Abstract
Bacterial photosynthetic membranes, also known as chromatophores, are tightly packed with integral membrane proteins that work together to carry out photosynthesis. Chromatophores display a wide range of cellular morphologies; spherical, tubular, and lamellar chromatophores have all been observed in different bacterial species, or with different protein constituents. Through recent computational modeling and simulation, it has been demonstrated that the light-harvesting complexes abundant in chromatophores induce local membrane curvatures via multiple mechanisms. These protein complexes assemble to generate a global curvature and sculpt the chromatophores into various cellular-scale architectures.
Introduction
Sunlight harvested via photosynthesis is the primary energy source of the biosphere. The multi-step process is carried out by a collection of proteins. In photosynthetic purple bacteria, the photosynthetic proteins aggregate and form distinct membrane indentations with a size scale of ~100 nm that can be observed under a light microscope. These membrane indentations, known as chromatophores, serve as the simplest prototype of a photosynthetic machinery, and have been studied intensely to characterize various aspects of photosynthesis. In this article we consider the overall architecture of chromatophores: how their distinct shapes arise and, in particular, how computational methods have helped to address this question. Studies on the formation of cellular-scale photosynthetic chromatophores coincide with a rising interest in the morphology of cellular membranes.[1, 2, 3, 4] The computational methods discussed here are not restricted to photosynthetic proteins and can be similarly applied to other membrane-bending molecular assemblies.[5, 6, 7, 8]
Chromatophores: the photosynthetic machineries
Using light and electron microscopy, microbiologists have long noticed the large protrusions populating photosynthetic bacterial membranes.[9, 10, 11, 12] The shape of these membrane protrusions were observed to vary significantly with the growth environment or upon the removal of certain proteins.[10, 11, 12, 13, 14] Later, through biochemical analyses and imaging techniques such as electron microscopy (EM) and atomic force microscopy (AFM), photosynthetic proteins were found to be the primary constituents of these membrane protrusions, termed chromatophores (for reviews see [15, 16]).
In photosynthetic bacteria, light is first absorbed by peripheral light-harvesting (LH) complexes, which transfer the excitation energy to a reaction center (RC). The reaction center, together with another membrane protein, cytochrome bc1, use the excitation energy to produce a transmembrane charge gradient, which is utilized by the protein complex, ATP synthase, to produce ATP. Each photosynthetic chromatophore is thought to be equipped with all these proteins at a certain stoichiometry. For example, in the case of Rhodobacter (Rba.) sphaeroides, on average, ten dimeric RC-light-harvesting complex I supercomplexes (RC-LH1),[17, 18] 100 light-harvesting complex II (LH2), five bc1,[19, 20] and 1–4 ATP synthase [21, 22, 23] per chromatophore are expected, the ratio depending heavily on environmental factors, like light intensity.
Although all these membrane-bound photosynthetic protein complexes (RC-LH1, LH2, bc1, and ATP synthase) are believed to be present in chromatophores, so far imaging studies have only identified RC-LH1 and LH2 (Figure 1a). In addition, studies have shown that the presence and concentration of RC-LH1 and LH2 are the dominant factors in determining the shape and size of the Rba. sphaeroides chromatophores. Indeed, the concentration of LH2 in Rba. sphaeroides increases with decreasing light intensity, leading to chromatophores with smaller radii.[13, 14] Also, deletion of LH2 in Rba. sphaeroides results in tubular chromatophores populated with orderly arranged dimeric RC-LH1 complexes, and the tubular chromatophores can extend and elongate the bacterial cell (Figure 1b).[10, 11, 24, 25, 26] These experiments prompt the consideration that both LH2 and RC-LH1 are curvature-inducing elements in chromatophores, with the former causing spherical curvature and the latter tubular curvature. The dynamic process of membrane remodeling is difficult to observe experimentally, but is possible with computational modeling even at atomistic resolution, given that structural information for many photosynthetic proteins has become available. Here we review computational studies elucidating the formation of photosynthetic chromatophores.
Figure 1.
Two examples of chromatophore shapes. a) Spherical chromatophore (radius 30 nm) found in wild-type Rba. sphaeroides containing LH2 (orange) and the RC-LH1-PufX supercomplex (RC: green, LH1: blue, PufX: red). Placement of the proteins was performed in [27] using a combination of structural and imaging data. b) Mutant Rba. sphaeroides lacking LH2 possesses tubular chromatophores, which are populated with helically ordered RC-LH1-PufX.[26] The tubular chromatophore shown here has a radius of 36 nm.
Curvature properties of LH2
LH2 is a ring-shaped membrane protein complex found in the chromatophores of photosynthetic bacteria. The function of LH2 is to absorb light via its protein-bound pigments, i.e., bacteriochlorophylls and carotenoids, and to pass the energy on to LH1 and the RC. As noted above, it is suspected that the aggregation of LH2 in the membrane plays a role in the development of the spherical chromatophore shape seen in Rba. sphaeroides, as this shape persists even in LH2-only mutants of Rba. sphaeroides.[13, 14]
To determine the membrane-curving properties of aggregates of LH2 complexes, arrays of seven hexagonally-arranged LH2s were modeled computationally at atomistic detail and were subjected to equilibrium molecular dynamics (MD) simulations.[28] The hexagonal arrangement of LH2 complexes was revealed in AFM images [29, 30, 23] and, therefore, is biologically realistic. While crystal structures of LH2 are available for two species, Rps. acidophila and Rs. molischianum,[31, 32] none are available for LH2 from Rba. sphaeroides. Because LH2s from different species are quite similar in sequence and structure, homology modeling is feasible and was applied to construct an all-atom model for Rba. sphaeroides LH2.[28] The resulting simulation system contains, in addition to the protein array, also the membrane environment and the solvent in order to mimic the physiological conditions as closely as possible, and consists of one million atoms in total, nearing current computational limits for all-atom MD.[28, 33] The membrane is decoupled from its periodic neighbors through additional water, eliminating resistance to curvature at the boundaries (Figure 2a). Equilibration of the system showed that the LH2 proteins pack together and tilt with respect to their neighbors to produce an overall curvature on a timescale of 20 ns (Figure 2b).[28] The observed tilt angle for Rba. sphaeroides LH2s of ~13° corresponds to a radius of curvature of approximately 32 nm, within the range of the size of native chromatophores (25–55 nm).[10, 11, 24, 25, 26]
Figure 2.

a) Top view (perpendicular to membrane plane) of a setup of an LH2 simulation containing seven LH2s packed hexagonally in a membrane patch. Each LH2 is colored differently for distinction. Membrane is shown in blue, and water box in transparent blue. b) Side view (along the membrane plane) of the Rba. sphaeroides LH2 patch before and after equilibration.[28, 34] LH2s are colored according to residue type, with red being negatively-charged residues and blue positively-charged. Here, as well as in all the following protein-membrane systems shown in side views, the proteins are orientated with their cytoplasmic sides pointing upward. The dashed red lines indicate the membrane center to illustrate the curvature arising during the simulation.
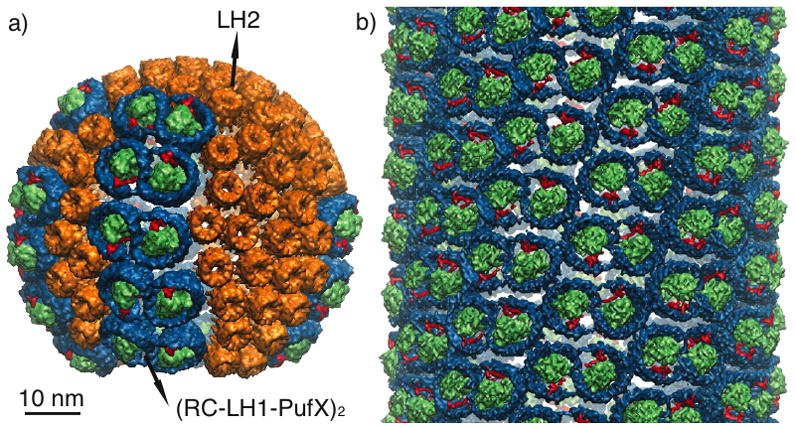
MD simulations of arrays of LH2s from Rps. acidophila and Rs. molischianum were also performed, permitting comparison of the curvature effects from different species of LH2.[28, 34] Such a comparison is particularly interesting because Rps. acidophila and Rs. molischianum possess lamellar, folded chromatophores, unlike the spherical chromatophores in Rba. sphaeroides. Surprisingly, the packing of Rps. acidophila and Rs. molischianum LH2s also resulted in a net membrane curvature (Figure 3).[28, 34] These results indicate that the distinct chromatophore shapes observed are not due to differences in LH2-LH2 interactions between species, but rather due to other factors, e.g., possibly presence of other proteins, membrane composition or ion concentration.[35, 34]
Figure 3.
Curvature induced by patches of LH2s from wild-type vs. alanine-replacement mutants. The mutated LH2s, in which the charged residues were changed to alanines, curved less than their wild-type counterparts.
The question of what molecular mechanisms drive the tilting of LH2s remains. Based on sequence alignments, highly-conserved charged residues at the cytoplasmic side of the LH2s were identified for all three species, the electrostatic interactions of which could induce neighboring LH2s to tilt away from each other on the cytoplasmic side (Figure 2b).[34] To determine their role in the curvature of LH2 patches, charged residues were mutated to either alanines or neutral analogs, and the same MD protocol was applied.[34] Indeed, for LH2s without the charged residues, the curvature of the patches was seen to be greatly reduced (Figure 3).[34] Further analysis of the simulations suggests that the presence of the charged residues causes the LH2s to pack less efficiently on the cytoplasmic side than on the periplasmic side, contributing to the overall curvature. In addition to the influence of charged residues, the physical shape of the LH2 also affects curvature, with the more wedge-shaped LH2s (Rs. molischianum) curving the membrane more than the more cylindrical LH2s (Rps. acidophila), and with the alanine-replacement mutants curving the membrane less than the neutralized versions due to the reduced bulk on the cytoplasmic side.[34] These studies suggest that LH2s in all species induce spherical curvature via a collective behavior, namely packing of the protein complexes mediated by electrostatic and steric interactions.
Curvature properties of Rba. sphaeroides RC-LH1-PufX
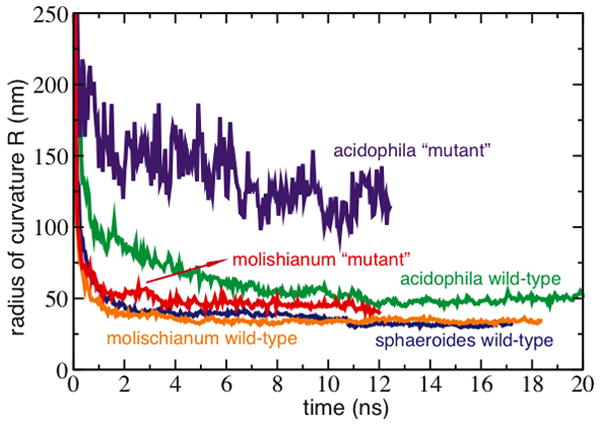
The RC-LH1 complex in Rba. sphaeroides is dimeric, and contains an additional polypeptide, PufX. The large RC-LH1-PufX molecular assembly is approximately 10 nm along the dimer’s short axis and 20 nm along the long axis (Figure 4a). MD simulations of RC-LH1-PufX first faced the obstacle of incomplete structural information. Unlike LH2, there is no high-resolution structure available for a complete RC-LH1. For this reason computational studies on RC-LH1-PufX made use of several modeling techniques, incorporating structural data at different resolutions. An initial Rba. sphaeroides RC-LH1-PufX model was constructed combining homology modeling, solution structures, and crystal structures. [28] The protein components were assembled into a dimeric complex guided by an 8.5-Å-resolution cryo-EM projection map reported in Qian et al., 2005,[36] the highest resolution structural information available for the complex (Figure 4a). All-atom MD was employed to equilibrate the resulting RC-LH1-PufX model. The simulation revealed that the dimeric complex spontaneously bends slightly at the dimerizing interface (θdimer ~ 172°) due to the relative orientation of the two RCs (Figure 4b).[28] The bent dimer was observed to pull its surrounding membrane along, thereby inducing a local curvature.
Figure 4.
Rba. sphaeroides RC-LH1-PufX model and equilibration.[28] a) Top and side view of the RC-LH1-PufX complex immersed in a membrane patch before MD equilibration. LH1 shown in blue, RC in green, PufX in red, and membrane in light blue. Water is not shown for clarity. For the side view, some lipid molecules are hidden to show better the protein complex. An alternative placement for PufX is discussed in Scheuring et al., 2004.[37] b) Simulation system after a 20-ns equilibration. Top panel shows the complex attaining a bending angle θdimer ~ 172°. On the bottom panel, some LH1 helices are hidden to show the relative orientation of the two RCs, colored by residue types, which form a bent hydrophobic region.
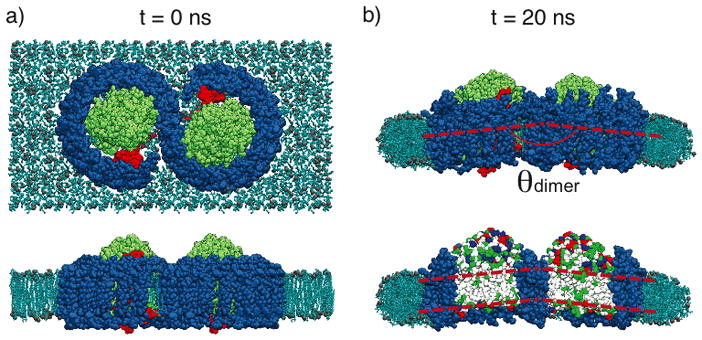
The bending reported in Chandler et al., 2008[28] rationalizes how the large RC-LH1-PufX dimer can fit into a curved chromatophore membrane, but the bending produced is actually insufficient to sustain the size of a Rba. sphaeroides chromatophore (θdimer ~ 172° corresponds to a radius of curvature of 72 nm, whereas a typical chromatophore has radius 25–55 nm [10, 11, 24, 25, 26]). A three-dimensional cryo-EM map of the Rba. sphaeroides RC-LH1-PufX dimer was published in a single-particle analysis study,[26] and revealed that the complex indeed is bent, but with a sharper bending angle (θdimer ~ 148°) than was observed computationally (Figures 5a and b). It became necessary to improve the atomistic model of the Rba. sphaeroides RC-LH1-PufX complex to better reflect the quaternary structure unveiled by the EM density map, even though the low resolution of the map disallows direct extraction of atomistic data.
Figure 5.
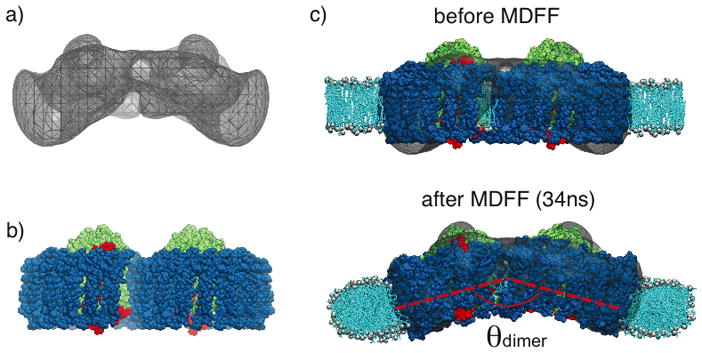
Application of MDFF for the membrane-bending properties of Rba. sphaeroides RC-LH1-PufX complex. a) EM envelope obtained in a single-molecule analysis study, showing a large bending.[26] b) Modeled RC-LH1-PufX complex from Chandler et al., 2008,[28] which is initially flat. c) RC-LH1-PufX complex fitted into the EM map employing MDFF, resulting in a much more prominent θdimer. The membrane curves along with the complex.[46]
In this situation computational methods combining structural information at different resolutions is useful, the particular methodology employed was the molecular dynamics flexible fitting (MDFF) method.[38, 39, 40] MDFF was developed to transform, in a realistic manner, an all-atom structure, obtained either through crystallography or modeling, to an alternative configuration defined by a lower-resolution EM density. In an MDFF simulation two potentials, in addition to the force field describing the interactions between atoms, are incorporated into the MD simulation. One of the potentials steers atoms into high-density regions of the EM map, while the other maintains the secondary structure of the protein. With properly tuned parameters for these two potentials, a biomolecule can be fitted into an EM map within ns timescales, revealing the atomic structures and interactions within protein complexes in physiologically relevant conformations.[41, 42, 43, 44]
In addition to aiding the generation of new atomistic structures, MDFF permits the inclusion of additional molecules not coupled to the EM map in simulations. This feature of MDFF is directly applicable for the case of protein-induced membrane curvature, since lipid molecules need to be described in the simulated systems but should not in general be coupled to the EM map. Employing MDFF, an all-atom RC-LH1-PufX model conforming to the geometry of the EM density map was obtained. [45, 46] The membrane patch surrounding the protein complex was found to also bend, with a radius of curvature (~45 nm) in agreement with the size of chromatophore vesicles (Figure 5c). In addition, the local membrane curvature property of Rba. sphaeroides RC-LH1-PufX was observed to be asymmetric; namely, while bending the membrane around its short axis, the RC-LH1-PufX complex also twists the membrane slightly around its long axis. This membrane-bending behavior of RC-LH1-PufX offers a microscopic rationalization for the helical packing of the dimers seen in EM images of tubular chromatophores comprised solely of RC-LH1-PufX (Figure 1b);[24, 25, 26] detailed analysis of local curvature properties and their relationship to the global arrangement of RC-LH1-PufX is described in Hsin et al., 2009.[46]
Membrane curvature due to Rba. sphaeroides RC-LH1-PufX is a direct result of its highly bent geometry, which is unique to dimeric RC-LH1-PufX. In PufX-deficient Rba. sphaeroides, or in other species lacking PufX, the RC-LH1 complex is monomeric and ring-like, and does not possess a bent geometry.[47, 48, 49] The question remains what is the molecular basis for the dimerization and the resulting bent geometry of the Rba. sphaeroides RC-LH1-PufX complex. Recently, computational methods were applied in an attempt to address this issue. The important structural element considered was the PufX protein, which is a single transmembrane helix known to play the determining role in RC-LH1 dimerization,[50, 16, 51] but its location within the complex has yet to be precisely established.
One proposed placement of PufX is at the dimerizing junction of RC-LH1-PufX.[37] Since two PufX helices are expected per RC-LH1-PufX complex,[52] this placement requires the dimerization of PufX, yet only monomeric structures of PufX have been solved.[53, 54] To test if a dimeric PufX is feasible, computational modeling and MD simulations were performed.[55] A model for PufX dimer was constructed based on the suggestion that there is a dimerization motif in the PufX sequence.[56, 57] Subsequent all-atom MD simulations of this PufX dimer model, placed in a membrane and water box, demonstrated that the dimer remained structurally stable over the 50 ns simulated, and that the PufX helices maintained a uniform crossing-angle of θPufX ~ 38° (Figure 6).[55] Assuming that such a PufX dimer indeed serves as the nucleation point of the RC-LH1-PufX assembly, and that the LH1 helices remain parallel to the PufX monomer on each side of the RC-LH1-PufX dimer, the overall bending of the RC-LH1-PufX complex arises naturally. This scheme explains the geometry of the Rba. sphaeroides RC-LH1-PufX dimer; the definitive answer to the question where PufX is located in the LH1-RC-PufX dimer, however, requires a full atomic-level structure of the complex.
Figure 6.
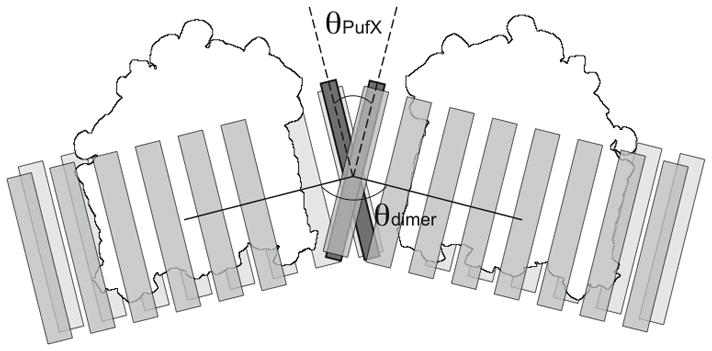
PufX dimerization and the arrangement of Rba. sphaeroides RC-LH1-PufX complex. A pair of dimerized PufX helices are shown to be located at the center of the RC-LH1-PufX complex. In this scheme, the crossing angle of the PufX helices, θPufX, leads to the bending angle of the RC-LH1-PufX, θdimer.[55] PufX shown in dark gray, LH1 in light gray, and RC in white.
Formation of chromatophores upon assembly of LH complexes
Computational studies have established that both Rba. sphaeroides LH2 and RC-LH1-PufX complexes are membrane-curving elements.[28, 34, 46] Assembly of these complexes should then produce chromatophores with a global curvature. Demonstration of the dynamic chromatophore formation process requires simulation of hundreds of proteins, and becomes computationally costly if performed at atomic resolution. The problem of computational cost can be avoided by using coarse-grained representations, assuming that the simplified model correctly replicates the membrane-curving properties of the protein complexes. An example of a successful application is the shape-based coarse-graining method used in demonstrating the membrane-sculpting effect of an array of BAR domain proteins.[5, 7, 8]
For the case of assembly of the photosynthetic chromatophores, a set of Monte Carlo simulations were performed.[58] In each simulation, a collection of membrane-curving “beads” mimicking the size and geometry of the LH2 and RC-LH1-PufX complexes were placed on a flexible surface. Each bead, as well as certain bead-to-bead interaction, was designed to produce a specific local curvature; bending of the membrane was ascribed an energetic cost calculated through the Helfrich free energy.[59, 2] An originally flat membrane consisting of a random mixture of LH2 and RC-LH1-PufX beads was seen to become vesicular as it approached equilibration, forming segregated domains.[58] A separate set of coarse-grained MD simulations similarly showed that in a membrane populated with curvature-inducing capsids, a vesicle spontaneously formed as attractive interactions between capsids were observed.[60, 61] Altogether, the formation of chromatophores is possibly a result of the spontaneous aggregation of the protein elements, which occurs to minimize the energy penalty of bending the membrane (Figure 7).
Figure 7.

Schematic diagram displaying curvature-mediated “attraction” between two membrane-bending proteins. In a), two proteins mimicking the curvature effect of the Rba. sphaeroides RC-LH1-PufX complex are placed far apart in the membrane, with each protein bending its surrounding membrane that requires energy ΔE. The shaded area in the membrane indicates the most prominent local curvature. In b), the two proteins are stacked together, and in this arrangement there is less membrane bending, indicated by fewer shaded areas. The energy required to bend the membrane is, therefore, less than the scenario depicted in a). Detailed theoretical calculations of the interactions between membrane-curving proteins can be found in several studies, e.g. Kim et al., 1998 and Chou et al., 2001.[62, 63]
Why do photosynthetic chromatophores self-assemble?
Computational studies over the past few years on the curvature properties of photosynthetic proteins [58, 28, 34, 46, 55] are providing a plausible mechanism for the self-assembly of the chromatophores that can be a general principle for the formation of cellular organelles. The protein components of the chromatophore not only serve photosynthetic functions, but each also possesses unique membrane-shaping properties. Since bending the membrane comes with an energetic cost, the optimal arrangement for these particular membrane-bending elements is a tightly-packed aggregate, resulting in the formation of a well-defined functional unit.
Formation of the photosynthetic chromatophore is not only relevant in terms of membrane architecture, but also provides functional advantages. For example, the electronic excitation resulting from light absorption has to be transferred to an RC for charge separation and further processing. A closer packing between the light-harvesting complexes and the RC facilitates this process by lowering the excitation transfer times between complexes, and as a result increasing the overall quantum efficiency of the chromatophore.[27, 58, 45] Another example concerns the subsequent steps of photosynthesis, which involves diffusions of quinones and cytochrome c2. These processes are also likely more efficient in a connected, compact compartment of the cell such as the chromatophore as opposed to having the constituent proteins loosely distributed over a large membrane area.[16] The advantages of compartmentalization become readily evident by a comparison with the plant and cyanobacterial systems.[64, 65] In evolutionarily more advanced oxygenic photosynthetic species,[66] not only do the individual light-harvesting complexes display a greater packing density of pigments (per amino acid), but also the constituent proteins are organized in a dense arrangement in the form of thylakoid membranes.[67] Formation of densely packed cellular compartments seem to arise as a result of evolutionary pressures. It appears that cellular functional units arise readily upon the assembly of their protein components, and the formation of functional units in turn facilitates the work of the proteins, interweaving structure and function into a complex relationship.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Health (P41-RR005969) and the National Science Foundation (MCB-0744057). All-atom MD simulations discussed here were performed using the package NAMD [68], with computer time provided by NCSA and TACC via Large Resources Allocation Committee grant MCA93S028, and resources of the Argonne Leadership Computing Facility at Argonne National Laboratory, supported by the Office of Science of the U.S. DOE (contract DE-AC02-06CH11357). The authors thank the following collaborators for many insightful and inspirational discussions and guidance: C. Neil Hunter, Pu Qian, Christophe Chipot, and John D. Stack. Molecular images in this article were rendered using the molecular visualization software VMD [69].
References
- 1.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 3.Shnyrova AV, Frolov VA, Zimmerberg J. Domain-driven morphogenesis of cellular membranes. Curr Biol. 2009;19:R772–R780. doi: 10.1016/j.cub.2009.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deserno Markus. Mesoscopic membrane physics: Concepts, simulations, and selected applications. Macromol Rapid Comm. 2009;30:752–771. doi: 10.1002/marc.200900090. [DOI] [PubMed] [Google Scholar]
- 5.Arkhipov Anton, Yin Ying, Schulten Klaus. Four-scale description of membrane sculpting by BAR domains. Biophys J. 2008;95:2806–2821. doi: 10.1529/biophysj.108.132563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein ML, Shinoda W. Large-scale molecular dynamics simulations of self-assembling systems. Science. 2008;321:798–800. doi: 10.1126/science.1157834. [DOI] [PubMed] [Google Scholar]
- 7.Yin Ying, Arkhipov Anton, Schulten Klaus. Simulations of membrane tubulation by lattices of amphiphysin N-BAR domains. Structure. 2009;17:882–892. doi: 10.1016/j.str.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arkhipov Anton, Yin Ying, Schulten Klaus. Membrane-bending mechanism of amphiphysin N-BAR domains. Biophys J. 2009;97:2727–2735. doi: 10.1016/j.bpj.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oelze J, Drews G. Membranes of photosynthetic bacteria. Biochim Biophys Acta. 1972;265:209–239. doi: 10.1016/0304-4157(72)90003-2. [DOI] [PubMed] [Google Scholar]
- 10.Kiley Patricia J, Varga Amy, Kaplan Samual. Physiological and structural analysis of light-harvesting mutants of Rhodobacter sphaeroides. J Bacteriol. 1988;170(3):1103–1115. doi: 10.1128/jb.170.3.1103-1115.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter CN, Pennoyer JD, Sturgis JN, Farrelly D, Niederman RA. Oligomerization states and associations of light-harvesting pigment protein complexes of Rhodobacter sphaeroides as analyzed by lithium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry. 1988;27:3459–3467. [Google Scholar]
- 12.Kiley Patricia J, Kaplan Samuel. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol Rev. 1988;52:50–69. doi: 10.1128/mr.52.1.50-69.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sturgis James N, Niederman Robert A. The effect of different levels of the B800-B850 light-harvesting complex on intracytoplasmic membrane development in Rhodobacter sphaeroides. Arch Microbiol. 1996;165:235–242. doi: 10.1007/s002030050321. [DOI] [PubMed] [Google Scholar]
- 14.Olsen JD, Tucker JD, Timney JA, Qian P, Vassilev C, Hunter CN. The organization of LH2 complexes in membranes from Rhodobacter sphaeroides. J Biol Chem. 2008;283:30772–30779. doi: 10.1074/jbc.M804824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Xiche, Ritz Thorsten, Damjanović Ana, Autenrieth Felix, Schulten Klaus. Photosynthetic apparatus of purple bacteria. Quart Rev Biophys. 2002;35:1–62. doi: 10.1017/s0033583501003754. [DOI] [PubMed] [Google Scholar]
- 16.Cogdell RJ, Gall A, Köhler J. The architecture and function of the light-harvesting apparatus of purple bacteria: from single molecules to in vivo membranes. Quart Rev Biophys. 2006;39:227–324. doi: 10.1017/S0033583506004434. [DOI] [PubMed] [Google Scholar]
- 17.Jamieson SJ, Wang P, Qian P, Kirkland JY, Conroy MJ, Hunter CN, Bullough PA. Projection structure of the photosynthetic reaction centre-antenna complex of Rhodospirillum rubrum at 8.5 Å resolution. J Mol Biol. 2002;21:3927–3935. doi: 10.1093/emboj/cdf410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Xiche, Schulten Klaus. A model for the light-harvesting complex I (B875) of Rhodobacter sphaeroides. Biophys J. 1998;75:683–694. doi: 10.1016/S0006-3495(98)77558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crofts Antony R, Hong Sangjin, Zhang Zhaolei, Berry Edward A. Physicochemical aspects of the movement of the Rieske iron sulphur protein during quinol oxidation by bc1 complex from mitochondria and photosynthetic bacteria. Biochemistry. 1998;38:15827–15839. doi: 10.1021/bi990963e. [DOI] [PubMed] [Google Scholar]
- 20.Axelrod HL, Abresch EC, Okamura MY, Yeh AP, Ress DC, Feher G. X-ray structure determination of the cytochrome c2: Reaction center electron transfer complex from Rhodobacter sphaeroides. J Mol Biol. 2002;319:501–515. doi: 10.1016/S0022-2836(02)00168-7. [DOI] [PubMed] [Google Scholar]
- 21.Feniouk BA, Cherepanov DA, Voskoboynikova NE, Mulkidjanian AY, Junge W. Chromatophore vesicles of Rhodobacter capsulatus contain on average one FoF1-ATP synthase each. Biophys J. 2002;82:1115–1122. doi: 10.1016/S0006-3495(02)75470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gubellinia Francesca, Franciaa Francesco, Turinaa Paola, Levy Daniel, Venturolia Giovanni, Melandria Andrea. Heterogeneity of photosynthetic membranes from Rhodobacter capsulatus: size dispersion and ATP synthase distribution. Biochim Biophys Acta. 2007;1767:1340–1352. doi: 10.1016/j.bbabio.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Sturgis James N, Tucker Jaimey D, Olsen John D, Hunter C Neil, Niederman Robert A. Atomic force microscopy studies of native photosynthetic membranes. Biochemistry. 2009;48:3679–3698. doi: 10.1021/bi900045x. [DOI] [PubMed] [Google Scholar]
- 24.Jungas C, Ranck JL, Rigaud JL, Joliot P, Verméglio A. Supramolecular organization of the photosynthetic apparatus of Rhodobacter sphaeroides. EMBO J. 1999;18(3):534–542. doi: 10.1093/emboj/18.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siebert CA, Qian P, Fotiadis D, Engel A, Hunter CN, Bullough PA. Molecular architecture of photosynthetic membranes in Rhodobacter sphaeroides: the role of PufX. EMBO J. 2004;23:690–700. doi: 10.1038/sj.emboj.7600092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian Pu, Bullough Per A, Hunter C Neil. Three-dimensional reconstruction of a membrane-bending complex: The RC-LH1-PufX core dimer of Rhodobacter sphaeroides. J Biol Chem. 2008;283:14002–14011. doi: 10.1074/jbc.M800625200. [DOI] [PubMed] [Google Scholar]
- 27.Sener Melih K, Olsen John D, Hunter C Neil, Schulten Klaus. Atomic level structural and functional model of a bacterial photosynthetic membrane vesicle. Proc Natl Acad Sci USA. 2007;104:15723–15728. doi: 10.1073/pnas.0706861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandler Danielle, Hsin Jen, Harrison Christopher B, Gumbart James, Schulten Klaus. Intrinsic curvature properties of photosynthetic proteins in chromatophores. Biophys J. 2008;95:2822–2836. doi: 10.1529/biophysj.108.132852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheuring S, Lévy D, Rigaud JL. Watching the components of photosynthetic bacterial membranes and their in situ organisation by atomic force microscopy. Biochim Biophys Acta. 2005;1712:109–127. doi: 10.1016/j.bbamem.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Scheuring S. AFM studies of the supramolecular assembly of bacterial photosynthetic core-complexes. Curr Opin Struct Biol. 2006;10:1–7. doi: 10.1016/j.cbpa.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Koepke Juergen, Hu Xiche, Muenke Cornelia, Schulten Klaus, Michel Hartmut. The crystal structure of the light harvesting complex II (B800-850) from Rhodospirillum molischianum. Structure. 1996;4:581–597. doi: 10.1016/s0969-2126(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 32.Papiz Miroslav Z, Prince Steve M, Howard Tina, Cogdell Richard J, Isaacs Neil W. The structure and thermal motion of the B800-850 LH2 complex from Rps. acidophila at 2.0 Å resolution and 100 K: New structural features and functionally relevant motions. J Mol Biol. 2003;326:1523–1538. doi: 10.1016/s0022-2836(03)00024-x. [DOI] [PubMed] [Google Scholar]
- 33.Lee Eric H, Hsin Jen, Sotomayor Marcos, Comellas Gemma, Schulten Klaus. Discovery through the computational microscope. Structure. 2009;17:1295–1306. doi: 10.1016/j.str.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandler Danielle E, Gumbart James, Stack John D, Chipot Christophe, Schulten Klaus. Membrane curvature induced by aggregates of LH2s and monomeric LH1s. Biophys J. 2009;97:2978–2984. doi: 10.1016/j.bpj.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varga Amy R, Staehelin L Andrew. Membrane adhesion in photosyntetic bacterial membranes. Light harvesting complex I (LH1) appears to be the main adhesion factor. Arch Microbiol. 1985;141:290–296. doi: 10.1007/BF00428839. [DOI] [PubMed] [Google Scholar]
- 36.Qian P, Hunter CN, Bullough PA. The 8.5 Å projection structure of the core RC-LH1-PufX dimer of Rhodobacter sphaeroides. J Mol Biol. 2005;349:948–960. doi: 10.1016/j.jmb.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 37.Scheuring Simon, Francia Francesco, Busselez Johan, Melandris Bruno Andrea, Rigaud Jean-Louis, Levy Daniel. Structural role of PufX in the dimerization of the photosynthetic core complex of Rhodobacter sphaeroides. J Biol Chem. 2004;279(5):3620–3626. doi: 10.1074/jbc.M310050200. [DOI] [PubMed] [Google Scholar]
- 38.Wells David, Abramkina Volha, Aksimentiev Aleksei. Exploring transmembrane transport through alpha-hemolysin with grid-steered molecular dynamics. J Chem Phys. 2007;127:125101–125101-10. doi: 10.1063/1.2770738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trabuco Leonardo G, Villa Elizabeth, Mitra Kakoli, Frank Joachim, Schulten Klaus. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trabuco Leonardo G, Villa Elizabeth, Schreiner Eduard, Harrison Christopher B, Schulten Klaus. Molecular Dynamics Flexible Fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villa Elizabeth, Sengupta Jayati, Trabuco Leonardo G, LeBarron Jamie, Baxter William T, Shaikh Tanvir R, Grassucci Robert A, Nissen Poul, Ehrenberg Måns, Schulten Klaus, Frank Joachim. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gumbart James, Trabuco Leonardo G, Schreiner Eduard, Villa Elizabeth, Schulten Klaus. Regulation of the protein-conducting channel by a bound ribosome. Structure. 2009;17:1453–1464. doi: 10.1016/j.str.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becker Thomas, Mandon Elisabet, Bhushan Shashi, Jarasch Alexander, Armache Jean-Paul, Funes Soledad, Jossinet Fabrice, Gumbart James, Mielke Thorsten, Berninghausen Otto, Schulten Klaus, Westhof Eric, Gilmore Reid, Beckmann Roland. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seidelt Birgit, Innis C Axel, Wilson Daniel N, Gartmann Marco, Armache Jean-Paul, Villa Elizabeth, Trabuco Leonardo G, Becker Thomas, Mielke Thorsten, Schulten Klaus, Steitz Thomas A, Beckmann Roland. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sener Melih K, Hsin Jen, Trabuco Leonardo G, Villa Elizabeth, Qian Pu, Hunter C Neil, Schulten Klaus. Structural model and excitonic properties of the dimeric RC-LH1-PufX complex from Rhodobacter sphaeroides. Chem Phys. 2009;357:188–197. doi: 10.1016/j.chemphys.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsin Jen, Gumbart James, Trabuco Leonardo G, Villa Elizabeth, Qian Pu, Hunter C Neil, Schulten Klaus. Protein-induced membrane curvature investigated through molecular dynamics flexible fitting. Biophys J. 2009;97:321–329. doi: 10.1016/j.bpj.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roszak AW, Howard TD, Southall J, Gardiner AT, Law CJ, Isaacs NW, Cogdell RJ. Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris. Science. 2003;302:1969–1972. doi: 10.1126/science.1088892. [DOI] [PubMed] [Google Scholar]
- 48.Fotiadis D, Qian P, Philippsen A, Bullough PA, Engel A, Hunter CN. Structural analysis of the reaction center light-harvesting complex I photosynthetic core complex of Rhodospirillum rubrum using atomic force microscopy. J Biol Chem. 2004;279:2063–2068. doi: 10.1074/jbc.M310382200. [DOI] [PubMed] [Google Scholar]
- 49.Richter Martin F, Baier Jürgen, Southall June, Cogdell Richard J, Oellerich Silke, Köhler Jürgen. Refinedment of the x-ray structure of the RC–LH1 core complex from Rhodopseudomonas palustris by single-molecule spectroscopy. Proc Natl Acad Sci USA. 2007;104:20280–20284. doi: 10.1073/pnas.0704599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frese RN, Olsen JD, Branvall R, Westerhuis WHJ, Hunter CN, van Grondelle R. The long-range supraorganization of the bacterial photosynthetic unit: A key role for PufX. Proc Natl Acad Sci USA. 2000;97:5197–5202. doi: 10.1073/pnas.090083797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holden-Dye K, Crouch LI, Jones MR. Structure, function and interactions of the PufX protein. Biochim Biophys Acta. 2008;1777:613–630. doi: 10.1016/j.bbabio.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 52.Abresch EC, Axelrod HLA, Beatty JT, Johnson JA, Nechushtai R, Paddock ML. Characterization of a highly puried, fully active, crystallizable RC-LH1-PufX core complex from Rhodobacter sphaeroides. Photosyn Res. 2005;86:61–70. doi: 10.1007/s11120-005-5106-z. [DOI] [PubMed] [Google Scholar]
- 53.Wang ZY, Suzuki H, Kobayashi M, Nozawa T. Solution structure of the Rhodobacter sphaeroides PufX membrane protein: Implications for the quinone exchange and protein-protein interactions. Biochemistry. 2007;46:3635–3642. doi: 10.1021/bi0618060. [DOI] [PubMed] [Google Scholar]
- 54.Tunnicliffe RB, Ratcliffe EC, Hunter CN, Williamson MP. The solution structure of the PufX polypeptide from Rhodobacter sphaeroides. FEBS Lett. 2006;580:6967–6971. doi: 10.1016/j.febslet.2006.11.065. [DOI] [PubMed] [Google Scholar]
- 55.Hsin Jen, Chipot Chris, Schulten Klaus. A glycophorin A-like framework for the dimerization of photosynthetic core complexes. J Am Chem Soc. 2009;131:17096–17098. doi: 10.1021/ja905903n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 57.Busselez J, Cottevielle M, Cuniasse P, Gubellini F, Boisset N, Lévy D. Structural basis for the PufX-mediated dimerization of bacterial photosynthetic core complexes. Structure. 2007;15:1674–1683. doi: 10.1016/j.str.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 58.Frese RN, Pàmies JC, Olsen JD, Bahatyrova S, van der Weij-de Wit CD, Aartsma TJ, Otto C, Hunter CN, Frenkel D, van Grondelle R. Protein shape and crowding drive domain formation and curvature in biological membranes. Biophys J. 2008;94:640–647. doi: 10.1529/biophysj.107.116913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z Naturforsch. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 60.Reynwar BJ, Illya G, Harmandaris VA, Müller MM, Kremer K, Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–464. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- 61.Murtola Teemu, Bunker Alex, Vattulainen Ilpo, Deserno Markus, Karttunen Mikko. Multiscale modeling of emergent materials: biological and soft matter. Phys Chem Chem Phys. 2009;11:1869–1892. doi: 10.1039/b818051b. [DOI] [PubMed] [Google Scholar]
- 62.Kim KS, Neu J, Oster G. Curvature-mediated interactions between membrane proteins. Biophys J. 1998;75:2274–2291. doi: 10.1016/S0006-3495(98)77672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chou Tom, Kim Ken S, Oster George. Statistical thermodynamics of membrane bending-mediated protein-protein attractions. Biophys J. 2001;80:1075–1087. doi: 10.1016/S0006-3495(01)76086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sener Melih K, Jolley Craig, Ben-Shem Adam, Fromme Petra, Nelson Nathan, Croce Roberta, Schulten Klaus. Comparison of the light harvesting networks of plant and cyanobacterial photosystem I. Biophys J. 2005;89:1630–1642. doi: 10.1529/biophysj.105.066464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nelson Nathan, Yocum Charles F. Structure and function of photosystems I and II. Annu Rev Plant Biol. 2006;57:521–565. doi: 10.1146/annurev.arplant.57.032905.105350. [DOI] [PubMed] [Google Scholar]
- 66.Xiong J, Fischer WM, Inoue K, Nakahara M, Bauer CE. Molecular evidence for the early evolution of photosynthesis. Science. 2000;289:1724–1730. doi: 10.1126/science.289.5485.1724. [DOI] [PubMed] [Google Scholar]
- 67.Dekker Jan P, Boekema Egbert J. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta – Bioener. 2005;1706:12–39. doi: 10.1016/j.bbabio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 68.Phillips James C, Braun Rosemary, Wang Wei, Gumbart James, Tajkhorshid Emad, Villa Elizabeth, Chipot Christophe, Skeel Robert D, Kale Laxmikant, Schulten Klaus. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Humphrey William, Dalke Andrew, Schulten Klaus. VMD – Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


