Abstract
TRC8/RNF139 encodes an ER-resident E3-ubiquitin ligase that inhibits growth in a RING- and ubiquitylation-dependent manner. TRC8 also contains a predicted sterol-sensing domain. Here we report that TRC8 protein levels are sterol-responsive, and that it binds and stimulates ubiquitylation of the ER-anchor protein, INSIG. Induction of TRC8 destabilized the precursor forms of the transcription factors, SREBP-1 and SREBP-2. Loss of SREBP precursors was proteasome-dependent, required a functional RING domain, occurred without generating processed nuclear forms and suppressed SREBP target genes. TRC8 knockdown had opposite effects in sterol-deprived cells. In Drosophila, growth inhibition by DTrc8 was genetically suppressed by loss of specific MPN domain-containing proteins found in the COP9 signalosome and eIF3. DTrc8 genetically and physicially interacted with two eIF3 subunits, eIF3f and eIF3h. Co-immunoprecipitation experiments confirmed these interactions in mammalian cells and TRC8 over-expression suppressed polysome profiles. Moreover, high molecular weight ubiquitylated proteins were observed in eIF3 immunoprecipitations from TRC8 over-expressing cells. Thus, TRC8 function may provide a regulatory link between the lipid and protein biosynthetic pathways.
Keywords: RNF139, tumor suppressor, RING-H2 domain, ubiquitylation, SREBP, INSIG, cholesterol, clear-cell RCC, eIF3, Minute genes
Introduction
Renal cell carcinoma (RCC) comprises 5% of epithelial cancers with over 38,000 new cases in the United States each year (1). Most are clear-cell RCCs and of these, a majority contain mutations or epigentic silencing of VHL with up-regulation of HIFα subunits and a constitutively activated hypoxic response (2). While HIF deregulation is a major factor in RCC development, it does not explain all features of the disease (3). Similarly, loss of VHL function alone is insufficient for tumor formation (4). Besides VHL, additional genes have been implicated in hereditary renal cancer. These include fumarate hydratase and succinate dehydrogenase(5, 6), which affect HIFα prolyl hydroxylation and stability (7), the hepatocyte growth factor receptor, MET (8), as well as Tuberous Sclerosis (TSC1/2) and Birt-Hogg-Dubé/Folliculin (BHD/FLCN), which affect mTOR activity and protein translation (9–11). However the control of protein translation initiation involves not only mTOR, which regulates cap-mediated translation, but also other factors including eIF3, which regulates ribosome and mRNA assembly, as well as IRES-mediated translation (12).
We identified the TRC8/RNF139 gene from its disruption by a constitutional translocation, t(3;8)(3p14.2;q24.1), in a family with hereditary RCC and thyroid cancer (13–15). Recently, a second family with hereditary RCC carrying an independent (3;8) translocation affecting TRC8 was described (16). In addition to disruption of TRC8 in RCC by chromosomal translocation and tumor-specific loss of the wild-type allele, a rare sporadic mutation was also identified, consistent with a proposed tumor suppressor function (15, 16).
TRC8 encodes a multi-membrane spanning protein, located in the endoplasmic reticulum (ER), and contains a C-terminal RING-H2 domain with E3-ubiquitin ligase activity (15, 17–19). Recent work has identified MHC class I molecules as one target for TRC8-mediated ubiquitylation and degradation in cells expressing the CMV protein, US2 (20), but other targets remain unknown. In HEK293 cells, TRC8 overexpression inhibits growth and tumorigenesis in a RING-dependent manner (19). In Drosophila, expression of DTrc8 in the wing suppressed growth, while targeting expression to the eye caused disappearance of retinal cells (17, 21). A yeast 2-hybrid screen with DTrc8 identified Csn5/Jab1(17), the catalytic component of the COP9 signalosome, which acts to remove NEDD8 from E3-ubiquitin ligase complexes (22). Moreover, genetic interaction studies demonstrated that Csn5 loss-of-function mutations partially reversed the effect of DTrc8 overexpression in the wing (21). The MPN domain of Csn5 was specifically required for interaction with DTrc8. This domain is found in only a limited number of proteins, including subunits in the COP9 signalosome (Csn5, Csn6), the 19S regulatory lid of the proteasome (Rpn8, Rpn11) and eIF3 (eIF3f, eIF3h) (23). Like Csn5, crosses with mutant Csn6 flies relieved TRC8-induced growth suppression, while those with Rpn8 and Rpn11 had no effect. Interactions with the eIF3 subunits were not tested.
In addition to the RING domain, TRC8 contains a predicted sterol-sensing domain in its N-terminus, suggesting that sterols may influence some aspect of TRC8 function (15). Interestingly, the clear-cell phenotype in RCC is due to abundant inclusions of endogenously synthesized cholesterol esters and other lipids, suggesting that lipid homeostasis may be deregulated in this disease (24–26). Sterol-sensing domains (SSDs) are found in a limited number of proteins, some of which (e.g., HMGCR, SCAP) are directly involved in sterol biosynthesis (27). When sterol levels are high, the SSD of HMG CoA reductase (HMGCR) interacts with the ER-anchor protein, INSIG-1 (INSulin-Induced Gene). The ERAD-associated E3-ubiquitin ligase, gp78, binds INSIG-1 despite the lack of a sterol sensing domain (28). As a result of this complex formation, HMGCR is polyubiquitylated by gp78, leading to its proteasome-mediated destruction (28, 29). Patched, Dispatched and Neimann-Pick proteins also contain SSDs, although the function of this domain in these proteins is not well understood and no interactions with INSIG have been reported (30).
The transcriptional regulation of cholesterol and lipid biosynthetic enzymes is under the control of two ER-tethered transcription factors, SREBP-1 and -2. When sterol levels are high, the SREBP precursor proteins (preSREBPs) are retained in the ER in a preSREBP-SCAP-INSIG complex. In low sterols, the SCAP-INSIG interaction is lost and SREBP precursors are transported to the Golgi where they undergo release from the membrane by proteolytic cleavage and subsequently enter the nucleus (27). The SREBPs regulate their own transcription, and that of INSIG-1, as part of positive and negative feedback loops (31). In fission yeast, SREBP activation occurs in response to hypoxia (32), although a similar response has not been described in mammalian cells. However, INSIG-2 is hypoxia regulated (33). Given the apparent disruption of lipid homeostatis in clear-cell RCC, this raises the question of whether other regulatory components of the SREBP pathway might be affected in the disease process.
With the presence of a putative SSD in TRC8, we asked whether TRC8 was affected by low sterol levels. We report here that TRC8 protein is stabilized when sterols are reduced and destabilized when sterols are replete. This effect is RING-dependent and accompanied by sterol-stimulated ubiquitylation. TRC8 interacts with and stimulates ubiquitylation of INSIG. In addition, it destabilized preSREBPs and reduced expression of SREBP target genes. TRC8 knockdown had opposite effects in sterol-depleted cells. In addition, we found that TRC8 interacts with the MPN domain containing subunits of the translation initiation factor, eIF3, induced ubiquitylation of the complex and reduced protein translation as assessed by polysome loading. Together, these observations suggest that TRC8 function may provide a regulatory link between the lipid and protein biosynthetic pathways. Of note, the hereditary kidney cancer gene BHD/FLCN may perform a parallel function affecting AMP kinase and HMG CoA reductase (see Discussion).
Results
TRC8 protein stability is regulated by sterols
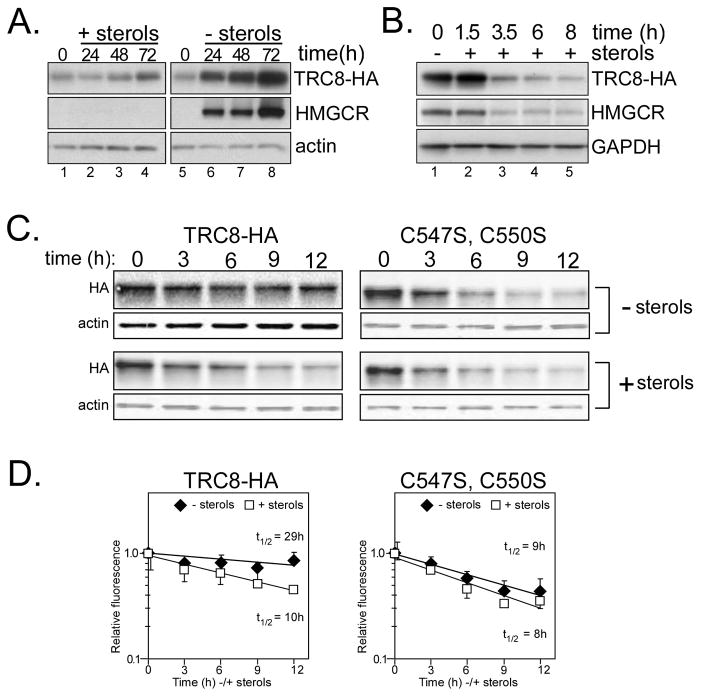
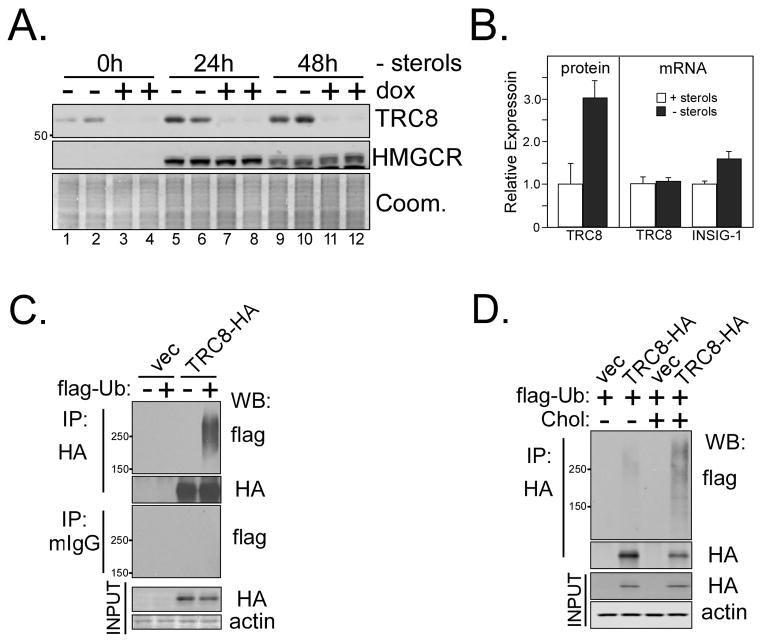
The presence of a putative sterol-sensing domain [Supplemental Figure S1, (15)] prompted us to determine if TRC8 was affected by sterols. We initially examined CHO cells stably transfected with an HA-tagged, tet-inducible TRC8 (TRC8-HA-FlpIn) (19), which is detectable at low levels in the absence of doxycycline (dox) because of leaky expression. When cells were grown in medium with 5% FCS containing abundant lipoproteins, TRC8 showed little change over time (Fig. 1A, lanes 1–4). However, under sterol-depleted conditions, there was substantial accumulation of both TRC8 (lanes 6–8) and the positive control, HMGCR (29). Following cholesterol depletion in HEK293 cells containing TRC8-HA, the addition of sterols caused disappearance of both TRC8 and HMGCR with similar kinetics (Fig. 1B). In the presence of cycloheximide (CHX), the decay of wild type TRC8 was substantially delayed in the absence of sterols (Fig 1C) yielding a t1/2 of 29h compared to 10h in the presence of sterols (Fig. 1D). Similar results were observed in CHO cells (Fig. S2). Analysis of endogenous TRC8 protein, detected in membrane fractions prepared from sterol-starved HEK293 cells, yielded similar results. Compared to sterol-replete cells (Fig. 2A, lanes 1,2), endogenous TRC8 accumulated when sterols were removed, as did the positive control, HMGCR (lanes 5, 6, 9, 10). Quantification of these results indicated ~3-fold more endogenous TRC8 in sterol-deprived cells after 24 h (Fig. 2B). Real-time RT-PCR indicated that sterol starvation did not change TRC8 mRNA levels, while increases were observed for the SREBP target gene, INSIG-1, as expected. Stable transfection of a tet- inducible knockdown construct (Fig. S3A–C) resulted in specific loss of TRC8 following dox treatment (Fig. 2A, lanes 3,4), and TRC8 in knockdown cells remained low following sterol depletion (lanes 7,8 and 11,12). Thus, endogenous TRC8 was increased by sterol-deprivation without concurrent effects on mRNA levels. We have not determined whether mutating the putative sterol-sensing domain affects these responses.
Figure 1. TRC8 levels are modulated by sterols.
(A) CHO FlpIn TRex cells containing dox-inducible TRC8-HA were cultured in the absence of dox with either abundant sterols (+ sterols; 5% FCS in F-12) or without (− sterols; 5% LPDS, 50 μM mevastatin, 50 μM mevalonate). The indicated time points were harvested, lysed and 10 μg aliquots analyzed by Western blot for TRC8-HA. Controls included HMGCR and actin. (B) HEK293 cells stably transfected with the TRC8-HA-FlpIn construct were cultured in sterol-depletion media for 18h to accumulate TRC8. Excess sterols were then added (+ sterols; 10μg/mL 25-HC and 50 μg/mL cholesterol) to all but the control (0h) in the absence of cycloheximide (CHX), and time points analyzed for decay of TRC8-HA and HMGCR; GAPDH indicates loading. (C) HEK293 cells containing wild type TRC8-HA or the RING mutant, C547S;C550S, were dox-induced for 24 h then sterol-starved for 1h in DMEM with 5% LPDS and 1% hydroxypropyl β-cyclodextrin (HPβCD). Media were then changed to DMEM with 5% LPDS containing either 10 μM statin and 50 μM mevalonate (sterols) or 50 μg/mL cholesterol/10 μg/mL 25-HC (+ sterols). CHX was then added to 10 μM and time points harvested at 3h intervals for analysis by Western blot using HA and actin antibodies. (D) Fluorescent band intensities from triplicate samples as in (C) were analyzed on an Odyssey IR fluorescence scanner and quantified. Points represent means, +/− s.d., from three replicates; gels and graphs are representative of 3 independent experiments.
Figure 2. Sterols regulate endogenous TRC8 and induce ubiquitylation.
(A) Membranes purified from HEK293 cells stably transfected with a tet-inducible shRNA targeting TRC8 permitted detection of endogenous TRC8 by Western blot (lanes 1, 2, −dox). Identity of the band was confirmed by 4 days of dox treatment (+dox) to induce knockdown. Parallel cultures were sterol-starved for 24 (lanes 5–8) or 48h (lanes 9–12) and membrane preparations analyzed for endogenous TRC8 and HMGCR (postive control). Coomassie staining verified equal loading. (B) Data for 24 h sterol starvation were densitized and quantified. Each bar represents the mean of TRC8 signal in triplicate samples, +/− s.d. RNA isolated from the same cultures was analyzed by realtime RT-PCR for endogenous TRC8 and INSIG-1 mRNAs. Delta Ct values were converted to fold expression and normalized to control samples (+sterols). (C) TRC8-HA or control (vec) HEK293 cells, grown in sterol-replete medium, were transfected with flag-tagged ubiquitin (1 μg), dox-induced and harvested 24 h later following MG132 addition (10 μM) during the final 2h. Lysates (200 μg aliquots) were immunoprecipitated with anti-HA or murine IgG (controls) and IP pellets analyzed on Western blots for flag-ubiquitin. HA immunoblots showed equal recovery of TRC8-HA; this and other HA blots were cropped to remove strong IgG background. Actin and HA blots verified equal input. (D) TRC8-HA and control cells were transfected with flag-ubiquitin and induced with dox as in (C). Cells were then sterol-starved for 15h followed by addition of cholesterol (12.5 μM) and 25-HC (10 μg/mL) for 3h to the indicated plates. MG132 was added 2h prior to harvest and lysates (200 μg aliquots) were immunoprecipated with anti- HA beads. Western blot analysis utilized the indicated antibodies.
Many E3 ubiquitin ligases are subject to auto-ubiquitylation, prompting us to examine TRC8-HA in FlpIn cells transfected with flag-tagged ubiquitin (flag-Ub). Anti-HA immunoprecipitations revealed high MW ubiquitin-conjugated products in cells expressing wild-type TRC8 (Fig. 2C). To determine if sterols influence the degree of TRC8 ubiquitylation, flag-Ub transfected cells were lipid-starved overnight then sterols were added. An increase in ubiquitylated TRC8 was observed in cells 3 h post sterol addition (Fig. 2D). Thus TRC8 is ubiquitylated in vivo. We then asked if a mutation in the RING domain, C547S, C550S, affected the response to sterols. These results are shown in Fig. 1C and D. Of note, although the sterol responsiveness was absent in the RING mutated protein, its half life was reduced to 8–9h regardless of sterols. This may be the result of protein instability due to mutation. Thus we cannot determine at the present time whether the effects of sterols on the half life of the wild type protein are RING-dependent.
TRC8 co-precipitates INSIGs and stimulates ubiquitylation
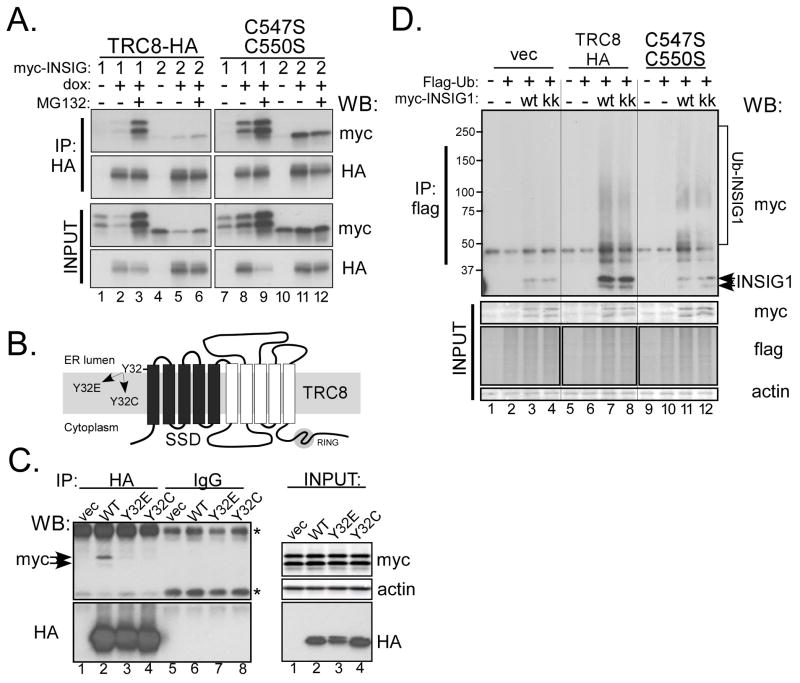
Some proteins with sterol-sensing domains, such as SCAP and HMGCR, interact with the ER-anchor proteins, INSIG-1/2 (27, 34, 35). To explore this, we transfected TRC8-HA-FlpIn cells with myc-tagged INSIG-1 or -2 and performed anti-HA (Fig. 3A) or reciprocal anti-myc immunoprecipitations (Fig. S4A). In the absence of TRC8 induction, neither INSIG-1 nor -2 were detectable in the IP pellets (Fig. 3A, lanes 1,4). However, with the addition of dox, both INSIG-1 and 2 were co-precipitated with TRC8 (lanes 2, 5). This interaction was observed regardless of the sterol status in HEK293 cells (Fig. S4B), and was not affected by mutation of INSIG residues K156R and K158R (Fig. S4A, kk), which abrogate INSIG-1 ubiquitylation by gp78 (37, 38). Interestingly, the TRC8 RING mutant increased levels of INSIG-1/2, both in lysates and IP pellets (Fig. 3A, lanes 8,11). Proteasome inhibition with MG132 increased the level of INSIG-1 in both lysates and IP pellets, while INSIG-2 levels were only marginally affected. INSIG binds SCAP and HMGCR through their respective SSDs, and this interaction is dependent upon a tyrosine residue within the conserved YIYF motif present in the first transmembrane (TM) segment (27, 39). The presence of a similar motif and conserved tyrosine (Y32) in the first TM of TRC8 (Fig. 3B and S1) prompted us to test appropriate mutants for INSIG interaction. Compared to the wild type control, Y32 mutations reduced TRC8’s ability to co-precipitate INSIG-1 (Fig. 3C). These results are consistent with INSIG-1 binding being mediated, at least in part, by TRC8’s SSD.
Figure 3. TRC8 binds and ubiquitylates INSIG.
(A) TRC8-HA-FlpIn cells (wt or C547S;C550S RING mutant), cultured in sterol-replete DMEM (10% FCS), were transfected overnight with 300ng of pCMV-INSIG-1-myc or pCMV-INSIG-2-myc, then dox-treated or not for 24 h. Where indicated, MG132 was added 4h prior to harvest. CHAPSO lysates (500μg) were immunoprecipitated with anti-HA and pellets analyzed by Western blot for INSIG (myc) and TRC8-HA (HA). (B) Diagram of TRC8 showing 10 TM segments (bars), the first five of which comprise the SSD (black bars). TRC8 also contains a RING-H2 domain in the C-terminus (shaded circle). The conserved tyrosine 32, in the first TM of the SSD, was mutated to glutamate or cysteine and used for the IP experiments in panel C. (C) Control HEK293 cells (vec) and stable transfectants expressing wild type or SSD mutant TRC8 (Y32E or Y32C) were transfected with INSIG-1-myc and harvested after 48h. TritonX-100 lysates containing 400ug of protein were immunoprecipitated using anti-HA or control beads. IP pellets were analyzed for co-precipitated INSIG-1 (myc) and TRC8 (HA). Input blots verified equal expression; asterisks indicate background IgG bands. (D) Control cells (vec) and stable transfectants expressing wild type or RING mutant TRC8 (C547S, C550S) were co-transfected with flag-ubiquitin and myc-INSIG-1 (wt) or the K156R, K158R mutant (kk), as indicated. Cells were dox-induced for 24 h and treated with MG132 for 2h prior to harvest. TritonX-100 lysates (200 μg aliquots) were immunoprecipitated with anti-flag beads and analyzed on Western blots with the indicated antibodies. Input blots verified equal expression; conjugated ubiquitin, detected with anti-flag antibodies, was used to verify equal expression because unconjugated ubiquitin was not visible.
Next, we co-transfected myc-INSIG-1 and flag-ubiquitin into TRC8-FlpIn cells, then induced TRC8 for 24 h and performed immunoprecipitations using anti-flag beads. We observed increased INSIG-1 in the IP pellet in wild-type TRC8 expressing cells accompanied by high molecular weight, Ub-conjugated INSIG-1(Fig 3B). In contrast, there was a marked decrease of ubiquitylated INSIG-1 in cells expressing the TRC8 RING mutant (lanes 11, 12), although some high molecular weight forms were detected. The INSIG-1-K156, 158R mutant was immunoprecipitated from wild type TRC8-expressing cells at levels comparable to normal INSIG-1 (lane 8), suggesting lysines other than K156 and K158 may be targeted for ubiquitylation by TRC8. Thus, we conclude that INSIG-1 and 2 physically interact with TRC8, and that TRC8 enhances ubiquitylation of INSIG-1 in a RING-dependent manner. We have not determined whether these effects also involve gp78.
Ectopic TRC8 destabilizes the precursor forms of SREBP-1/2 in a proteasome-dependent manner
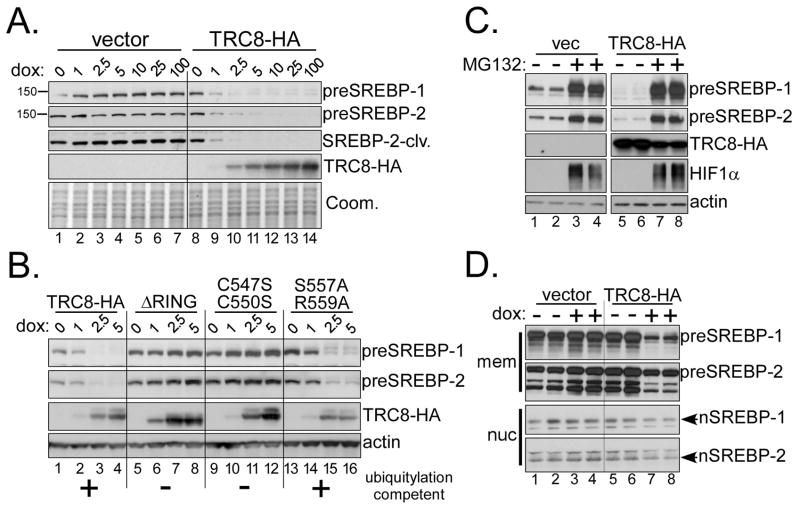
In the presence of sterols, INSIG forms a complex in the ER with SCAP and the SREBP precursor proteins (27). To determine if TRC8 affected preSREBPs, we analyzed total lysates before and after TRC8-HA induction (Fig. 4A). Increasing amounts of dox-induced TRC8 led to a dose-dependent reduction of both preSREBP-1 and -2. In control cells, dox treatment had no effect. A time-course experiment demonstrated that loss of preSREBP-1 was detectable after 6 h of TRC8 induction and reduced levels of pre-SREBP-2 were evident after 18 h (Fig. S5).
Figure 4. TRC8 destablizes SREBP precursors in a RING and proteasome-dependent manner.
(A) TRC8-HA and control HEK293 cells (vector) were treated for 24 h with increasing dox (ng/ml). RIPA buffer lysates (10 μg/lane) were analyzed for preSREBP-1 (2A4 antibody), preSREBP-2 (1C6 antibody) and TRC8-HA; Coomassie stain verified equal loading. (B) HEK293 cells containing TRC8-HA or three RING mutations (ΔRING, C547S;C550S and S557A;R559A) were induced for 24 h with increasing doses of dox (ng/ml) and analyzed as in (A). Ubiquitylation activity is only present in wild type TRC8 and in the S557A;R559A mutation, as indicated (19). The ΔRING mutation removes 39 amino acids, or about 4kDa, and detectably increases mobility on SDS/PAGE. (C) Duplicate plates of TRC8-HA or control HEK293 cells were dox-induced (100 ng/ml) for 12 h, then MG132 (10 μM) was added as indicated. After 9 h, cells were harvested and analyzed for the indicated proteins. HIF1α accumulation verified proteasome inhibition by MG132 (2); actin verified loading. (D) Duplicate plates of TRC8-HA or vector HEK293 cells were cultured in excess sterols (12.5 μM cholesterol, 10 μg/mL 25-HC), with or without dox, as indicated. After 24 h, cells were processed into membrane and nuclear fractions [Sever et al., (2003) (29), see Methods]. Fractions were analyzed for precursor and nuclear forms of SREBP-1/2 using 2A4 (SREBP-1) and 1D2 (SREBP-2) antibodies.
We previously reported that growth and cell-cycle inhibition by TRC8 were dependent upon a ubiquitylation-competent RING domain (19). As shown in Fig. 4B, a RING deletion mutant (ΔRING) failed to down-regulate SREBP-1/2 (lanes 5–8) and similar results were obtained with the C547S;C550S RING mutant. In contrast, expression of the ubiquitylation competent mutation S557A;R559A caused loss of the preSREBPs nearly as well as wild-type TRC8. An analysis of additional mutations (data not shown) consistently demonstrated that both preSREBPs were sensitive only to ubiquitylation-competent TRC8. Furthermore, the loss of preSREBPs was completely blocked by MG132 (Fig. 4C), verifying that the effect of TRC8 is proteasome-dependent. Interestingly, MG132 treated cells consistently contained less TRC8, suggesting that destabilization of ectopic TRC8-HA can still occur in the presence of proteasome inhibition.
To confirm that the preSREBP loss was not due to proteolyic activation and nuclear accumulation, and after verifying that the precursors were appropriately processed in HEK293 cells (Fig. S6), nuclear and membrane fractions were analyzed after 24 h of TRC8-induction (Fig. 4D). As expected, preSREBP-1/2 levels were reduced in the membrane fraction of TRC8-induced cells (lanes 7,8) compared to controls. Nuclear SREBPs did not increase upon TRC8 expression; rather, the levels appeared reduced (lanes 7,8). These experiments were performed with excess cholesterol to accumulate SREBP precursors; similar results were obtained in standard culture medium (Fig. S7). In addition, levels of the cleaved C-terminus of preSREBP-2 (SREBP-2-clv), which are known to increase during processing, were reduced by TRC8 (Fig. 4A). Together, these results indicate that TRC8-induced loss of preSREBPs is proteasome-dependent and not the consequence of enhanced processing.
Knockdown of endogenous TRC8 upregulates SREBPs
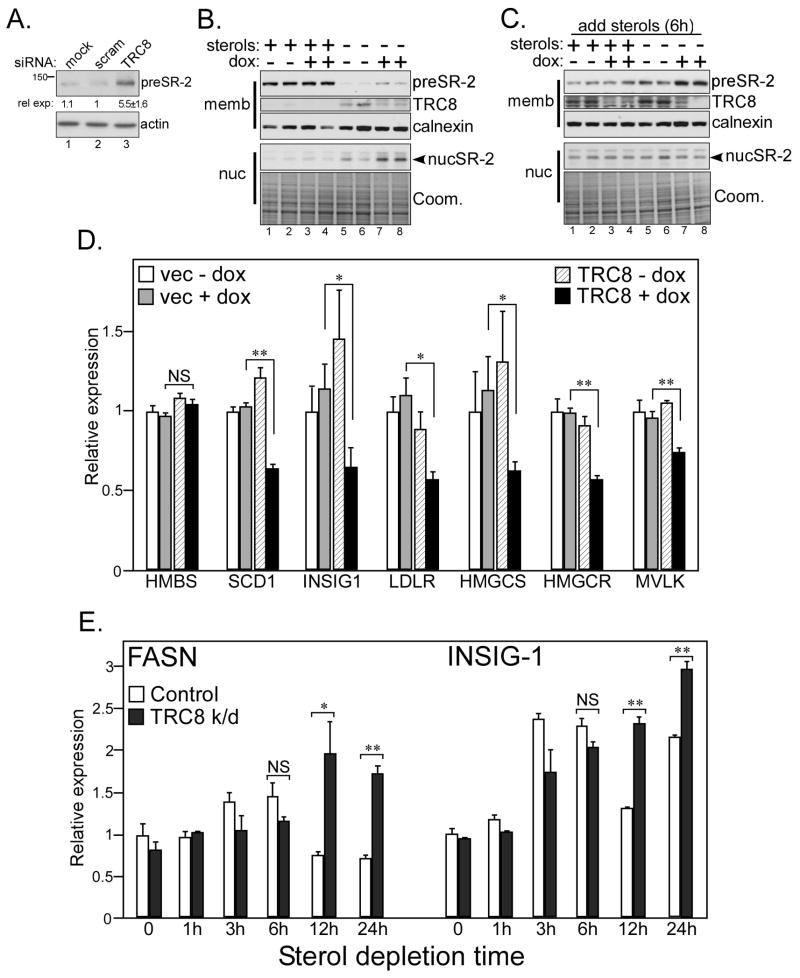
To determine if the modulation of preSREBPs was physiological, we used RNAi to knockdown endogenous TRC8. Transiently transfected siRNAs targeting TRC8 variably increased preSREBP-2 levels compared to mock-transfected or scrambled siRNA controls following several days of culture (Fig. 5A). We reasoned that depletion of serum lipoproteins with effects on TRC8 might account for this variability. In addition, sterol starvation would induce SREBP processing. To separate these effects, we analyzed membrane and nuclear fractions from stably transfected TRC8 knockdown cells cultured with or without sterols (Fig. 5B). In medium containing abundant sterols, knockdown of TRC8 had little effect on either the precursor or nuclear forms of SREBP-2 (lanes 1–4). However, after 24 h of sterol-deprivation (lanes 5–8), TRC8 knockdown resulted in higher levels of preSREBP-2 (lanes 7,8) compared to controls (lanes 5,6). Similarly, nuclear fractions from TRC8 knockdown cells contained more processed SREBP-2. Furthermore, in sterol-starved cells, when SREBP processing was subsequently blocked acutely by the addition of excess sterols, TRC8 knockdown cells accumulated higher levels of preSREBP-2 (Fig. 5C, lanes 7,8). Thus, inhibition of endogenous TRC8 affected SREBPs in a manner opposite to its over-expression, particularly in cultures subjected to prior sterol-deprivation.
Figure 5. TRC8 knockdown upregulates SREBP-2.
(A) HEK293 cells cultured in DMEM (10% FCS) were transfected with the indicated siRNAs (mock, scrambled or TRC8-specific [Ambion #-136327]). Detergent lysates were analyzed 72h post-transfection for pre-SREBP-2 and actin. Densitometry was performed on serial dilutions of replicate (n = 6) transfected lysates, relative expression values are means, +/− s.d. Representative blots are shown. (B) Duplicate cultures of HEK293 cells stably transfected with the inducible shRNA-TRC8 construct were treated for 4 days with vehicle (−) or dox (+), as indicated, to knockdown endogenous TRC8 (Fig. S2C). Cells were then incubated with fresh 5% FCS (+ sterols) or sterol-depletion media (sterols) for 20 h, harvested and processed into membrane and nuclear fractions. Western blots were analyzed for precursor and nucSREBP-2 (1D2); calnexin and Coomassie stain provided loading controls; knockdown of TRC8 was verified using anti-RNF139 antibody. (C) Cell cultures identical to (B) were treated with excess sterols (12.5 μg/mL cholesterol/10 μg/mL 25-HC) for the final 6h prior to harvest to arrest processing and accumulate SREBP precursors. Membrane and nuclear fractions were analyzed for SREBP-2 (1D2); knockdown of TRC8 was verified using anti-RNF139 antibody. Calnexin and a Coomassie stained gel provided loading controls. (D) Cultures of TRC8-HA or control (vec) HEK293 cells in DMEM (10% FCS) were treated with vehicle or dox (100 ng/mL) to induce TRC8-HA and harvested for RNA 24 h later. qRT-PCR was conducted for the indicated genes. Ct values were normalized geometric means of 4 controls; HMBS is graphed as a negative control. Bars represent means +/− SEM; *p < 0.05; **p < 0.01 by Student’s t-test (n = 6). Vector – dox (open bars); vector + dox (grey bars); TRC8-HA – dox (striped bars); TRC8-HA + dox (black bars). (E). TRC8 knockdown cells, treated with dox as in (B, C) to reduce TRC8, were sterol-deprived for the indicated times. Quadruplicate samples were analyzed by qRT-PCR for SREBP target genes. Bars represent means +/− SEM; *p < 0.05; **p < 0.01 by Student’s t-test (n = 4). TRC8 k/d cells – dox (open bars); + dox (black bars).
TRC8 modulates SREBP target gene expression
We next examined the consequence of TRC8 expression on SREBP target genes. Our initial microarray analysis compared cells expressing wild-type TRC8 to the C547S;C550S RING mutation. Significant differential expression was observed for several hundred genes (GC-RMA, see Supplemental Methods), including 21 associated with lipid metabolism and 16 known SREBP targets (31) (Table S1). All SREBP targets were down-regulated by wild-type TRC8. Eight genes (SCD1, INSIG-1, LDLR, HMGCS, HMGCR, MVLK, DHCR7 and SQLE) were further analyzed by qRT-PCR. After 24 h of TRC8 induction, six were significantly repressed from controls (Fig. 5D), while the remaining two trended lower without achieving statistical significance (data not shown). FASN, SREBF1 and MAC30, although not detected by microarray analysis, were also analyzed and found to be suppressed after 48 or 72 h of TRC8 induction. There was no apparent distinction between SREBP1 and SREBP2 target genes, as examples of each [e.g., SCD1 (SREBP1a) and MVLK (SREBP2)] were modulated by TRC8.
In contrast to overexpression, knockdown of endogenous TRC8 was associated with higher levels of SREBP target genes (Fig. S8). However, in time course studies, this effect was only observed after cells were sterol-starved for 12 to 24 h (Fig. 5E and S9). In control cells, acute sterol deprivation consistently led to SREBP target gene induction by 3 h, typically returning to baseline by 12 h. However, 12 and 24 h sterol-deprivation led to significantly higher expression of FASN and INSIG-1 in TRC8 knockdown cells (Fig. 5E). Similar results were obtained for LDLR (Fig. S9), SCD1 and SREBP-1 (data not shown). Together, these observations indicate that accumulation of TRC8 following longer term sterol deprivation negatively regulates preSREBP levels with corresponding effects on nucSREBPs and their target genes.
TRC8 interacts with the eukaryotic translation initiation factor, eIF3
We previously reported that Drosophila Trc8 genetically and physically interacted with the MPN domain of the COP9 signalosome subunit, Csn5/Jab1(17). To pursue this observation, we generated mitotic recombinant clones that ectopically expressed myc-tagged DTrc8 within a background of normal cells (Fig. S10). In the cuticle of adult flies, such clones were associated with aberrant bristle formation (Fig. S11). Microchaetes were smaller and deformed while macrochaetes were shortened. In addition, the bases of affected macrochaetes were thinner and had lost their characteristic fluting pattern. These effects are highly suggestive of Minute mutations, which primarily involve ribosomal protein genes and are associated with reduced translation efficiency (40).
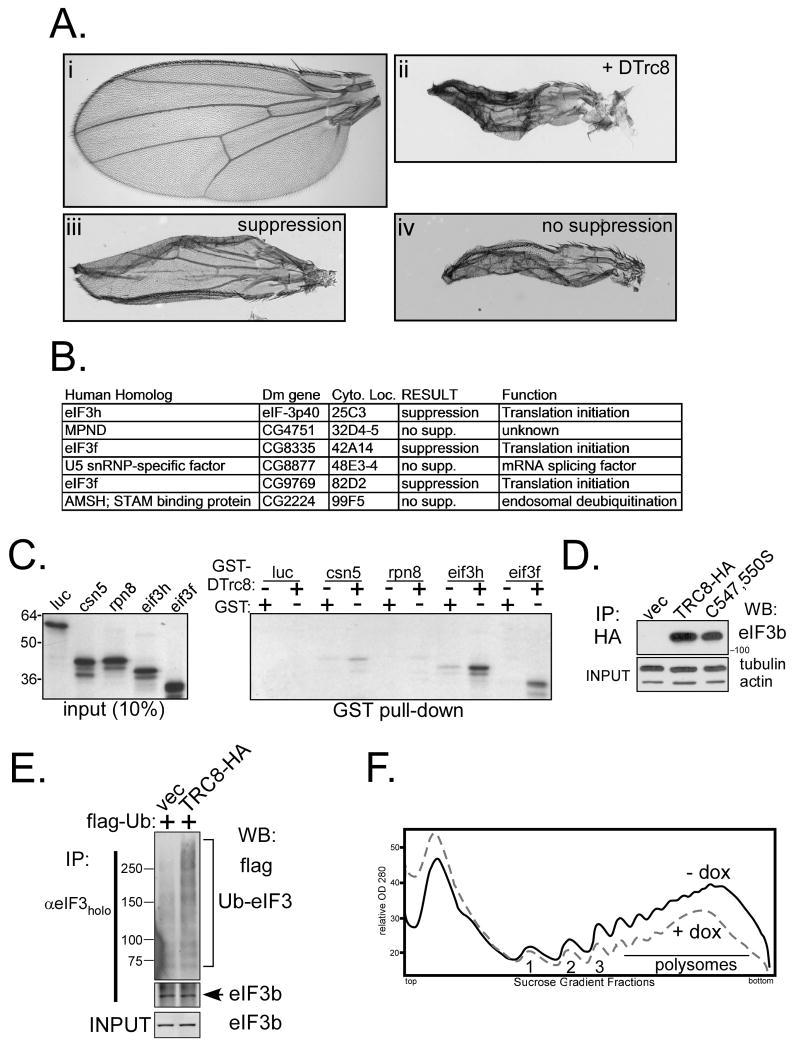
We previously reported that loss-of-function mutations or hemizygous deletions of two MPN domain genes, Csn5 and Csn6, partially restored growth to DTrc8-inhibited tissues in flies (21). The Drosophila genome contains 10 proteins with MPN domains, leading us to test each in a genetic interaction screen for suppression of the DTrc8 phenotype. Besides Csn5 and 6, mutations in three other MPN domain genes were suppressive; eIF3f (two genes) and eIF3h. All three encode subunits of the eukaryotic translation initiation factor, eIF3 (41) and hemizygous loss of each restored some growth to DTrc8 inhibited wings (Fig. 6A, B).
Figure 6. Genetic and physical interaction of TRC8 with subunits of eIF3.
(A; i–iv) Examples of male fruit fly wings; (i) wild type; (ii) DTrc8 over-expression; (iii) suppressor locus with heterozygous loss-of-function mutation (partially restores growth to DTrc8-inhibited wing); (iv) equivalent mutation in non-suppressor locus (fails to restore growth). (B) Results of suppressor screen among 6 genes containing MPN domains. All three loci encoding subunits of eIF3 suppressed the DTrc8 wing phenotype. Note there are two eIF3f homologs in flies. (C) GST pull-down assays of labeled TNT products. Purified GST and GST-DTrc8562–809 fusion proteins (21) were mixed with [35S] met-labeled in vitro translation products, as indicated. Bound proteins were identified by SDS-PAGE and autoradiography. Negative controls included luciferase (luc) and the proteasome subunit Rpn8 (21); CSN5 provided a positive control. The eIF3f isoform shown derived from the Drosophila gene CG9769. (D) HEK293 cells carrying TRC8-HA, vector or RING-mutant (C547S;C550S) were dox-induced for 24 h. Lysates were immunoprecipitated with anti-HA beads and bound proteins analyzed by Western blot for eIF3b, a core eIF3 subunit. Tubulin and actin established equal inputs. (E) TRC8-HA and control 293 cells were transfected with flag-ubiquitin (1 μg) and dox-induced for 24 h. Cells were harvested following MG132 addition (10 μM) during the final 2h. Triton X 100 lysates (200 μg aliquots) were immunoprecipitated with antibodies directed against the eIF3 holocomplex (54). IP pellets were analyzed by Western blot for flag-ubiquitin conjugated proteins and eIF3b as an indicator of eIF3 complex recovery and for equal input. (F) Polysome suppression by TRC8. TRC8-HA cells were treated with dox or vehicle for 24 h and harvested (see Methods). Polysomes were analyzed on 10 to 50% sucrose density gradients (53) to generate A280 gradient profiles; the first three polysome peaks are labeled 1–3. Results shown are representative of three repetitions.
In vitro GST-pulldown experiments confirmed that DTrc8 specifically interacted with eIF3f and eIF3h in addition to Csn5 (Fig. 6C). To validate the interaction in mammalian cells, TRC8-HA immunoprecipitations were performed from HEK293 cells expressing wild-type or the C547S;C550S mutant. Both wild type and RING mutant TRC8 co-immunoprecipitated eIF3 (Fig. 6D). When cells were transfected with flag-tagged ubiquitin and immunoprecipitated for eIF3, we observed high MW ubiquitylated products that were increased by expression of TRC8 (Fig. 6E). These results suggest that one or more components of the eIF3 complex may be ubiquitylated by TRC8.
eIF3 contains 13 subunits and represents part of a larger translation initiation complex. In addition, the availability of suitable antibodies against multiple components is limiting. Therefore, we used sucrose gradients to analyze polysome profiles from control and TRC8-inducible cells to determine if TRC8 affected translation (Fig. 6F). In three independent experiments, TRC8 induction suppressed the polysome profile, consistent with an inhibition of protein translation. In contrast, the ΔRING mutant had no demonstrable effect (data not shown). Taken together, these results are consistent with TRC8 inhibiting protein translation in a ubiquitylation-dependent manner through its interaction with eIF3 subunits. Further investigations will be required to determine which eIF3 subunits, or components of the larger complex, are affected and whether this is modulated by sterol levels.
Discussion
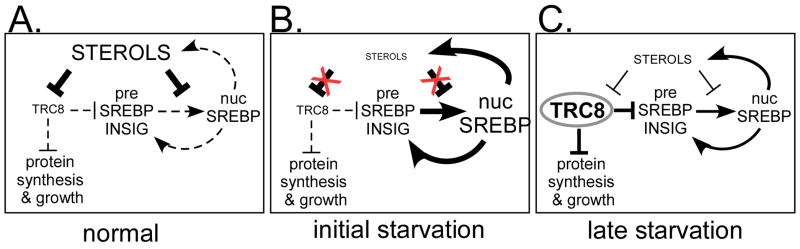
The results reported here suggest that TRC8, an E3-ubiquitin ligase, may provide a regulatory link between pathways of lipid homeostasis and protein translation initiation (Fig. 7). The involvement of TRC8 in cholesterol homeostasis is based on its predicted sterol-sensing domain and demonstrated sterol-dependent protein stability, its interaction with and ubiquitylation of INSIGs, and the regulation of SREBP precursor proteins with corresponding effects on SREBP target genes. Our results suggest that upregulation of TRC8 during prolonged periods of sterol deprivation may serve to dampen, or fine-tune, the SREBP response.
Figure 7. Model for TRC8 interaction with sterols and protein translation.
(A) Cells cultured in media with 10% FCS (normal lipoprotein/sterol content) undergo basal preSREBP processing (dashed arrows) and contain low levels of TRC8, which has only modest effects on growth, translation and SREBPs (indicated by the dashed lines). (B) Acute removal of sterols leads to rapid activation of preSREBPs with increases in SREBP target gene expression, including SREBPs themselves in a positive feed-forward loop (bold arrows). Production of lipid biosynthetic enzymes initiates restoration of lipid homeostasis. Initially, TRC8 does not restrain this process due to low levels, but the accumulation process begins so that by 12 to 24h of sterol deprivation (panel C), TRC8 causes a reduction of growth, reduction of translation and specific reduction of SREBPs (and other ER proteins like INSIG). This action may limit the stress of over-accumulating transmembrane proteins in an ER compartment compromised by loss of sterols and other lipids. Normalization of lipid levels occurs because TRC8-mediated reductions are not complete, permitting lipid synthesis to continue even as membrane protein accumulation becomes restricted. Moreover, reduced proliferative rates caused by accumulating TRC8 would permit restoration of lipid homeostasis by reducing requirements for membrane biogenesis. Restored lipids would then destabilize TRC8, releasing its inhibition of growth and translation.
TRC8 protein stability was found to vary in a sterol-dependent manner (Fig. 1, 2) with kinetics similar to those of HMG-CoA reductase, which contains a sterol-sensing domain and is polyubiquitylated by gp78 upon binding INSIG-1/2 (27, 28). Both wild-type and RING mutant TRC8 proteins bound INSIG-1/2 (Fig. 3) without apparent regard to sterol levels, at least in over-expression experiments, and TRC8 stimulated the polyubiquitylation of INSIG-1 in a RING-dependent manner. Mutation of conserved tyrosine 32 in the TRC8 SSD inhibited this interaction (Fig. 3C). Tyrosine 32 appears homologous to tyrosine 298 of SCAP, mutations of which impair INSIG binding (27). Thus the SSD of TRC8 is the likely interaction site for INSIG. These results are consistent with TRC8 mediating the ubiquitylation of both INSIG-1 and 2, in contrast to gp78, which acts only on INSIG-1 (38). Polyubiquitylation of TRC8 itself was observed (Fig. 2C) and this increased with added sterols (Fig. 2D), consistent with its destabilization by excess cholesterol. Since TRC8 mRNA levels did not change upon sterol starvation, these results suggest that a primary regulator of TRC8 activity is the cholesterol content of the ER membrane.
Ectopic TRC8 caused a substantial reduction in pre-SREBP levels, an effect that was RING dependent (Fig. 4). Moreover, pre-SREBP loss was completely blocked by proteasome inhibition, consistent with the ubiquitylation function of TRC8. However, it is currently unknown if the SREBP precursors are direct targets of this activity. Reduction of SREBP target gene expression demonstrated that induced TRC8 inhibits these transcription factors (Fig. 5D, Table S1). In contrast, TRC8 knockdown resulted in higher levels of precursor and nuclear SREBPs and increased target genes following chronic sterol-deprivation (Fig. 5). Therefore, our results indicate that this ubiquitin ligase acts, at least in part, to down-modulate the SREBP pathway, especially after TRC8 accumulates during prolonged sterol-deprivation.
As noted, the Drosophila genome encodes only a limited number of proteins with MPN domains. Among these are the COP9 signalosome subunits, Csn5 and Csn6, the proteasome lid subunits Rpn8 and Rpn11, and the eIF3 subunits, eIF3f and eIF3h. We previously reported that Drosophila Trc8 genetically and physically interacts with Csn5 and Csn6, but not Rpn8 or Rpn11 (21). The interaction with Csn5 was specifically shown to involve the MPN domain. Here, we demonstrate that Drosophila and human TRC8 proteins also interact with eIF3f and eIF3h (Fig. 6). Mitotic recombinant clones over-expressing DTrc8 resulted in a striking phenocopy of Minute mutations, which characteristically affect ribosomal protein genes and cause reduced protein translation (40). Reducing the copy number of eIF3f or eIF3h reduced the growth inhibitory phenotype of DTrc8 over-expression, much like reduction of the COP9 signalosome subunits, CSN5 and 6 (21). Genetic interaction between eIF3 subunits and DTrc8 was substantiated by demonstrating direct physical binding using GST pull-downs. In fact, interactions between DTrc8 and eIF3f or eIF3h were stronger than observed with Csn5. In HEK293 cells, TRC8 and eIF3 co-precipitate. Over-expression of TRC8 and immunoprecipitation of the eIF3 complex was associated with an increase in high molecular weight ubiquitylated products. In addition, TRC8 over-expression caused a reproducible reduction of the polysome profile in a RING-dependent manner, consistent with inhibition of protein translation. Further studies will be required to identify which eIF3 subunits or interacting factors are affected. Because downregulation of eIF3f or eIF3h suppresses DTrc8-induced growth inhibition in flies, we suspect these components form a bridge by which TRC8 affects its targets. Similarly, we suspect that INSIG forms a bridge by which TRC8 can access the precursor SREBPs, although this has not been formally examined.
Taken together, these findings suggest that TRC8 is positioned to coordinate lipid homeostasis and protein translation during extended periods of cholesterol limitation (Fig. 7). We propose that when TRC8 levels are low, under normal growth conditions (Fig. 7A) and during early times of sterol-deprivation (Fig. 7B), SREBP activation occurs with minimal hindrance. However, during periods of chronic sterol loss (Fig. 7C), TRC8 will accumulate, exerting a suppressive effect on growth (21), on protein translation and on synthesis of excessive ER transmembrane proteins, including the SREBPs. Activation of TRC8 will then be reversed by the accumulation of sterols, which would occur through the continued action of nuclear SREBPs and by stabilized HMGCR protein. We note that the effects of TRC8 on SREBPs are never absolute; some precursor always remains upon TRC8 expression (Fig. 4A–D), while nuclear forms and transcription targets are only modestly reduced (Fig. 4D, 5D). Thus lipid biosynthesis will continue in the presence of TRC8, although it may be slowed, while the reduction of growth following TRC8 accumulation would provide ample opportunity for cells to restore necessary lipids and achieve homeostasis.
The need for cells to coordinately inhibit growth during times of lipid-depletion is implicit, since membrane biogenesis depends on available phospholipids and cholesterol. We suspect that TRC8 accumulation also serves to protect the ER from the stress of accelerating transmembrane protein synthesis during a time of cholesterol insufficiency. The fact that SREBP activation results in a positive feedback loop (31, 42) may necessitate such a countermeasure. Intriguingly, crosstalk between lipid and protein translation pathways may also be a function of the kidney cancer-associated Birt-Hogg-Dubé protein, folliculin (FLCN) (11). FLCN forms a complex with FNIP1 and AMPK, which regulates mTOR activity through TSC1/2. Loss of FLCN leads to higher mTOR activity under certain stress conditions, including serum starvation (11). Nearly 20 years ago, AMPK was identified as the kinase that phosphorylates and inhibits HMGCR and acetyl CoA carboxylase (ACC) (43, 44), rate limiting steps for cholesterol and fatty acid biosynthesis, respectively (45, 46). FLCN, via interaction with AMPK, is positioned to influence lipid and protein biosynthetic pathways at the level of HMGCR, ACC and mTOR. Our results suggest that TRC8, which causes another form of hereditary kidney cancer, links these same pathways, at the level of the preSREBPs and eIF3. How interrupting these points of coordination affects kidney cancer development represents a new dimension in this disease and a new investigational direction.
Methods
Cell lines, Tissue Culture and Reagents
HEK293 FlpIn TRex (HEK293 FlpIn) cells were maintained in DMEM media containing 50 μg/ml Zeocin and 5 μg/ml blasticidin. CHO FlpIn TRex lines were generated as described by the supplier (Invitrogen). Stably transfected FI lines were selected in 50 μg/ml hygromycin B. Sterol-depletion medium contained 5% de-lipidized fetal bovine serum (LPDS) (47), 10μM mevastatin and 50 μM mevalonate. Sterol-replete media contained 5% FCS, cholesterol (12.5 μM) and 25-hydroxycholesterol (25-HC; 10 μg/mL), or as noted in figure legends. Sodium mevalonate was prepared by saponification of mevalonolactone (Sigma-Aldrich, St. Louis, MO). Antibody sources included; TRC8/RNF139 [H00011236-A01] (Abnova/Novus Biologicals, Littleton, CO); SREBP-1 [IgG-2A4], SREBP-2 [IgG-1C6, C-terminal specific] (Labvision/NeoMarkers, Fremont, CA), SREBP-2 [IgG-1D2, N-terminal specific] (MBL International, Woburn, MA). Anti-FLAG M2 beads and antibody were from Sigma. HA [Y-11] and eIF3b (p116) antibodies, anti-myc 9E10 beads and protein A/G PLUS beads were from Santa Cruz, Inc., (Santa Cruz, CA); anti-eIF3-complex antibody was kindly provided by J.W. Hershey. HIF1α antibody was from BD Biosciences (San Jose, CA). HMGCR mAb was purified from A9 hybridoma (CRL-1811, ATCC, Manassas, VA).
Expression Constructs and Western blots
TRC8 expression constructs have been described (19). MYC-INSIG-1 and -2 were obtained from ATCC; the K156R, K158R double mutation was generated by site-specific mutagenesis using the QuikChange kit (Stratagene, La Jolla, CA), as was the Y32E and Y32C mutations of TRC8-HA; pcDNA3-flag-ubiquitin was a gift from R. Wojcikiewicz and D. Sliter. Cells were lysed on ice using specific buffers (see Supplemental Methods for formulas). In general, ten μg of protein were resolved by SDS/PAGE using 4–15% gradient gels (BioRad, Hercules, CA) and analyzed by Western blot using PVDF membranes and standard protocols (Millipore, Bedford, MA). Antibody detection used HRP-conjugated secondary antibodies and chemiluminescence reagents (Pierce, Rockford, IL) or AlexaFluor 488 conjugated secondary antibodies and a Typhoon 9400 Variable Mode Imager or CW800 and CW680 IRDye conjugated secondary antibodies and a LiCor Odyssey fluorescence scanner.
Immunoprecipitations, GST-pulldown and subcellular fractionations
Immunoprecipitations were performed using published protocols (48, 49). Briefly, washed cells were suspended in 0.5 ml of CHAPSO buffer (Supplemental Methods) and ruptured by passage 15x through a 22-ga needle. Cleared supernatants (500 μg protein) were incubated with beads and primary antibodies overnight, washed 3x and analyzed by Western blot. TritonX-100 and RIPA lysates were sonicated and clarified by centrifugation prior to immunoprecipitation. GST-pulldowns were performed as previously described using radiolabeled TNT reaction products and a GST-DTrc8 fusion protein (17, 21). Subcellular fractionation utilized a protocol (29) adopted from Sever et al., (2003). Washed cell pellets were suspended in 0.5 ml of Cell Disruption Buffer and passed 15x through a 22-ga needle. Lysates were cleared at 1000 x g and the supernatant centrifuged at 100,000 x g for 20 min to recover membranes. The pellet was resuspended in 50 μL of Membrane Solubilization Buffer. The low-speed pellet was resuspended in 100μL Nuclear Extraction Buffer and rotated for 4h in the cold. Following centrifugation at 100,000xg for 20 min in a TLA 120.1 rotor, the supernatant was recovered as the nuclear fraction.
Real-time qRT-PCR and polysome analysis
Preparation of RNA, cDNA and performance of qRT-PCR using hot-start conditions have been previously described (50, 51), except that the assays were carried out in a 10 μL volume using SYBR green in ABI 7500 instrument with 9600 emulation. Primer sequences used for real-time PCR (5′ to 3′) are listed in Supplemental Methods. Primers specific for TRC8 have been described (19). Ct values were normalized by subtracting the geometric mean of four control genes, GAPDH, HMBS, UBC1 and HPRT1 (52). Affymetrix microarray data analysis methods are in Supplemental Methods. Polysome profiling was performed on cultures of HEK293 FlpIn cells induced or not to express wild type or mutant TRC8-HA for 24h with 100ng/mL dox. Sucrose gradient separations of polysomes were performed according to protocols from P. Sarnow (53).
siRNA and shRNA
Multiple anti-TRC8 siRNAs (Ambion, Austin, TX) were tested for efficacy and Ambion ID# 136327 (TRC8-Amb-27, sense-strand sequence, GCUUGACGAUUAUGUCUACtt) was most effective. Cells were transfected with Lipofectamine 2000 according to manufacturer’s directions (Invitrogen, Inc., Carlsbad, CA) and assays performed 72 hrs post treatment. The tet-inducible anti-TRC8 shRNA construct was prepared by annealing complementary oligonucleotides using the following sequences: Forward- GATCCGCTTGACGATTATGTCTACTTCAAGAGAGTAGACATAATCGTCAAGCTTTTTTGGCCC; Reverse: TCGAGGGCCAAAAAAGCTTGACGATTATGTCTACTCTC TTGAAGTAGACATAATCGTCAAGCG. Annealed oligos were ligated into BamHI/XhoI cleaved pSuperior.puro vector, propagated in E. coli, sequence verified and stably transfected into HEK293 FlpIn TRex cells using puromycin (3 μg/mL) and blasticidin selection (5 μg/mL).
Drosophila methods
Fly culture, genetic manipulations and evaluation for genetic suppression were performed as previously described (21) using the following strains for testing MPN domains; y[1]w[67c23];P{w[+mC]=lacW}eIF-3p40[k09003]/CyO for eIF3h (eIF-3p40); FCK-20 dp bw/SM5q-TM6b for MPND (CG4751); Df(2R)nap1/In(2LR)Gla, Dp(2;2)BG, wg[Gla-1] for eIF3f (CG8335); Df(2R)CB21/CyO;ry[506] for U5 snRNP-specific factor (CG8877); Df(3R)110, ru[1]th[1]st[1]kni[ri-1]rn[roe-1]p[p]e[s]ca[1]/TM3, Sb[1] for eIF3f (CG9769); and Df(3R)tll-g, ca[1]/TM6B, Tb[1] for AMSH (CG2224). Mitotic recombinant clones were prepared by generating embryos containing the actin promoter-FRT-yellow-FRT-Gal4 cassette (AYG) along with heat-shock-inducible FLP recombinase(FLP122), UasGFP and UasDTrc87.1(17). Resulting larvae were heat shocked for 20 min (24h post egg laying) then development continued until adults eclosed. Patches of adult cuticle containing y −, DTrc8+ clones were identified by color, recorded and analyzed using a Zeiss DSM940A digital scanning electron microscope. Flies were mounted on standard stubs with double-sided carbon SEM tape and a silver colloidal suspension. Samples were vacuum-dried at room temperature and coated with 50 microns of gold using a sputter coater prior to examination.
Supplementary Material
Acknowledgments
We thank K. Lorick and A.M. Weissman for numerous discussions and suggestions, E. Robbins, S. McNamara and J. Nair-Menon for technical assistance and lab members for critical review of the manuscript. This investigation used the DNA Sequencing and cDNA Microarray Cores of the Univ. of Colorado Cancer Center. This work was supported by NIH RO1 CA076035.
Abbreviations
- CHO
chinese hampster ovary cells
- FCS
fetal calf serum
- LPDS
lipoprotein-deficient serum
- CHX
cycloheximide
- dox
doxycycline
- ER
endoplasmic reticulum
- IP
immunoprecipitation, RING, really interesting new gene, a zinc-binding domain that recruits ubiquitin conjugating enzymes
- ERAD
ER-associated degradation
- qRT-PCR
quantitative realtime reverse transcriptase-polymerase chain reaction
- COP9
constitutive photomorphogenesis 9
- MPN
Mpr1p, Pad1p N-terminal
- eIF3
eukaryotic translation initiation factor 3
- 25-HC
25-hydroxycholesterol
- HPβCD
hydroxypropyl β-cyclodextrin
- RIPA
radioimmune precipitation assay buffer
- k/d
knockdown
- IR
infrared
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2006;56(2):106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Kaelin WG., Jr The von Hippel-Lindau protein, HIF hydroxylation, and oxygen sensing. Biochemical and biophysical research communications. 2005;338(1):627–38. doi: 10.1016/j.bbrc.2005.08.165. [DOI] [PubMed] [Google Scholar]
- 3.Hughes MD, Kapllani E, Alexander AE, Burk RD, Schoenfeld AR. HIF-2alpha downregulation in the absence of functional VHL is not sufficient for renal cell differentiation. Cancer cell international. 2007;7:13. doi: 10.1186/1475-2867-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandriota SJ, Turner KJ, Davies DR, et al. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer cell. 2002;1(5):459–68. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nature genetics. 2002;30(4):406–10. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 6.Vanharanta S, Buchta M, McWhinney SR, et al. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am J Hum Genet. 2004;74(1):153–9. doi: 10.1086/381054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isaacs JS, Jung YJ, Mole DR, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer cell. 2005;8(2):143–53. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nature genetics. 1997;16(1):68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 9.Bjornsson J, Short MP, Kwiatkowski DJ, Henske EP. Tuberous sclerosis-associated renal cell carcinoma. Clinical, pathological, and genetic features. The American journal of pathology. 1996;149(4):1201–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–90. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 11.Baba M, Hong SB, Sharma N, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(42):15552–7. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science (New York, NY) 2005;310(5753):1513–5. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 13.Cohen AJ, Li FP, Berg S, et al. Hereditary renal-cell carcinoma associated with a chromosomal translocation. The New England journal of medicine. 1979;301(11):592–5. doi: 10.1056/NEJM197909133011107. [DOI] [PubMed] [Google Scholar]
- 14.Li FP, Decker HJ, Zbar B, et al. Clinical and genetic studies of renal cell carcinomas in a family with a constitutional chromosome 3;8 translocation. Genetics of familial renal carcinoma. Ann Intern Med. 1993;118(2):106–11. doi: 10.7326/0003-4819-118-2-199301150-00005. [DOI] [PubMed] [Google Scholar]
- 15.Gemmill RM, West JD, Boldog F, et al. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(16):9572–7. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poland KS, Azim M, Folsom M, et al. A constitutional balanced t(3;8)(p14;q24.1) translocation results in disruption of the TRC8 gene and predisposition to clear cell renal cell carcinoma. Genes, chromosomes & cancer. 2007;46(9):805–12. doi: 10.1002/gcc.20466. [DOI] [PubMed] [Google Scholar]
- 17.Gemmill RM, Bemis LT, Lee JP, et al. The TRC8 hereditary kidney cancer gene suppresses growth and functions with VHL in a common pathway. Oncogene. 2002;21(22):3507–16. doi: 10.1038/sj.onc.1205437. [DOI] [PubMed] [Google Scholar]
- 18.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(20):11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brauweiler A, Lorick KL, Lee JP, et al. RING-dependent tumor suppression and G2/M arrest induced by the TRC8 hereditary kidney cancer gene. Oncogene. 2007;26(16):2263–71. doi: 10.1038/sj.onc.1210017. [DOI] [PubMed] [Google Scholar]
- 20.Stagg HR, Thomas M, van den Boomen D, et al. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. The Journal of cell biology. 2009;186(5):685–92. doi: 10.1083/jcb.200906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gemmill RM, Lee JP, Chamovitz DA, Segal D, Hooper JE, Drabkin HA. Growth suppression induced by the TRC8 hereditary kidney cancer gene is dependent upon JAB1/CSN5. Oncogene. 2005;24(21):3503–11. doi: 10.1038/sj.onc.1208509. [DOI] [PubMed] [Google Scholar]
- 22.Cope GA, Suh GS, Aravind L, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science (New York, NY) 2002;298(5593):608–11. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 23.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC biochemistry. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebhard RL, Clayman RV, Prigge WF, et al. Abnormal cholesterol metabolism in renal clear cell carcinoma. Journal of lipid research. 1987;28(10):1177–84. [PubMed] [Google Scholar]
- 25.Clayman RV, Bilhartz LE, Buja LM, Spady DK, Dietschy JM. Renal cell carcinoma in the Wistar-Lewis rat: a model for studying the mechanisms of cholesterol acquisition by a tumor in vivo. Cancer Res. 1986;46(6):2958–63. [PubMed] [Google Scholar]
- 26.Tosi MR, Tugnoli V. Cholesteryl esters in malignancy. Clin Chim Acta. 2005;359(1–2):27–45. doi: 10.1016/j.cccn.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Molecular cell. 2005;19(6):829–40. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Molecular cell. 2003;11(1):25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 30.Epand RM. Cholesterol and the interaction of proteins with membrane domains. Progress in lipid research. 2006;45(4):279–94. doi: 10.1016/j.plipres.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Horton JD, Shah NA, Warrington JA, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):12027–32. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120(6):831–42. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 33.Abdulrahman M, Maina EN, Morris MR, et al. Identification of novel VHL targets that are associated with the development of renal cell carcinoma. Oncogene. 2006 doi: 10.1038/sj.onc.1209932. [DOI] [PubMed] [Google Scholar]
- 34.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(20):12753–8. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang T, Espenshade PJ, Wright ME, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110(4):489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 36.Chang TY, Chang CC, Ohgami N, Yamauchi Y. Cholesterol sensing, trafficking, and esterification. Annual review of cell and developmental biology. 2006;22:129–57. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- 37.Gong Y, Lee JN, Lee PC, Goldstein JL, Brown MS, Ye J. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell metabolism. 2006;3(1):15–24. doi: 10.1016/j.cmet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 38.Lee JN, Song B, DeBose-Boyd RA, Ye J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. The Journal of biological chemistry. 2006;281(51):39308–15. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- 39.Nohturfft A, Brown MS, Goldstein JL. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(22):12848–53. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marygold SJ, Roote J, Reuter G, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome biology. 2007;8(10):R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M, Sandercock AM, Fraser CS, et al. Special Feature: Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proceedings of the National Academy of Sciences of the United States of America; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. The Journal of clinical investigation. 2002;109(9):1125–31. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beg ZH, Stonik JA, Brewer HB., Jr Characterization and regulation of reductase kinase, a protein kinase that modulates the enzymic activity of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proceedings of the National Academy of Sciences of the United States of America. 1979;76(9):4375–9. doi: 10.1073/pnas.76.9.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carling D, Clarke PR, Zammit VA, Hardie DG. Purification and characterization of the AMP-activated protein kinase. Copurification of acetyl-CoA carboxylase kinase and 3-hydroxy-3-methylglutaryl-CoA reductase kinase activities. European journal of biochemistry/FEBS. 1989;186(1–2):129–36. doi: 10.1111/j.1432-1033.1989.tb15186.x. [DOI] [PubMed] [Google Scholar]
- 45.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8(10):774–85. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 46.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763–77. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 47.Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods in enzymology. 1983;98:241–60. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 48.Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. The Journal of biological chemistry. 1997;272(32):20213–21. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 49.Feramisco JD, Radhakrishnan A, Ikeda Y, Reitz J, Brown MS, Goldstein JL. Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(9):3242–7. doi: 10.1073/pnas.0500206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roche J, Zeng C, Baron A, et al. Hox expression in AML identifies a distinct subset of patients with intermediate cytogenetics. Leukemia. 2004;18(6):1059–63. doi: 10.1038/sj.leu.2403366. [DOI] [PubMed] [Google Scholar]
- 51.Drabkin HA, Parsy C, Ferguson K, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16(2):186–95. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 52.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3(7):1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA (New York, NY) 1998;4(12):1500–13. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer LJ, Milburn SC, Hershey JW. Immunochemical characterization of mammalian protein synthesis initiation factors. Biochemistry. 1982;21(18):4206–12. doi: 10.1021/bi00261a003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.