouse cerebellum, including cell bodies and processes, were visualized using a modified Golgi-Cox staining procedure (FD Rapid GolgiStain Kit; FD NeuroTechnologies, Inc. Ellicot City, MD). After initial perfusion with NaCl, the brains were stained following the kit instructions, and sectioned at 120 µm on a vibratome, mounted on slides and stained, then cleared and coverslipped as described above.
Cell Identification and Evaluation of Intranuclear Inclusions
The sections were analyzed using a Nikon ECLIPSE E600 microscope with epifluorescence attachment and digital camera. Images were converted to a file format for processing as an Adobe Photoshop document. Images were analyzed to verify the presence of ubiquitin-positive intranuclear inclusions in different cell types identified with various neuronal and glial cell markers in brains of CGG KI and wildtype mice. Cresyl violet and/or H&E-stained sets of serial sections from wildtype and CGG KI mouse brains at different ages were used for comparison and evaluation of gross anatomical differences. Identification of different cell types in the brain was carried out based on standard morphological criteria, using Nissl cell staining, and neuronal and glial cell markers (Greco et al., 2006; McKhann et al., 2003). Neurons were identified by their size, large round nuclei, single or multiple nucleoli, and their abundant cytoplasm, as well as by using specific neuronal markers (e.g., GAD67, Kv2.1 channel protein). Astrocytes were identified by their round/ovoid nuclei with light euchromatin, and absence of nucleoli and cytoplasm. In addition, GFAP- and S100β-immunocytochemistry and/or immunofluorescence were used to identify subpopulations of astrocytes based on their differing immunoreactivities (i.e, protoplasmic and/or fibrous astrocytes) in different brain regions. To estimate the percentage of astrocytes with intranuclear inclusions, the number of astrocytes and intranuclear inclusions were counted manually in 10 fields on two GFAP- and ubiquitin-immunoreacted sections from 3 CGG KI mice within layer I of the neocortex (including sensory and motor areas) at 400× magnification using a microscope and optical imaging system. Oligodendrocytes were identified based on their typically small, round, hyperchromatic nuclei (which did not allow differentiation between nucleus, nucleoli, or cytoplasm), localization in white and grey matter, and confirmed by immunocytochemistry for MBP. Microglia were identified primarily on cellular morphology obtained from Iba1 immunostaining which displayed small cell bodies with a round nucleus and fine, ramified processes that are characteristic of resting microglia. Microglial cells with retracted and/or hypertrophic processes were defined as activated (but non-phagocytic) microglia which can be transformed into phagocytotic cells (i.e., brain macrophages) (Graeber and Moran, 2002). Immunocytochemical staining for ubiquitin was used to specifically label intranuclear inclusions in combination with cell-specific markers to identify the cell type (i.e., neuronal and/or non-neuronal cells). Whenever possible, experiments using immunocytochemistry using DAB and immunofluorescence were carried out in parallel to verify staining patterns as well as to allow for future quantification.
Acknowledgments
We are grateful to Dr. Mareike Wenzel for her excellence in technical support. This research was supported by grants NINDS RL1 NS 062411, NIH Roadmap Consortium NIDCR UL1 DE 19583, and NIA AG032119.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare that they have no conflict of interest.
REFERENCES
- Arocena DG, Iwahashi CK, Won N, Beilina A, Ludwig AL, Tassone F, Schwartz PH, Hagerman PJ. Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum Mol Genet. 2005;14:3661–3671. doi: 10.1093/hmg/ddi394. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Bergink S, Severijnen LA, Wijgers N, Sugasawa K, Yousaf H, Kros JM, van Swieten J, Oostra BA, Hoeijmakers JH, Vermeulen W, Willemsen R. The DNA repair-ubiquitin-associated HR23 proteins are constituents of neuronal inclusions in specific neurodegenerative disorders without hampering DNA repair. Neurobiol Dis. 2006;23:708–716. doi: 10.1016/j.nbd.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Berman RF, Willemsen R. Mouse Models of Fragile X-Associated Tremor Ataxia. J Investig Med. 2009 doi: 10.231/JIM.0b013e3181af59d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis E, Abrams L, Coffey SM, Hall DA, Greco C, Gane LW, Grigsby J, Bourgeois JA, Finucane B, Jacquemont S, Brunberg JA, Zhang L, Lin J, Tassone F, Hagerman PJ, Hagerman RJ, Leehey MA. Fragile X-associated tremor/ataxia syndrome: clinical features, genetics, and testing guidelines. Mov Disord. 2007;22:2018–2030. doi: 10.1002/mds.21493. quiz 2140. [DOI] [PubMed] [Google Scholar]
- Bontekoe CJ, de Graaff E, Nieuwenhuizen IM, Willemsen R, Oostra BA. FMR1 premutation allele (CGG)81 is stable in mice. Eur J Hum Genet. 1997;5:293–298. [PubMed] [Google Scholar]
- Brouwer JR, Huizer K, Severijnen LA, Hukema RK, Berman RF, Oostra BA, Willemsen R. CGG-repeat length and neuropathological and molecular correlates in a mouse model for fragile X-associated tremor/ataxia syndrome. J Neurochem. 2008;107:1671–1682. doi: 10.1111/j.1471-4159.2008.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer JR, Mientjes EJ, Bakker CE, Nieuwenhuizen IM, Severijnen LA, Van der Linde HC, Nelson DL, Oostra BA, Willemsen R. Elevated Fmr1 mRNA levels and reduced protein expression in a mouse model with an unmethylated Fragile X full mutation. Exp Cell Res. 2007;313:244–253. doi: 10.1016/j.yexcr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer JR, Willemsen R, Oostra BA. The FMR1 gene and fragile X-associated tremor/ataxia syndrome. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:782–798. doi: 10.1002/ajmg.b.30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunberg JA, Jacquemont S, Hagerman RJ, Berry-Kravis EM, Grigsby J, Leehey MA, Tassone F, Brown WT, Greco CM, Hagerman PJ. Fragile X premutation carriers: characteristic MR imaging findings of adult male patients with progressive cerebellar and cognitive dysfunction. AJNR Am J Neuroradiol. 2002;23:1757–1766. [PMC free article] [PubMed] [Google Scholar]
- Entezam A, Biacsi R, Orrison B, Saha T, Hoffman GE, Grabczyk E, Nussbaum RL, Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber MB, Moran LB. Mechanisms of cell death in neurodegenerative diseases: fashion, fiction, and facts. Brain Pathol. 2002;12:385–390. doi: 10.1111/j.1750-3639.2002.tb00452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, Trapp BD, Iwahashi C, Brunberg J, Grigsby J, Hessl D, Becker EJ, Papazian J, Leehey MA, Hagerman RJ, Hagerman PJ. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- Greco CM, Hagerman RJ, Tassone F, Chudley AE, Del Bigio MR, Jacquemont S, Leehey M, Hagerman PJ. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- Hagerman PJ, Hagerman RJ. Fragile X-associated tremor/ataxia syndrome (FXTAS) Ment Retard Dev Disabil Res Rev. 2004;10:25–30. doi: 10.1002/mrdd.20005. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Coffey SM, Maselli R, Soontarapornchai K, Brunberg JA, Leehey MA, Zhang L, Gane LW, Fenton-Farrell G, Tassone F, Hagerman PJ. Neuropathy as a presenting feature in fragile X-associated tremor/ataxia syndrome. Am J Med Genet A. 2007;143A:2256–2260. doi: 10.1002/ajmg.a.31920. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- Hessl D, Tassone F, Loesch DZ, Berry-Kravis E, Leehey MA, Gane LW, Barbato I, Rice C, Gould E, Hall DA, Grigsby J, Wegelin JA, Harris S, Lewin F, Weinberg D, Hagerman PJ, Hagerman RJ. Abnormal elevation of FMR1 mRNA is associated with psychological symptoms in individuals with the fragile X premutation. Am J Med Genet B Neuropsychiatr Genet. 2005;139:115–121. doi: 10.1002/ajmg.b.30241. [DOI] [PubMed] [Google Scholar]
- Hsu SM, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem. 1981;29:577–580. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- Hunsaker MR, Wenzel HJ, Willemsen R, Berman RF. Progressive spatial processing deficits in a mouse model of the fragile X premutation. Behavioral Neuroscience. 2009 doi: 10.1037/a0017616. (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Leehey M, Grigsby J, Zhang L, Brunberg JA, Greco C, Des Portes V, Jardini T, Levine R, Berry-Kravis E, Brown WT, Schaeffer S, Kissel J, Tassone F, Hagerman PJ. Fragile X premutation tremor/ataxia syndrome: molecular, clinical, and neuroimaging correlates. Am J Hum Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- Lehman NL. The ubiquitin proteasome system in neuropathology. Acta Neuropathol. 2009;118:329–347. doi: 10.1007/s00401-009-0560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann GM, 2nd, Wenzel HJ, Robbins CA, Sosunov AA, Schwartzkroin PA. Mouse strain differences in kainic acid sensitivity, seizure behavior, mortality, and hippocampal pathology. Neuroscience. 2003;122:551–561. doi: 10.1016/s0306-4522(03)00562-1. [DOI] [PubMed] [Google Scholar]
- Oostra BA, Willemsen R. FMR1: a gene with three faces. Biochim Biophys Acta. 2009;1790:467–477. doi: 10.1016/j.bbagen.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL, Chan-Palay V. Cytology and Organization. Berlin, Heidelberg, New York: Springer Verlag; 1974. Cerebellar Cortex. [Google Scholar]
- Primerano B, Tassone F, Hagerman RJ, Hagerman P, Amaldi F, Bagni C. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. Rna. 2002;8:1482–1488. [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Schnell SA, Staines WA, Wessendorf MW. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem. 1999;47:719–730. doi: 10.1177/002215549904700601. [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Garcia-Arocena D, Khandjian EW, Greco CM, Hagerman PJ. Intranuclear inclusions in neural cells with premutation alleles in fragile X associated tremor/ataxia syndrome. J Med Genet. 2004;41:e43. doi: 10.1136/jmg.2003.012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tydlacka S, Wang CE, Wang X, Li S, Li XJ. Differential activities of the ubiquitin-proteasome system in neurons versus glia may account for the preferential accumulation of misfolded proteins in neurons. J Neurosci. 2008;28:13285–13295. doi: 10.1523/JNEUROSCI.4393-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam D, Errijgers V, Kooy RF, Willemsen R, Mientjes E, Oostra BA, De Deyn PP. Cognitive decline, neuromotor and behavioural disturbances in a mouse model for fragile-X-associated tremor/ataxia syndrome (FXTAS) Behav Brain Res. 2005;162:233–239. doi: 10.1016/j.bbr.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Wenzel HJ, Patel LS, Robbins CA, Emmi A, Yeung RS, Schwartzkroin PA. Morphology of cerebral lesions in the Eker rat model of tuberous sclerosis. Acta Neuropathol. 2004;108:97–108. doi: 10.1007/s00401-004-0865-8. [DOI] [PubMed] [Google Scholar]
- Wenzel HJ, Vacher H, Clark E, Trimmer JS, Lee AL, Sapolsky RM, Tempel BL, Schwartzkroin PA. Structural consequences of Kcna1 gene deletion and transfer in the mouse hippocampus. Epilepsia. 2007;48:2023–2046. doi: 10.1111/j.1528-1167.2007.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R, Hoogeveen-Westerveld M, Reis S, Holstege J, Severijnen LA, Nieuwenhuizen IM, Schrier M, van Unen L, Tassone F, Hoogeveen AT, Hagerman PJ, Mientjes EJ, Oostra BA. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum Mol Genet. 2003;12:949–959. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- Woulfe JM. Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol Appl Neurobiol. 2007;33:2–42. doi: 10.1111/j.1365-2990.2006.00819.x. [DOI] [PubMed] [Google Scholar]