Abstract
Cancer genomes contain many aberrant gene fusions—a few that drive disease and many more that are nonspecific passengers. We developed an algorithm (the concept signature or ‘ConSig’ score) that nominates biologically important fusions from high-throughput data by assessing their association with ‘molecular concepts’ characteristic of cancer genes, including molecular interactions, pathways and functional annotations. Copy number data supported candidate fusions and suggested a breakpoint principle for intragenic copy number aberrations in fusion partners. By analyzing lung cancer transcriptome sequencing and genomic data, we identified a novel R3HDM2-NFE2 fusion in the H1792 cell line. Lung tissue microarrays revealed 2 of 76 lung cancer patients with genomic rearrangement at the NFE2 locus, suggesting recurrence. Knockdown of NFE2 decreased proliferation and invasion of H1792 cells. Together, these results present a systematic analysis of gene fusions in cancer and describe key characteristics that assist in new fusion discovery.
Gene fusions resulting from chromosomal rearrangements often define molecular subtypes of cancers and appear as initial events in oncogenesis1. The discovery of recurrent fusions in common epithelial cancers2,3 has stimulated a widespread search for novel gene fusions. Yet, new fusion discovery and molecular targeting of known fusions is complicated by the complex biological behavior displayed by fusion genes. First, most genes involved in fusions recombine with many different partners, forming interrelated gene fusion networks4. Second, recurrent gene fusions in carcinomas are often found in the background of many nonspecific gene fusions, which illustrates the karyotypic complexity of solid tumor evolution. Distinguishing nonspecific (passenger) fusions from recurrent (driver) fusions is a formidable task. In this study, we sought to investigate the functional and genetic landscape of fusion genes and characterize fundamental principles to help facilitate new gene fusion discovery from large-scale genomic data and next-generation sequencing data.
RESULTS
Understanding the recombination of fusion partners
To determine common characteristics of fusion gene recombinations, we explored the hypothesis that fusion genes sharing a common partner might share common domain architectures. Using GenBank, we extracted core nucleotide sequences of chimeras representing known fusions. Open reading frames and their domain architectures were determined using the Entrez Gene conserved domain database. The resulting unique domain architectures were clustered by domain similarities, enabling the global analysis of domain recombination in gene fusions (Supplementary Fig. 1, Supplementary Table 1). Interestingly, the domain architectures of fusion proteins are very diverse, especially for 5′ partners. In addition, clustering gene fusions according to their domain architectures resulted in few pathologically related clusters; the majority of the clusters did not show tumor-entity specificity. This suggested the possible existence of other major factors influencing fusion gene recombinations, such as preferential selection for shared pathways or gene ontologies.
We compiled pathway data from Reactome5, Kyoto Encyclopedia of Genes and Genomes (KEGG)6 and Biocarta, and analyzed the shared pathways within fusion partner groups. However, most fusion genes with a mutual partner are involved in distinct cell signaling pathways (data not shown).
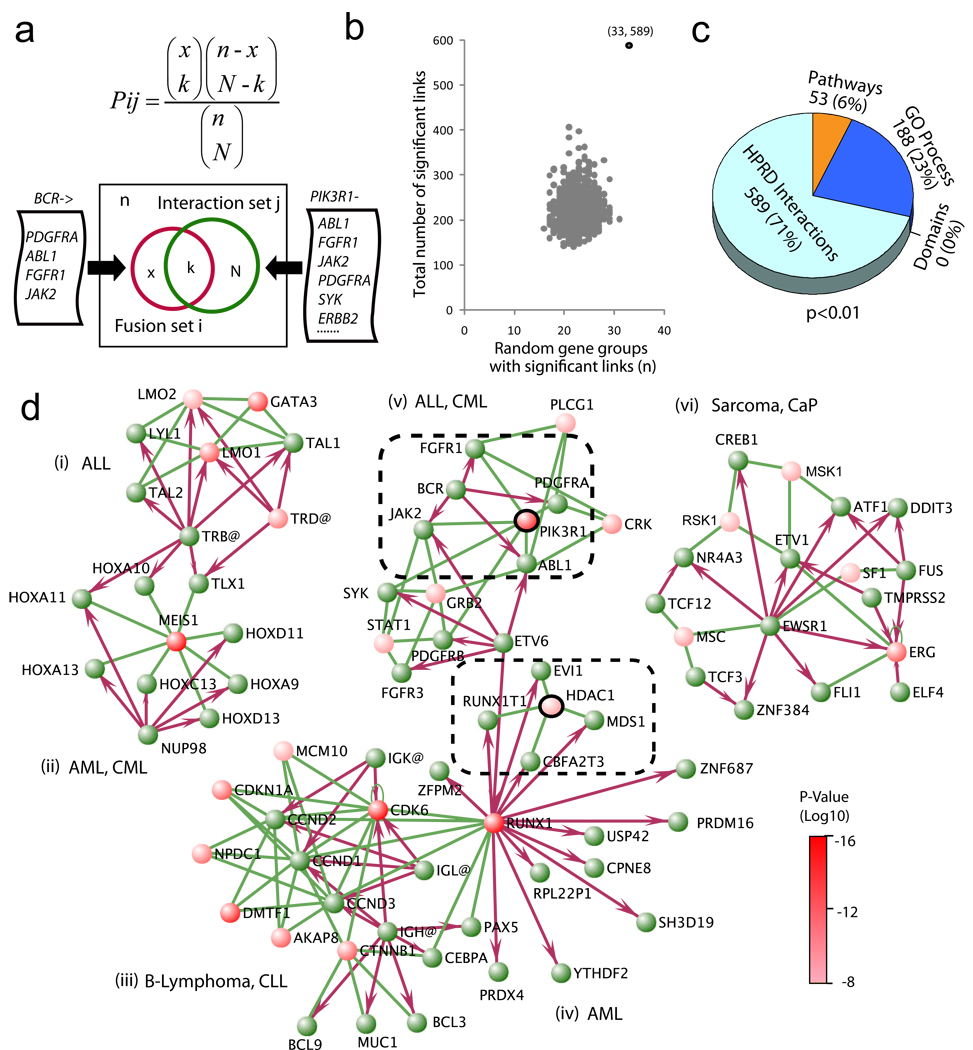
Yet, because canonical pathways may not encompass the complexities of cell biology, we interrogated a molecular interaction database to generate a comprehensive view of cancer signaling. We derived 90 fusion partner groups from the Mitelman database and mapped these to the molecular interaction network extracted from the Human Protein Reference Database (HPRD)7. For all human genes in the database, we defined the interaction gene set J to be all genes that interact with gene j. If we denote a given fusion gene and its fusion partners as i and I, respectively, we can then individually test the significance of overlap between every set of fusion partners I with every gene interaction set J using the hypergeometric distribution (Fig. 1a). In aggregate, this analysis yielded a total of 589 genes whose interacting genes were enriched for genes in 33 out of 90 fusion partner groups in the Mitelman database (P < 0.01). The top shared interacting genes are supplied in Supplementary Table 2.
Figure 1. Exploring cancer-related gene fusions in the context of known molecular interaction networks.
(a) The hypergeometric statistics for assessing whether a group of fusion gene partners (e.g., all BCR partners) contains an unexpected number of genes that physically interact with the same gene (e.g., all genes that interact with PIK3R1). (b) The total number of significant links (589) and the number of fusion partner groups having these links (33) were plotted with the distribution calculated from randomly chosen gene sets with an equal amount of connectivity (1,000 permutations). (c) Analysis of the fusion partner groups with a compendium of molecular concepts by hypergeometric statistics. The numbers in the pie chart represent the number of significant concepts in each functional category (P ≤ 0.01). (d) Network visualization of the most significant (P < 10−7) instances where many fusion partners also interact with a shared gene. Fusion genes are green nodes; shared interacting genes are red (with color intensity indicating P-value). Red arrows designate gene fusions (from 5’ partners to 3’ partners); green lines represent molecular interactions. For simplicity, genes and proteins are both given in roman type in the diagram. For each fusion partner set, the shared interacting gene having the most significant P-value was designated as a fusion-interaction hub. Clusters are limited to known fusions joined by established molecular interactions. (i) Acute lymphoblastic lymphoma (ALL) fusions with a hub of GATA3. (ii) Acute/chronic myelogenous leukemia (AML and CML) fusions with a hub of MEIS1. (iii) B-cell lymphoma and chronic lymphoblastic lymphoma (CLL) fusions through the hubs of CDK6 and CTNNB1. (iv) AML fusions partially focusing on HDAC1; RUNX1 is the hub of immunoglobulin fusions, and also involved in multiple fusions in AML, and thus links clusters iii and iv. (v) ALL and CML fusions through the hub of PIK3R1. (vi) Sarcoma and prostate cancer fusions around ERG and MSK1. The PI3K3R1 and HDAC1 hubs are within dashed lines.
To test whether fusion genes are significantly enriched for mutual interacting genes, we randomly chose 90 gene sets with an equivalent level of connectivity as the fusion partner groups (Online Methods), and determined the extent to which they were linked by mutual interacting genes. This process was repeated 1,000 times, and then the total number of significant links and the number of gene groups having these links were plotted (Fig. 1b). The number of links generated is significantly greater for fusion genes, validating our observation (P < 0.001).
To systematically evaluate the importance of shared interacting genes in fusion gene recombinations, we applied these statistics to the pooled domains, pathways, Gene Ontology (GO) database biological process and HPRD interactions data. We refer to these data sets collectively as ‘molecular concepts’ (Table 1). We benchmarked each of these classes by statistically assessing the number of molecular concepts shared by fusion partner groups (Fig. 1c). By setting the P value threshold to 0.01, the HPRD interactions data yielded 589 unique explanations, while pathway data and GO general process terms gave only 53 and 188, respectively.
Table 1. The compendia of molecular concepts for integrative functional analysis of fusion genes.
Four classes of molecular concepts were compiled from 6 sources. Connectivity represents the total number of concept to gene connections in each concept type.
| Class | Source | Web link | Type | Concepts (n) |
Connectivity (n) |
|---|---|---|---|---|---|
| Annotation | Gene Ontology | http://www.geneontology.org | Biologic process | 3920 | 46530 |
| Cellular component | 732 | 42463 | |||
| Molecular function | 2561 | 47026 | |||
| Pathways | Biocarta | http://cgap.nci.nih.gov/Pathways | Signaling pathways | 263 | 4459 |
| KEGG | http://www.genome.jp/kegg | Metabolic pathways | 112 | 2985 | |
| Signaling pathways | 2456 | 52238 | |||
| Reactome | http://www.reactome.com | Biochemical reactions | 5450 | 44347 | |
| Interactions | HPRD | http://www.hprd.org | Protein interaction sets | 7819 | 37206 |
| Domains | Entrez Gene | http://www.ncbi.nlm.nih.gov/gene | Conserved domains | 5650 | 5693 |
We next focused on the network of the most significant fusion-interaction (FI) links. We visualized fusion-interaction networks using the VisANT program8 and found six major clusters of interactions that connected gene fusions from similar tumor entities (Fig. 1d). The shared interacting genes with the greatest statistical significance in each subset of connected fusions were designated as ‘fusion-interaction hubs’ in each cluster. For example, BCR has four 3′ partners (ABL1, FGFR1, JAK2 and PDGFRA), all of which interact with PIK3R1, one of the fifteen subunits encoding PI3K (P = 9.54 × 10−11). This finding suggested that BCR fusion partners interact with and presumably activate PIK3R1 as part of leukemogenesis, which we confirmed by mining the literature9–14. These results show the utility of the fusion-interaction networks in elucidating fusion biology by distinguishing key genes that serve as network hubs with functional importance in mediating fusion signaling (See Supplementary Discussion).
Quantification of concept signatures
The fact that cancer-related fusion partner groups tend to cluster around shared interacting genes or share common gene ontologies prompted us to generalize this finding to develop a method that could filter out nonspecific gene fusions. We hypothesized that such ‘signatures’ of molecular concepts frequently found in fusion genes may be used to define biologically meaningful gene fusions underlying cancer, similar to signature genes defining certain phenotypes. This requires a systematic characterization of all fusion genes as a coherent group from multiple functional perspectives.
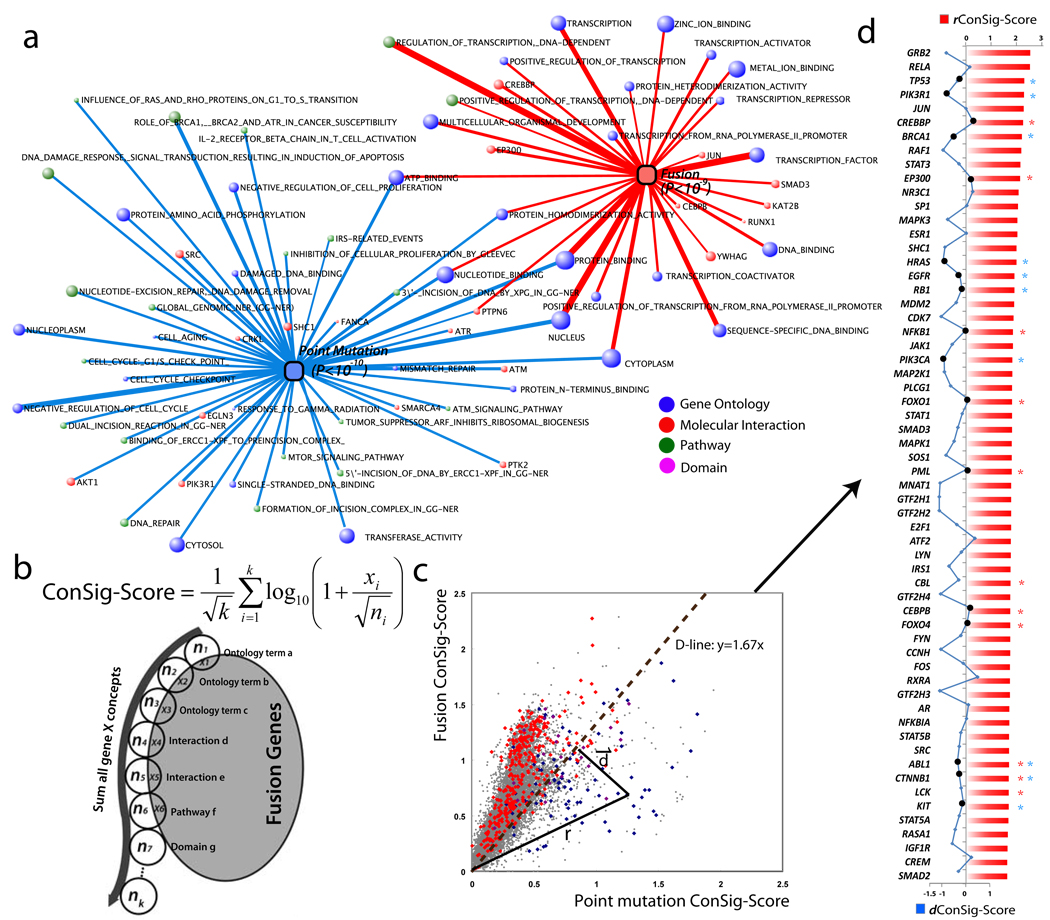
To benchmark the functional characteristics of fusion genes, we compared fusion genes to point mutation genes in cancer. We used Fisher’s exact test to identify molecular concepts enriched for fusion genes and concepts enriched for point mutations, generating two sets of minimally overlapping concepts (Fig. 2a). Fusion genes were enriched for molecular concepts related to signal transduction and transcription activation; in contrast, mutation genes were enriched for molecular concepts related to DNA repair and cell cycle checkpoints. Thus, we defined these two sets as ‘concept signatures’—a fusion concept signature and a mutation concept signature.
Figure 2. Distinguishing biological features of gene fusions and point mutations in cancer.
(a) Enrichment analysis with a compendium of molecular concepts generates two sets of minimally overlapping signature concepts for fusion and point mutation genes. Molecular concepts are depicted as nodes with the size of each node corresponding to the number of genes in each concept. The thickness of the lines correlates with the significance of overlap tested by hypergeometric statistics. (b) The ConSig algorithm. Circles represent concepts associated with gene X. k is the total number of concepts associated with gene X, xi = number of fusion genes in concept i, ni = total genes in concept i. A corresponding figure for mutation genes is not shown. (c) Fusion and mutation ConSig scores for known fusion genes (red dots), cancer point mutation genes (blue dots) and all other human genes (gray dots). Genes known to be both fusion and mutation genes are purple. r, r.ConSig score; d, d.ConSig score, D-line, distinction line. (d) Identifying the top 60 genes rated by r.ConSig score (red column chart) produced a list highly enriched for established cancer genes (known fusion genes labeled with red stars; mutation genes blue stars). The d.ConSig scores are depicted by a blue line chart, with positive and negative values indicating the dots above or below the D-line. Known mutation or fusion genes matching the prediction by d.ConSig scores are marked by black dots in the line chart.
Using these two concept signatures, we hypothesized that genes involved in fusions or point mutations could be distinguished from each other and from the remaining human genes. We designed an algorithm, termed concept signature score (ConSig score), to quantitatively rank genes underlying cancer by the strength of their association with the two concept signatures (Fig. 2b). The algorithm first determines the ‘relevance’ of each concept in a signature, where relevance is defined as log10 of the number of fusion (or point mutation) genes that are associated with that concept divided by the square root of the total number of genes in the concept. Then, the ‘fusion ConSig score’ of a gene is calculated by summing the relevances of the fusion signature concepts associated with the gene, normalized for the total number of assigned concepts k. The ‘mutation ConSig score’ is similarly calculated except using the concepts in the mutation signature.
An important step in this analysis was to remove the redundant information from the calculation of the ConSig score. First, to avoid redundant representation in the GO database, we subtracted the genes that appeared in the child ontologies from the parents. Second, to eliminate the bias from the gene itself in the overlap, we subtracted the seeding genes from the signature concepts during the calculation of their own ConSig score. Finally, to minimize the redundant information in the interactome and pathway databases, we have attempted to remove the pathways significantly overlapping with the molecular interactions (Fisher’s exact test, P < 0.01). However, this adjustment did not show an advantage over the unadjusted score (Supplementary Figure 2), therefore was not applied in the calculation of the ConSig score.
We calculated fusion and mutation ConSig scores for all known human genes. Plotting the fusion and mutation ConSig scores separated known fusion genes from mutation genes (Fig. 2c). The distinction line (D-line), y = 1.67x, was determined by testing optimal separation capacity, which separates 85% of mutation genes from 80% of fusion genes (Supplementary Figure 3). In this setting, the radius to the zero point is defined as the radial ConSig score of a gene (rConSig score), which indicates the strength of association with signature concepts of both fusion and mutation genes, thus implies the functional relevance of candidate genes in cancer. The distance vector from the node to the D-line, which illustrates a distinction between fusion and mutation genes, is defined as the distinction ConSig score (dConSig score). Rating all human genes by the rConSig score produced enrichment of established cancer genes in top-scoring genes, with the majority of fusion or mutation genes matching the prediction from the dConSig score (Fig. 2d). Replacing the fusion or mutation gene sets with random gene sets produced no enrichment of the randomly selected genes. Although the ConSig algorithm is able to segregate fusion genes and mutation genes, we propose that its main utility is in the identification of biologically important gene fusions from next-generation sequencing data, where a large number of candidate gene fusions hinder a quick discussion evaluation of their functional importance (See Supplementary Discussion).
Genetic characteristics of unbalanced fusion genes
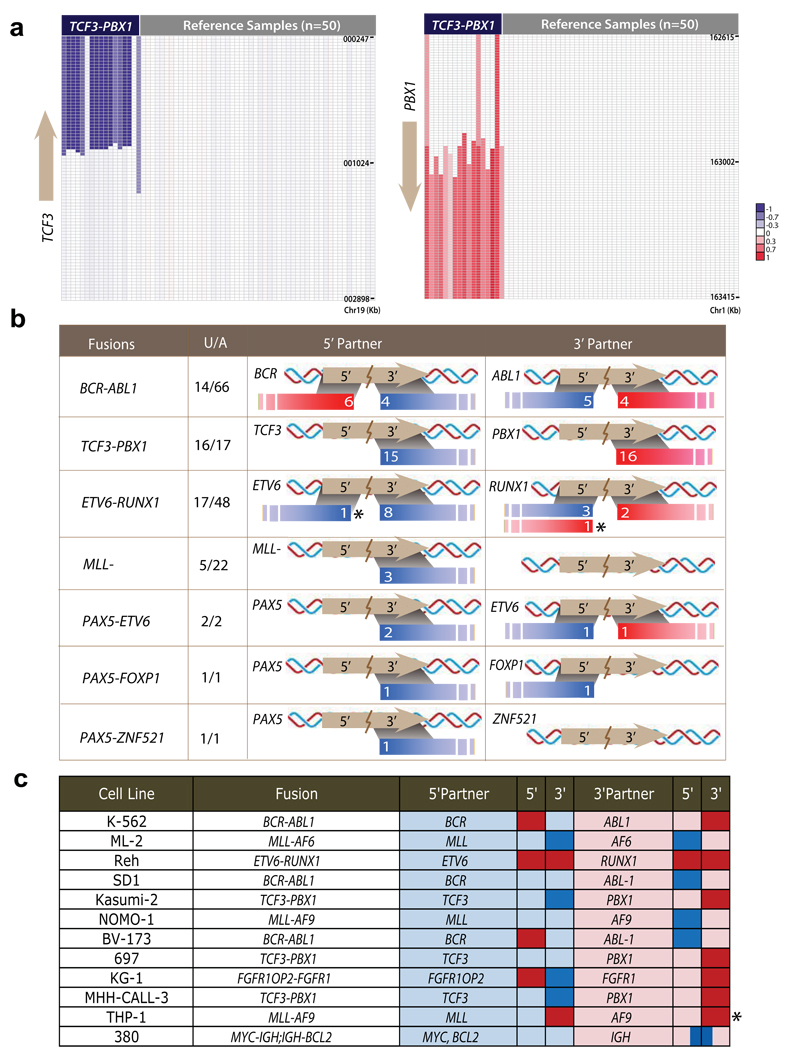
Having evaluated fusion genes by functional traits, we next used high-throughput copy number data to explore the genomic imbalance pattern that could inform unidentified gene fusions. Using leukemia as a genetic model, we studied the recurrent fusion genes in a high-resolution single nucleotide polymorphism (SNP) microarray data set with 304 leukemia samples15,16. A total of 157 samples are annotated with seven gene fusions in this data set (Supplementary Table 4). The percentage of unbalanced fusions ranged from 21.2–94.1% for different fusions, with most TCF3-PBX1 fusions identifiable by unbalanced breakpoints (Fig. 3a). The physical lengths of amplifications or deletions associated with fusion genes were 0.08–84.21 Mb (averaging 19.7 Mb). We observed a surprising heterogeneity in the genomic aberrations generating gene fusions. Often two fusion partners were found to possess different degrees of copy number gain or loss; elsewhere one fusion partner harbors a balanced translocation whereas the other partner has an unbalanced translocation.
Figure 3. Characterizing the genomic imbalances of recurrent gene fusions in acute lymphocytic leukemia.
An SNP array data set was used to evaluate genomic aberrations associated with gene fusions in acute lymphocytic leukemia (ALL). (a) The recurrent TCF3-PBX1 fusion (n = 17) was associated with deletion of the 3’ region of TCF3 and duplication of the 3’ region of PBX1. Color scales indicate log2 transformed relative copy number. (b) Of the 56 samples with unbalanced gene fusions in this data set, 55 samples conformed to the fusion breakpoint principle. Fusion genes on their corresponding chromosomes were aligned with the unbalanced breakpoints. “U/A” represents the number of samples with unbalanced fusions out of the total samples with gene fusions. Colored bars in the 5’ partner and 3’ partner columns indicate copy number increases (red) or deletions (blue), and given in white is the number of samples having the DNA aberration. For example, the first row shows that out of 66 cases of ALL patients with BCR-ABL1 fusions, 14 cases are unbalanced, of which 6 have 5’ BCR duplication and 4 have 3’ BCR deletion. ABL1 can be interpreted in a similar fashion. (c) Unbalanced fusions in the 12 leukemia cancer cell lines follow the fusion breakpoint principle. The exceptions to the principle are marked by asterisks.
Despite this diversity, an association analysis of unbalanced breakpoints with fusion gene placements revealed a consistent genetic pattern: copy number increases generally affect the 5′ region of 5′ partners and the 3′ region of 3′ partners, whereas deletions generally remove the 3′ region of 5′ fusion partners and the 5′ region of 3′ partners. Of 56 samples with 7 unbalanced fusions in this data set, 55 samples follow this pattern (Fig. 3b and Supplementary Table 5). We further analyzed the data for 36 leukemia cell lines15 and associated gene fusions from published sources17; 11 of 12 unbalanced fusions from these cell lines were found to follow this pattern (Fig. 3c and Supplementary Table 6). We termed this pattern the ‘fusion breakpoint principle’. Based on this reasoning, we can deduce an inferred principle for the unbalanced gene fusions within the same chromosome (Supplementary Figure 5a). For gene fusions having two partners on the same DNA strand, we define the fusion as ‘consistent’ if the genomic location of the two partners parallels their positions within the fusion transcript (that is, 5′ partner at the 5′ side of the 3′ partner), or ‘inconsistent’ if the two partners display the opposite genomic positioning (that is, 5′ partner at the 3′ side of the 3′ partner). Then, consistent fusions cannot be generated by a copy number increase, whereas inconsistent fusions cannot be generated by a deletion. For gene fusions having two partners on different strands (inversion), the fusion cannot be generated by simple interstitial deletions or copy number increases.
Although the fusion breakpoint principle can be inferred based on conventional cytogenetics, it should be stressed that unlike G-banding and fluorescence in situ hybridization (FISH), array-based high-throughput genomic data loses balanced genomic translocation information, and may misrepresent individual cases of complex genomic rearrangements (See Supplementary Discussion on the MLL-AF9 fusion). For this reason, extensive evidence from large numbers of malignancies is required to confirm the applicability of this principle to high-throughput genomic data.
To confirm the breakpoint principle, we performed a large-scale meta-analysis of recurrent gene fusions based on high-resolution array comparative genomic hybridization (array-CGH) and SNP array data sets annotated with gene fusions, as well as literature curation (Supplementary Table 8). In total, 276 tumor samples were identified as having unbalanced fusions, including 85 leukemia, 15 lymphoma, 23 sarcoma and 153 epithelial tumor samples. Although diverse breakpoint patterns were observed on these samples (Supplementary Fig. 5b), the unbalanced fusions from 273 samples conformed to the principle (98.9%). Furthermore, we also confirm the inferred principle by analyzing the reports for all unbalanced intrachromosome fusions from the Mitelman database (Supplementary Table 9).
An integrative approach to new fusion discovery
To demonstrate the application of those principles to new fusion discovery, we analyzed next-generation sequencing data and large-scale genomic data from lung cancer. First, we used the ConSig score to nominate biologically important fusion candidates from paired-end transcriptome data from lung cancer cell lines run in a single lane on an Illumina Genome Analyzer II flow cell. We extracted the chimeric paired reads from the paired-end libraries, and then ranked the 3′ partners by rConSig score. Second, the DNA breakpoints at the genomic loci of top candidate fusion genes were evaluated on the basis of the fusion breakpoint principle using publicly available lung cancer SNP array data encompassing a large number of tumor samples to search for recurrent rearrangements.
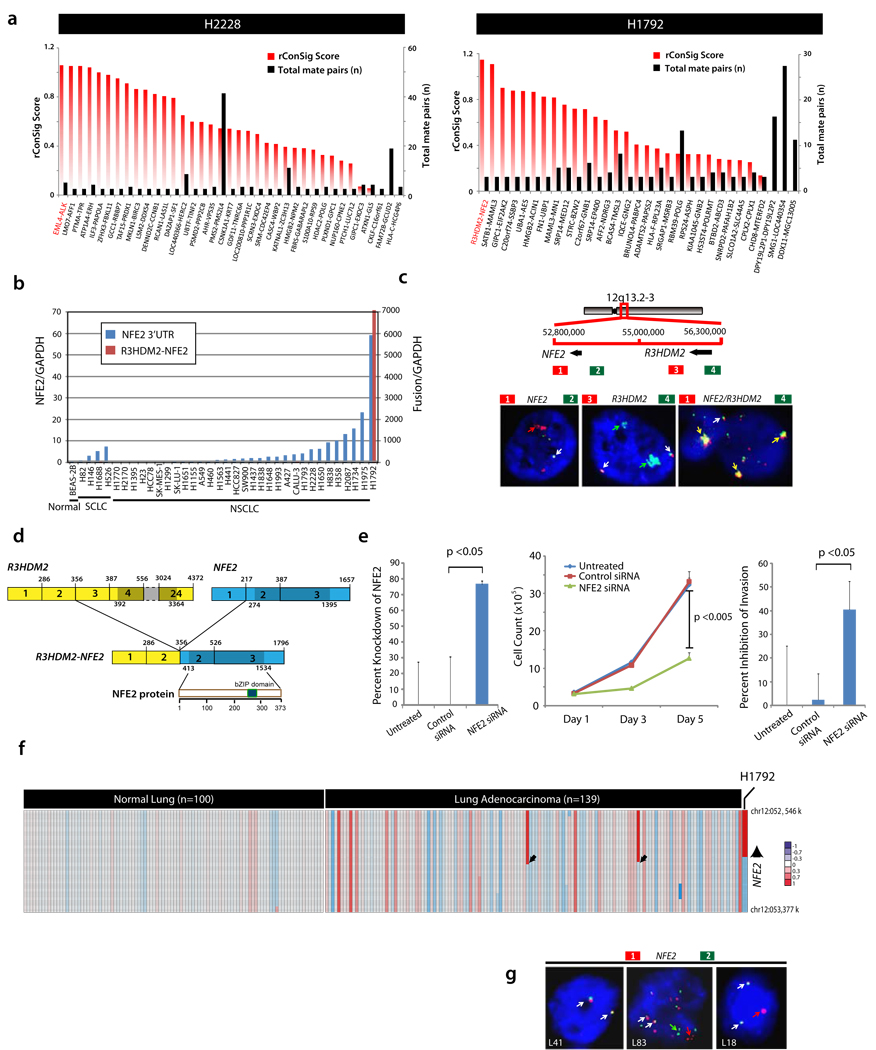
We first tested the ConSig approach on the H2228 cell line known to harbor the recurrent EML4-ALK fusion. Rating 3′ partners of paired-end chimeras by rConSig score revealed EML4-ALK as the top-ranked candidate on the H2228 cell line, which was supported by six mate pairs (Fig. 4a, left). This showed the effectiveness of the rConSig score in preferentially nominating driver gene fusions from numerous paired-end chimeras.
Figure 4. Discovery and validation of the R3HDM2-NFE2 fusion using the ConSig algorithm and the fusion breakpoint principle.
(a) Pair-end transcriptome sequencing of 12 lung cancer cell lines, followed by prioritizing the 3′ partners of paired-end chimeras (≥3 paired reads) by rConSig score (red bars). Left, EML4-ALK is a candidate fusion, and is supported by six paired reads (black bars), in the H2228 lung cancer cell line (known to harbor this fusion). Right, ConSig analysis nominates the R3HDM2-NFE2 fusion as the top candidate in the H1792 lung cancer cell line. (b) The R3HDM2-NFE2 fusion was confirmed by RT-PCR and sequencing of the PCR product. qRT-PCR of wild-type NFE2 revealed overexpression of NFE2 in a subset of lung cancer cell lines; only H1792 cells express the chimeric R2HDM2-NFE2. (c) Top, schematic of the genomic organization of R3HDM2-NFE2 fusion, with red and green bars indicating the location of BAC clones. This fusion was generated by an intrachromosomal translocation. Bottom, interphase FISH analysis showing amplification signals of 3′ NFE2 and 5′ R3HDM2 (left, middle) and R3HDM2-NFE2 fusion (right) on H1792 cell line. Normal signals are indicated by white arrows; aberrant colocalizing signals by yellow arrows; aberrant split signals by green or red signals. (d) Schematic of the R3HDM2-NFE2 fusion mRNA and protein. Structures for the R3HDM2 and NFE2 genes are derived from GenBank reference sequences. The numbers above the exons indicate the last base of each exon. Open reading frames are shown in darker shades. The exons of R3HDM2-NFE2 fusion are numbered from the original reference sequence. The lower schematic shows wild-type NFE2 protein and its domain architecture. (e) siRNA knockdown of NFE2 in H1792 cells leads to decreased cell proliferation (middle graph) and invasion (right graph). Percent knockdown of the fusion transcript revealed by qRT-PCR is shown in the left graph. (f) Analysis of SNP array data from 139 lung adenocarcinoma tissues revealed recurrent copy number aberrations in two patients at the 3′ NFE2 locus, as well as the focal amplification of R3HDM2-NFE2 fusion on H1792. (g) As in c, except the data are from three lung adenocarcinoma patients. L41 is a negative case with two colocalizing signals; L83 has split and high copy number gain at NFE2 locus; L18 showed one additional 3′ NFE2 signal (red).
We then applied this method to reveal driver gene fusions from the transcriptome sequencing data of 12 lung cancer cell lines. Although there were 530 gene fusions in total supported by more than two paired reads, the 3′ rConSig score prioritized R3HDM2-NFE2 as the lead in the H1792 lung cancer cell line (supported by three paired reads, Fig. 4a, right), and this fusion was confirmed by quantitative RT-PCR (qRT-PCR) (Fig. 4b), conventional capillary sequencing and interphase FISH, the latter of which showed high copy number gain of R3HDM2-NFE2 in H1792 (Fig. 4c). Consistent with previous microarray data on lung cancer cell lines (Supplementary Figure 7), qRT-PCR also revealed marked overexpression of NFE2 on H1792 and several additional lung adenocarcinoma cell lines (Fig. 4b); however, no rearrangements were detected in these samples by FISH, suggesting other mechanisms activating NFE2 expression (Supplementary Figure 9).
The R3HDM2-NFE2 fusion was predicted to encode the full-length open reading frame of NFE2, with only untranslated promoter sequences contributed from R3HDM2 (Fig. 4d), and exon-walking qRT-PCR demon-strated the specific overexpression of the NFE2 coding exons 2–3 under the regulation of the R3HDM2 promoter (Supplementary Figure 8). In H1792, knockdown of NFE2, which encodes a transcription factor normally expressed during erythropoiesis, resulted in a marked decrease in cell proliferation and to a lesser extent cell invasion (Fig. 4e), whereas no effect was seen in H460, which has low levels of endogenous NFE2 (Supplementary Figure 10).
Analysis of SNP array data for 139 lung adenocarcinoma tissues revealed copy number gain consistent with the fusion breakpoint principle at the 3′ NFE2 locus in two people with lung cancer (Fig. 4f), suggesting possible recurrent aberrations involving the NFE2 locus in this cancer. We therefore performed FISH analysis on a lung cancer tissue microarray comprised of a cohort of 76 lung adenocarcinoma samples, which confirmed recurrent NFE2 rearrangements in two individuals (Fig. 4g).
DISCUSSION
The complex biological events contributing to tumorigenesis are frequently driven by chromosomal rearrangements. Although previous studies have observed the generation of recurrent fusions at the edges of genomic imbalances18, efforts to identify cancer-promoting fusions from unbalanced breakpoints have been met with limited success, often discovering nonfunctional aberrations that appear to be biological by-products. This also holds true for next-generation sequencing transcriptome data, which routinely generates a large number of putative chimeras, most of which are nonfunctional19,20. Here, we describe a methodology to nominate biologically important fusions from an integrative analysis of next-generation transcriptome data and high-throughput genomic data.
By undertaking a comprehensive analysis of the biological associations of all genes contributing to gene fusions, we demonstrate that, although analysis of domain architectures and shared pathways was less informative, cancer-related fusion genes tend to engage distinct interaction networks or share common gene ontologies. Using such information, we generalized this finding to a genomic scale and developed an algorithm, ConSig score, to assay the probability that any given gene may contribute to a driving gene fusion based on the strength of that gene’s association with biological concepts characteristic of cancer genes. Although ConSig analysis can nominate putative cancer genes, the association of a gene with a specific tumor type requires additional evidence compiled from other biological data sets. To integrate use of high-throughput genomic data, we characterized the chromosomal imbalances associated with gene fusions, finding that recurrent gene fusions exhibit distinctive patterns of copy number alteration corresponding to differential portions of fusion partners. To our knowledge, this is the first evidence that integrative bioinformatics may be able to predict which genes are preferentially subject to chromosomal rearrangements important in tumorigenesis.
We applied the ConSig score to next-generation sequencing transcriptome data to benchmark fusion candidates, which were then assessed for chromosomal aberrations complying with the fusion breakpoint principle by integrating high-quality copy number data. We found that the ConSig score was able to identify the known EML4-ALK fusion as the top-ranked candidate in the H2228 lung cancer cell line, and in addition, we found further evidence of a R3HDM2-NFE2 fusion in H1792 cell line. We show that the R3HDM2-NFE2 fusion, which results in overexpression of wild-type NFE2, promotes cell proliferation and invasion. Moreover, through analysis of SNP arrays and lung tissue microarrays, we find that chromosomal rearrangements at the NFE2 locus are recurrent in a small subset of patient tumors, suggesting that NFE2 may contribute to a new class of lung cancer molecular biology. These data suggest that such approaches may have broad applicability to the analysis of multidimensional cancer genomic data.
The methodology described here can filter the large number of fusion candidates generated by paired-end next-generation sequencing data and preferentially identify driver gene fusions in cancer. The ConSig technology suggests the functional importance of putative fusions in cancer, whereas the breakpoint principle helps interpret large-scale cancer genomic data sets to explore potential recurrence. Although we have not applied this methodology to the discovery of novel mutations, we hypothesize that a similar computational schematic may yield insights in this area as well. Ultimately, we hope that this integrative methodology will elucidate key aspects of tumor biology as well as facilitate the development of targeted therapy of human cancers.
Supplementary Material
Acknowledgments
We thank F. Mitelman for offering the fusion genes list from the Mitelman database; T.W. Glover for the important comments for improving the manuscript; J. Granger for help with editing the manuscript; S. Qin for useful discussions about biostatistics; NCIBI colleagues L. Ke and A. Ade for helping in the implementation of tools and technologies; C. Yang for the guidance in drug informatics; Z. Hu from Boston University for help with network visualization. This work was supported by the National Institutes of Health (NIH; U54 DA021519) and a National Institutes of Health Cancer Biology Training Grant (CA009676-18 to J.R.P). J.R.P. is a Fellow of the University of Michigan Medical Scientist Training Program. A.M.C. is supported by NIH early detection network U01 CA111275, DOD W81XWH-09-2-0014, the Doris Duke Foundation and the American Cancer Society.
Footnotes
AUTHOR CONTRIBUTIONS
X-S.W., G.S.O. and A.M.C. designed the study. X-S.W., J.R.P. and M.A.S. performed bioinformatics analyses. X-S.W., J.R.P., G.C., Q.C., S.M.D., R.P., X.C. and S.V. performed experimental studies. B.H. and N.P. performed FISH analysis. D.G.T., T.J.G. and D.G.B. coordinated the clinical and pathology components. X-S.W., J.R.P., G.S.O. and A.M.C. wrote the manuscript.
References
- 1.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 3.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 4.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat. Rev. Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vastrik I, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad TS, et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z, et al. VisANT 3.0: new modules for pathway visualization, editing, prediction and construction. Nucleic Acids Res. 2007;35:W625–W632. doi: 10.1093/nar/gkm295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, et al. Leptin induces proliferation and anti-apoptosis in human hepatocarcinoma cells by up-regulating cyclin D1 and down-regulating Bax via a Janus kinase 2-linked pathway. Endocr. Relat. Cancer. 2007;14:513–529. doi: 10.1677/ERC-06-0027. [DOI] [PubMed] [Google Scholar]
- 10.Chen GJ, Weylie B, Hu C, Zhu J, Forough R. FGFR1/PI3K/AKT signaling pathway is a novel target for antiangiogenic effects of the cancer drug fumagillin (TNP-470) J. Cell. Biochem. 2007;101:1492–1504. doi: 10.1002/jcb.21265. [DOI] [PubMed] [Google Scholar]
- 11.Vantler M, et al. PI3-kinase/Akt-dependent antiapoptotic signaling by the PDGF alpha receptor is negatively regulated by Src family kinases. FEBS Lett. 2006;580:6769–6776. doi: 10.1016/j.febslet.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 12.Walz C, Cross NC, Van Etten RA, Reiter A. Comparison of mutated ABL1 and JAK2 as oncogenes and drug targets in myeloproliferative disorders. Leukemia. 2008;22:1320–1334. doi: 10.1038/leu.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrer DK, Yang YC. Complex formation of JAK2 with PP2A, P13K, and Yes in response to the hematopoietic cytokine interleukin-11. Biochem. Biophys. Res. Commun. 1996;224:289–296. doi: 10.1006/bbrc.1996.1023. [DOI] [PubMed] [Google Scholar]
- 14.Kharas MG, et al. Ablation of PI3K blocks BCR-ABL leukemogenesis in mice, and a dual PI3K/mTOR inhibitor prevents expansion of human BCR-ABL+ leukemia cells. J. Clin. Invest. 2008;118:3038–3050. doi: 10.1172/JCI33337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 16.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 17.Drexler HG. The Leukemia-Lymphoma Cell Line Factsbook. San Diego: Academic Press; 2000. [Google Scholar]
- 18.Mitelman F, Mertens F, Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat. Genet. 1997;15(Spec No):417–474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 19.Maher CA, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc. Natl. Acad. Sci. USA. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bashir A, Volik S, Collins C, Bafna V, Raphael BJ. Evaluation of paired-end sequencing strategies for detection of genome rearrangements in cancer. PLoS Comput. Biol. 2008;4:e1000051. doi: 10.1371/journal.pcbi.1000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent WJ. BLAT-the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 24.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richard W. National Cancer Institute; 2009. Overall experiment characteristics. ‹ https://array.nci.nih.gov/caarray/project/woost-00041›. [Google Scholar]
- 26.Roth RB, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubin MA, et al. Overexpression, amplification, and androgen regulation of TPD52 in prostate cancer. Cancer Res. 2004;64:3814–3822. doi: 10.1158/0008-5472.CAN-03-3881. [DOI] [PubMed] [Google Scholar]
- 29.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 30.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.