Abstract
DNA methyltransferase (DNMT) inhibitors are currently the standard of care for myelodysplastic syndrome and are in clinical trials for leukemias and solid tumors. However, the molecular basis underlying their activity remains poorly understood. Here we studied the induction and long-term stability of gene reactivation at three methylated tumor suppressor loci in response to the DNMT inhibitor 5-aza-2′-deoxycytidine (5-azaCdR)in human breast cancer cells. At the TMS/ASC locus, treatment with 5-azaCdR resulted in partial DNA demethylation, the re-engagement of RNA polymerase II (Pol II), and a shift from a repressive chromatin profile marked with H3K9me2 and H4K20me3 to an active profile enriched in H3ac and H3K4me2. Using a single molecule approach coupling chromatin immunoprecipitation with bisulfite sequencing, we show that H3ac, H3K4me2, and Pol II selectively associated with the demethylated alleles, whereas H3K9me2 preferentially marked alleles resistant to demethylation. H4K20me3 was unaffected by DNA demethylation and associated with unmethylated and methylated alleles. After drug removal, TMS1 underwent partial remethylation yet a subset of alleles remained stably demethylated for over three months. These alleles remained selectively associated with H3K4me2, H3ac, and Pol II and correlated with a sustained low level of gene expression. TMS1 alleles reacquire H3K9me2over time and those alleles that became remethylated retained H3ac. In contrast, CDH1and ESR1 were remethylated and completely silenced within ~1 week of drug removal, and failed to maintain stably unmethylated alleles. Our data suggest that the ability to maintain Pol II occupancy is a critical factor in the long-term stability of drug-induced CpG island demethylation.
Keywords: DNA methylation, epigenetics, TMS1/ASC, chromatin, Decitabine, breast cancer
Introduction
Cell-type specific gene expression patterns are established and maintained in part by epigenetic mechanisms including DNA methylation and post-translational histone modifications. In mammals, DNA methylation occurs at cytosine residues found within the context of CpG dinucleotides, which are under represented in the genome but can be found in clusters called CpG islands that are associated with the regulatory regions of approximately 70% of human genes (1–3). In normal cells, most CpG islands are unmethylated and transcriptionally competent. Methylation of CpG islands is associated with stable and heritable gene silencing (4). In addition to DNA methylation, post-translational modifications of the histone tails contribute to epigenetic regulation. Unmethylated CpG islands tend to be packaged into nucleosomes marked with ‘active’ histone modifications including, histone H3 acetylation of lysines 9 and 14 (H3ac), and histone H3 lysine 4 di-and tri-methylation (H3K4me2/3). Methylated CpG islands are generally depleted of these active marks, and are alternatively marked by a subset of ‘repressive’ histone modifications including histone H3 lysine 9 di-and tri-methylation (H3K9me2/3), and histone H3 lysine 27 tri-methylation (H3K27me3) (5, 6).
Genome-wide epigenetic alterations occur in cancer. There is an overall hypomethylation of the genome concurrent with the aberrant hypermethylation of a subset of CpG islands (7). This aberrant methylation of CpG islands in cancer is accompanied by a shift from a permissive to a more repressive histone modification profile generally characterized by the loss of H3ac and H3K4me2/3 and the acquisition of H3K9me2/3 (8–10). Recent work suggests that the epigenetic silencing of some loci also involves a shift in histone H4 modifications including loss of histone H4 acetylated at K16 (H4K16Ac) and gain of histone H4 tri-methylated at lysine 20 (H4K20me3) (11). The combination of CpG island methylation and accompanying histone modifications is associated with stable gene repression and is one mechanism leading to the heritable inactivation of tumor suppressor genes during tumor progression (12, 13).
The finding that tumor suppressor genes are often inactivated by epigenetic means and that these events can play a direct role in cancer initiation and progression provides a compelling rationale for the utilization of inhibitors of DNA methyltransferases (DNMTs) and histone modifying enzymes as a therapeutic strategy (6, 14). Over the last decade, there has been considerable effort in the clinical development of DNMT and histone deacetylase (HDAC) inhibitors in cancer therapy. For example, 5-aza-2′deoxycytidine (5-azaCdR) (Decitabine) and 5-aza-cytidine (Vidaza) have become the standard of care for myelodysplastic syndrome (MDS), and have shown promise in the treatment of leukemias(6, 15). In these cases, DNMT inhibitors have demonstrated both higher response rates and increased survival when compared to more traditional cytotoxic chemotherapeutic agents (14, 16). There is hope that such agents might also be useful in the treatment of solid tumors, if not as single agents then in combination with other epigenetic inhibitors (eg. HDAC inhibitors) or as a strategy to sensitize cells to conventional chemotherapy (for example, see (17)).
Despite their clinical success, there is still much to understand about the molecular mechanisms underlying the clinical activity of these agents and the durability of response. Studies in cell culture have shown that 5-azaCdR treatment induces DNA demethylation and reactivation of epigenetically silenced tumor suppressor genes. This reactivation is accompanied by the loss of some repressive histone modifications (e.g. H3K9me2) and the reappearance of active histone modifications (e.g. H3ac and H3K4me2) (18–20). However, the chromatin structure of CpG islands does not return to a fully active configuration due to the preservation of some repressive histone modifications unaffected by DNA demethylation such as H3K27me3 and H3K9me3, leaving open the potential for re-silencing after drug removal (20). Molecular analyses from biopsy-driven clinical trials indicate that global and gene-specific DNA demethylation is achievable in vivo. However, in cases where specific gene demethylation has been detected, remethylation is often observed within a few weeks of treatment(14).
To further understand the long-term effects of transient 5-azaCdR treatment on tumor suppressor gene reactivation, we studied the dynamics of DNA methylation, gene expression, and histone modifications at TMS1/ASC (Target of Methylation-induced Silencing 1), a CpG island-associated proapoptotic gene that is frequently silenced in conjunction with DNA hypermethylation in human breast, prostate and lung cancers (21). We find that 5-azaCdR-induced reactivation of TMS1 is accompanied by DNA demethylation and a shift from a repressive histone profile to a more active profile that includes the re-association of RNA polymerase II (Pol II) with the TMS1 promoter. Although a fraction of TMS1 alleles are re-methylated after drug removal, there is a subpopulation that remained stably unmethylated for at least 27 passages in culture (~ 3 months). This subpopulation is associated with both active (H3ac, H3K4me2,) and repressive histone marks (H4K20me3), and remains selectively occupied by Pol II. Our data suggest that the ability to attain and to maintain Pol II occupancy is a critical factor in the long-term stability of DNA demethylation and gene expression after drug-induced reactivation.
Materials and Methods
Cell culture and 5-azaCdR treatments
MDA-MB231 cells were obtained from the American Type Culture Collection and cultured in DMEM supplemented with 10% FBS and 2 mM L-glutamine. For 5-azaCdR treatments, 5×104MDA-MB231 cells were plated in a 10 cm dish 24 hours prior to treatment with 0.5 μM5-azaCdR. Medium containing fresh 5-azaCdR was applied every other day for six days. Following treatment, cells were maintained in the absence of 5-azaCdR and split 1:10 every three days for 27 passages (~ 3 months). Cells were harvested and DNA, RNA, and chromatin were collected at 0, 3, 6, 9, and 27 passages post-treatment.
Methylation Specific PCR
Genomic DNA (2 μg) was bisulfite-modified using the EZ DNA methylation kit (Zymo) and ~50 ng of modified DNA was utilized as template for Methylation-Specific PCR (MSP) as previously described (22). PCR conditions used were5 min. at 95°C, followed by 35 cycles of 30 s at 95°C;45 s at 58° C, and 45 at 72°C; with a final 5 min extension at 72°C. PCR products were resolved on a 1.5% agarose gel and stained with ethidium bromide. MSP primers are described in Supplemental Table 1.
COmbined Bisulfite Restriction Analysis(COBRA)
Bisulfite-modified DNA was amplified using primers devoid of any CpGs. Amplified products were purified with the PCR Purification kit (Qiagen), digested overnight at 37 °C with either FNU4HI or XmnI, precipitated, and resolved on a 2.0% agarose gel (23). Relative intensities of digested and undigested bands were quantified with Image Quant 5.2 and percent methylation was determined as the combined intensity of the digested bands relative to that of all bands (undigested and digested). Primer sequences are in Supplemental Table 1.
Genomic Bisulfite Sequencing
Genomic bisulfite sequencing (GBS) was performed as previously described (24). Briefly, bisulfite-modified DNA was amplified using primers devoid of CpGs as described above. PCR products were TA-cloned (Invitrogen), transformed into chemically competent E. Coli and plasmid DNA isolated from 10–17 individual colonies was sequenced. Bisulfite sequencing data were analyzed using the BiQ Analyzer software (25). Primer sequences are listed in Supplemental Table 1.
Reverse-Transcriptase PCR
RNA was isolated using the RNeasy kit (Qiagen) and was reverse-transcribed with random hexamer primers and M-MLV reverse transcriptase (24). Quantitative real-time PCR (q-PCR) was used to analyze gene-specific transcripts and their levels were normalized to 18s rRNA as previously described (11). Primer sequences are listed in Supplemental Table 1.
Chromatin Immunoprecipitation and ChIP-Bisulfite Sequencing
Chromatin Immunoprecipitation (ChIP) was performed as previously reported (11). The following antibodies were used for specific immunoprecipitations, H3ac (Millipore, 06-599), H3K4me2 (Millipore, 07-030), H3K9me2 (Millipore, 07-441), H4K20me3 (Abcam, ab9053), and Pol II (Santa Cruz, SC-9001x). Immunoprecipitated DNA was analyzed by qPCR with primers for TMS1, CDH1, or ESR1 (11). ChIP-bisulfite Sequencing (ChIP-bis) was adapted from the ChIP-MSP protocol initially reported by Zinn et. al. (26). Immunoprecipitated DNA from the ChIP procedure was subject to bisulfite conversion as described above. Modified DNA was then amplified using TMS1-specific bisulfite sequencing primers listed in Supplemental Table 1. PCR products were purified, cloned, and plasmid DNA from individual colonies was sequenced.
Results
TMS1 is a pro-apoptotic tumor suppressor gene containing a promoter-associated CpG island that is frequently methylated and silenced in human cancers (Figure 1A) (21). Previously, we compared the epigenetic landscape of TMS1 in breast cancer cell lines in which the TMS1 gene is either unmethylated and the gene is expressed (MCF7) or methylated and silenced (MDA-MB231). We find that in MCF7 cells and other normal cell lines that express TMS1 (e.g. IMR90), the CpG island is unmethylated, with DNase I hypersensitive sites (HSs), positioned nucleosomes, and distinct peaks of H4K16ac marking the boundaries between the unmethylated CpG island domain and surrounding methylated DNA (11, 27). In this state, the unmethylated CpG island is enriched for H3K4me2 and H3ac, and depleted for H3K9me2. Conversely, MDA-MB231 cells have a densely methylated CpG island in which the CpG island-associated DNase I HSs have been lost, nucleosomes are randomly positioned, and the active histone marks (H3K4me2, H4K16ac, and H3ac) have been replaced by repressive marks including H3K9me2 and H4K20me3. In this transcriptionally repressed state, H4K20me3 is localized to a prominent peak just upstream of transcription start, whereas H3K9me2 is enriched throughout the CpG island (11).
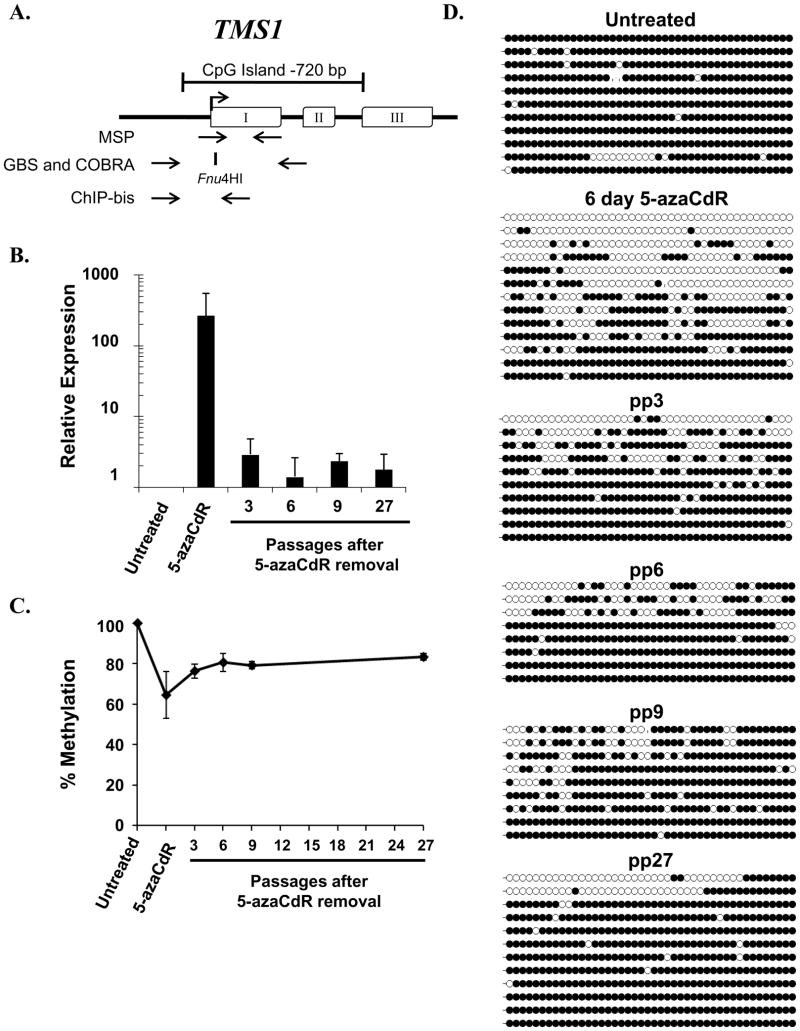
Figure 1. TMS1 expression and DNA methylation following the removal of 5-azaCdR.
A. Diagram of the TMS1 gene. Exons, open boxes; transcription start site, arrow. The CpG island is indicated by the bracket. Primers utilized for MSP, GBS, and COBRA analyses are indicated below the gene, as is the position of the Fnu4HI restriction site used in COBRA analysis. B. TMS1 mRNA abundance was measured immediately after treatment (5-azaCdR) or at the indicated time points after drug removal (pp, passages post 5-azaCdR) by reverse transcriptase qPCR and normalized to 18S rRNA. Shown is the fold change in expression (mean ± standard deviation) relative to untreated cells from three independent time-course experiments assayed in triplicate. C. COBRA analysis of DNA methylation following the removal of 5-azaCdR at the TMS1 locus. Data represents the mean percent methylation ( ± standard deviation) from three independent time course experiments D. DNA methylation was analyzed by bisulfite sequencing at the indicated time points. Each line represents a single colony isolate (8–13 isolated per sample). Open circles, unmethylated CpG; filled circles, methylated CpG.
Long-term effects of transient 5-azaCdR treatment at the TMS1 locus
To investigate the long-term effects of a transient exposure to a DNA de-methylating agent on the chromatin architecture at the TMS1 locus, MDA-MB231 cells were treated with 0.5 μM of 5-azaCdR every other day for 6 days and then maintained in culture for 27 passages (~80 days) in the absence of 5-azaCdR. This protocol resulted in the inhibition of cell growth (1.8-fold decrease in doubling time over six days) but little DNA damage as determined by γH2Ax focus formation (data not shown). Consistent with previous work, treatment of MDA-MB231 cells with 5-azaCdR induced the re-expression of TMS1 mRNA (Figure 1B)(24). However, while TMS1 expression was induced an average of 494-fold immediately following 5-azaCdR treatment, expression levels returned to ~3-fold over untreated cells within 3 passages after drug removal (Figure 1B). This low level of TMS1 expression was then maintained for at least three months (27 passages) in the absence of 5-azaCdR.
Treatment with 5-azaCdR resulted in an average demethylation of the TMS1 CpG island from 100% methylated to 64% methylated as determined by COBRA analysis (Figure 1C). After the removal of 5-azaCdR, there was an initial burst of remethylation (from 64% to 76% methylation) after 3 passages, which leveled off at ~83% and was maintained at this level for 27 passages in culture in the absence of drug (Figure 1D).
To distinguish between the possibilities that all TMS1 alleles exhibit a partial remethylation or that distinct subpopulations of unmethylated and methylated alleles persist after the removal of 5-azaCdR, DNA was bisulfite modified and individual alleles were sequenced. While all alleles were densely methylated in untreated MDA-MB231 cells, treatment with 5-azaCdR induced a heterogeneous methylation pattern consisting of alleles that were densely methylated, predominantly unmethylated, and those with a mixed pattern (Figure 1D). Removal of 5-azaCdR resulted in the partial remethylation of the locus that, over time, resolved into two distinct subpopulations; one comprised predominantly of methylated alleles and the other comprised predominantly of unmethylated alleles (Figure 1D). Interestingly, whereas there was a more checkered methylation profile seen in early post 5-azaCdR passages, the unmethylated alleles observed 27 passages later were almost entirely devoid of methylation.
Effects of 5-azaCdR on the histone modifications at TMS1
To examine the effects of 5-azaCdR on chromatin at the TMS1 locus, we utilized ChIP to map the histone modification profile before, immediately following, and over the course of 27 passages after treatment with 5-azaCdR. In untreated MDA-MB231 cells, the TMS1 locus was enriched for H3K9me2 and H4K20me3 (Figure 2). Treatment with 5-azaCdR led to the accumulation of H3ac and H3K4me2, as well as a decrease in H3K9me2 at TMS1 compared to untreated cells. This change was further accompanied by the re-association of Pol II (Figure 2). In contrast, the peak of H4K20me3, observed in untreated MDA-MB231 cells was unaffected by 5-azaCdR. Additionally, H4K16Ac, which is found at the unmethylated TMS1 locus in cells that express the gene (e.g. MCF7 cells,) (11), was not observed (data not shown). Thus, while 5-azaCdR treatment led to a shift to a more transcriptionally permissive chromatin configuration, it did not fully recapitulate the conformation observed in cells that normally express TMS1.
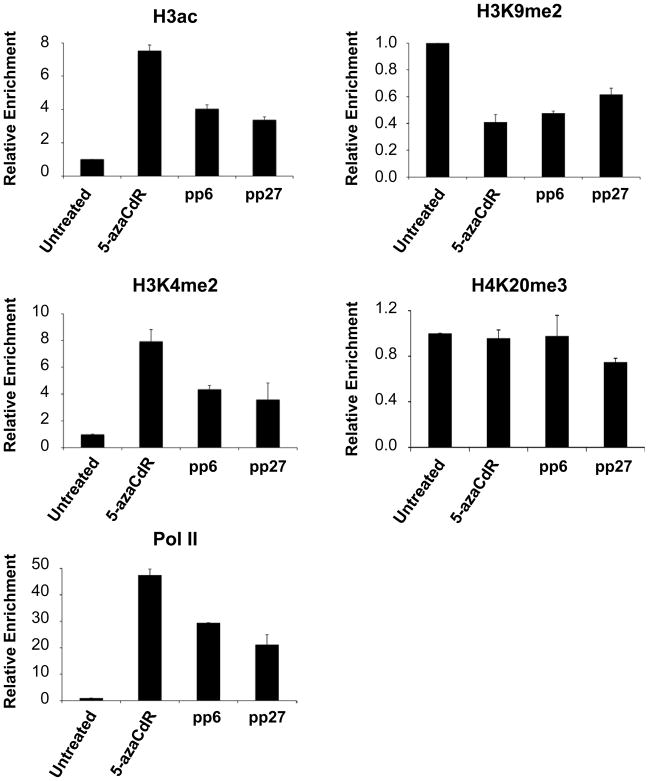
Figure 2. Histone modifications and RNA Pol II occupancy at TMS1 after the removal of 5-azaCdR.
MDA-MB231 cells were left untreated or treated with 0.5uM 5-azaCdR for six days. Chromatin was isolated immediately after treatment (5-azaCdR) or at the indicated time after drug removal (pp, passages post 5-azaCdR). Histone modifications and RNA Pol II occupancy were analyzed by ChIP followed by qPCR. Percent enrichment was determined by comparison of immunoprecipitated DNA relative to input DNA at each time point using primer set 3 of the TMS1 locus (11). Plotted is the mean (±standard deviation) of the fold change in enrichment relative to untreated MDA-MB231 cells from a single time course experiment assayed in triplicate. Similar results were obtained from a second independent time course (Supplemental Figure 1).
Over the course of 27 passages following the removal of 5-azaCdR the enrichment of H3ac, H3K4me2, and Pol II were partially depleted at TMS1, but still remained three to twenty-one fold higher than that of untreated MDA-MB231 cells (Figure 2). H3K9me2 levels were partially restored over the same time frame, but remained at levels lower than the untreated MDA-MB231 cells. This shift in the histone profile after the cessation of treatment coincided with the partial remethylation of DNA. The peak of H4K20me3 found upstream of the TMS1 transcription start remained unchanged throughout the time course. Thus, although there was a partial return to the pre-treatment chromatin state over time (DNA methylation and H3K9me2), a low level of active modifications and Pol II occupancy were maintained. Similar results were obtained from a second independent time course (Supplemental Figure 1) as well as at a second primer set (#5, (11)) in the TMS1 CpG island (data not shown).
Co-existing methylated and demethylated TMS1 alleles are packaged with differentially modified histones
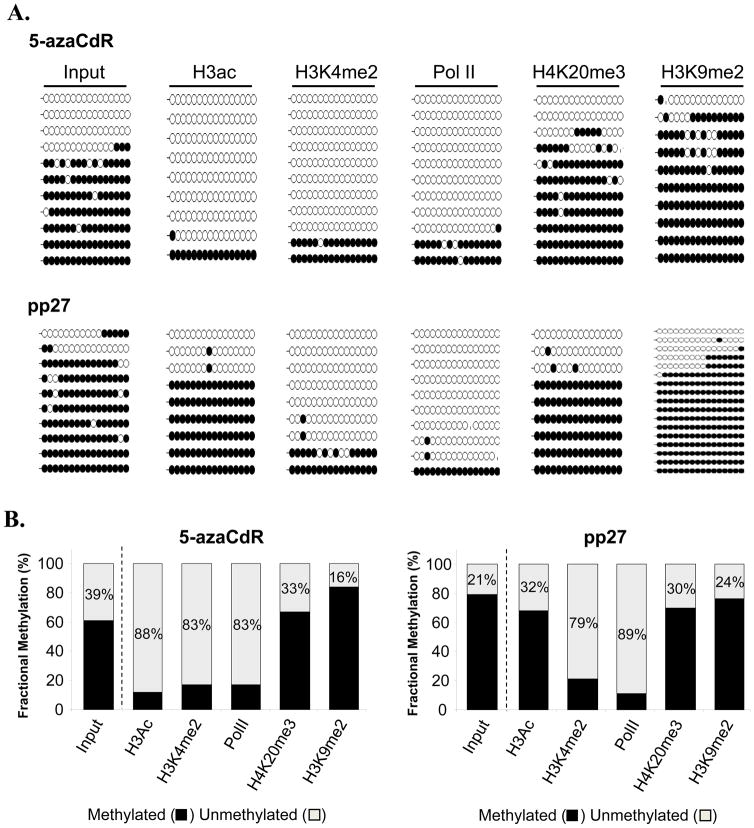
Previous work has suggested that treatment with 5-aza-CdR induces a “bivalent” chromatin signature at epigenetically reactivated tumor-suppressor genes(20, 28), implying that both active (e.g. H3K4me2) and repressive (e.g. H3K27me3) modifications co-exist on the same nucleosomes. So far, these studies have examined histone modifications at a population level within a mixed cell sample. We similarly show here that at a population level, both active and repressive epigenetic marks were present at the TMS1 CpG island after treatment with 5-azaCdR. However, at the level of individual alleles, two distinct subpopulations with different DNA methylation patterns were observed. To address the relationship between DNA methylation and histone modifications at the single molecule level, we employed a ChIP-bisulfite sequencing (ChIP-bis) approach, in which DNA was immunoprecipitated with antibodies specific to Pol II or the various histone modifications was eluted, bisulfite-modified and individual alleles were analyzed for their methylation status by bisulfite sequencing. As a control, DNA isolated from fixed and sheared total chromatin in the absence of antibody was also analyzed. Consistent with naked DNA (Figure 1D), DNA isolated from total chromatin directly after 5-azaCdR treatment showed a mixed DNA methylation pattern consisting of ~60% overall methylation (Figure 3A). ChIP-bis showed that H3Ac, H3K4me2, and Pol II were selectively associated with the unmethylated alleles in this population, as indicated by the relative enrichment of unmethylated DNA (decreased methylation density)in the immunoprecipitated DNA relative to input chromatin (Figure 3A). In contrast, H3K9me2 was selectively associated with the methylated alleles immediately following treatment with 5-azaCdR (84% methylation density compared to 61% in input) (Figure 3B). The distribution of alleles associated with H4K20me3 had a methylation profile similar to that of input DNA (67% methylation density compared to 61%), suggesting that the nucleosomes marked by this modification associate with both methylated and unmethylated DNA, consistent with our observation that total levels of H4K20me3 levels at TMS1 were unaltered by treatment with 5-azaCdR (Figure 2). Overall, these data suggest that treatment with 5-azaCdR leads to the coordinated demethylation of DNA and loss of H3K9me2, and that the chromatin associated with these demethylated alleles was selectively marked by H3Ac and H3K4me2, and bound by Pol II.
Figure 3. Single molecule association between DNA methylation and chromatin modifications at the TMS1 locus.
MDA-MB231 cells were left untreated or treated with 0.5uM 5-azaCdR for six days (5-azaCdR) A. Chromatin was isolated immediately after (5-azaCdR) or at the indicated time after drug removal and immunoprecipitated with the indicated antibodies. Precipitated DNA was eluted, bisulfite modified, and amplified with the bisulfite sequencing primers indicated in Figure 1A. For each immunoprecipitation, 9–17 individual clones were sequenced. Open circles, unmethylated CpGs; filled circles, methylated CpGs. Vertical hash marks represents missing data. B. Overall methylation density was determined as the total number of methylated CpGs relative to total number of CpGs in all alleles analyzed.
To investigate the stability of the associated marks after the removal of 5-azaCdR, we next performed ChIP-bis in cells 27 passages after drug removal. The DNA associated with total chromatin at this point had undergone a partial remethylation (from 61% to 79% methylation), similar to that observed for naked DNA analyzed by genomic bisulfite sequencing (compare Figures 1D and 3A). After 27 passages, H3K4me2-modified histones and Pol II remained predominantly associated with the unmethylated subpopulation of alleles (21% and 11% methylation density, respectively; Figure 3B). However, H3Ac, which selectively associated with the unmethylated alleles immediately after treatment, was now associated with both methylated and unmethylated alleles(from 12% to 68% methylation density), suggesting that the presence of H3Ac does not prevent the remethylation of TMS1 alleles. H3K9me2 marked histones, enriched on the methylated alleles immediately after treatment, was associated with both unmethylated and methylated TMS1 alleles at a ratio that was similar to that of input chromatin (76% total methylation compared with 79%) and to that immediately following treatment. This, together with the data showing that TMS1 alleles overall recover H3K9me2 over time (Figure 2) indicate that alleles that become remethylated also re-acquire H3K9me2 over time. H4K20me3, on the other hand, remained associated with both unmethylated and methylated alleles at a ratio similar to input both immediately after 5-azaCdR and 27 passages later. Thus, following the removal of 5-azaCdR in culture, we observe the preservation of a subset of unmethylated alleles uniquely marked by both active and repressive histone marks.
ESR1 and CDH1 are completely remethylated and silenced following transient exposure to 5-azaCdR
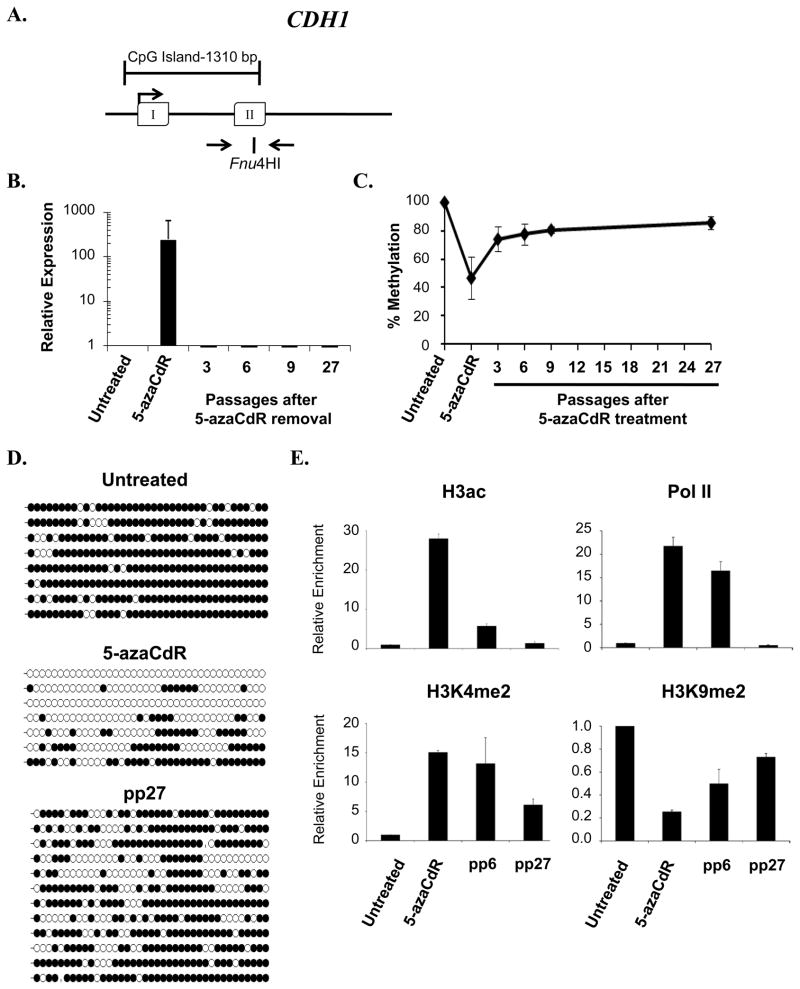
A comparison of the data presented above to that of other single gene studies examining the remethylation following 5-azaCdR treatment suggest that there may be gene specific variations in the remethylation kinetics following the removal of 5-azaCdR (19, 20, 29). This may reflect intrinsic differences in their underlying epigenetic regulation or could be a consequence of differences between the cell lines and treatment protocols utilized in different studies. To address this question, we monitored two additional tumor suppressor genes, CDH1 and ESR1 (Figures 4A, 5A), both of which are CpG island-associated genes that are aberrantly methylated and silent in human breast cancers and the MDA-MB231 cells used here (30). Like TMS1, the expression of CDH1 and ESR1was substantially induced after 5-azaCdR treatment (Figures 4B, 5B). In contrast to TMS1, which maintained a low, yet stable expression level several fold above untreated cells, both CDH1 and ESR1 were completely re-silenced within three passages (~9 days) after drug removal (Figures 4B, 5B). Analysis of a single CpG site via COBRA analysis showed that CDH1 and ESR1exhibited similar remethylation kinetics to TMS1 (Figures 1C, 4C and 5C), in that all three genes were initially demethylated by 40–50% during treatment and then ultimately remethylated to 82–85%. However, a more detailed methylation analysis by genomic bisulfite sequencing revealed distinctions in the patterns of their remethylation. After 27 passages in the absence of 5-azaCdR, both CDH1 and ESR1exhibited more uniform patterns of methylation, in that nearly all alleles were exhibited some level of remethylation( Figures 4D, 5D). Unlike TMS1, neither locus retained a subset of predominantly unmethylated alleles after 5-azaCdR treatment.
Figure 4. Analysis of CDH1 during and following treatment with 5-azaCdR.
A. Diagram of CDH1 CpG island. Open boxes, exons; arrow, transcription start site. The position of primers used for methylation analyses and the Fnu4H1 restriction enzyme site used for COBRA analysis are indicated. B. CDH1 mRNA expression was determined by reverse transcriptase qPCR and was normalized to 18S rRNA. Shown is the fold change in expression (mean ± standard deviation) relative to untreated cells from three independent experiments assayed in triplicate. C. COBRA analysis of DNA methylation following the removal of 5-azaCdR at the CDH1 locus. Data represents the mean percent methylation ( ± standard deviation) from three independent time course experiments D. DNA methylation was further analyzed by bisulfite sequencing at the indicated time points. Each line represents a single colony isolate (8–11 isolated per sample). Open circles, unmethylated CpG; filled circles, methylated CpG. pp, passages post 5-azaCdR treatment. E. Histone modifications and RNA Pol II occupancy were determined by ChIP as described in the legend to Figure 2. Plotted is the mean (±standard deviation) of the fold change in enrichment relative to untreated MDA-MB231 cells from a single time course experiment assayed in triplicate. Similar results were obtained from a second independent time course (Supplemental Figure 2).
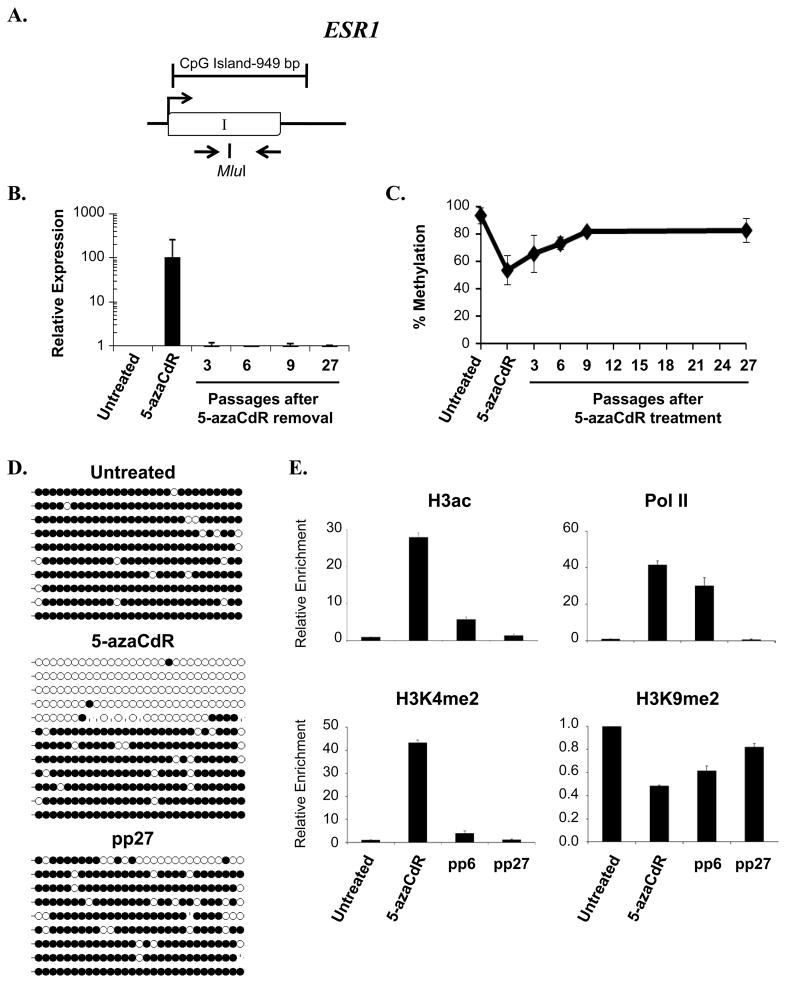
Figure 5. Analysis of ESR1 during and following treatment with 5-azaCdR.
A. Diagram of ESR1 CpG island. Open boxes, exons; arrow, transcription start site. The position of primers used for methylation analyses and the Mlu1restriction enzyme site used for COBRA analysis are indicated. B. mRNA expression was determined using primers specific to ESR1. Shown is the fold change in expression (mean ± standard deviation) relative to untreated cells after internal normalization to 18S rRNA from three independent experiments assayed in triplicate. C. COBRA analysis of DNA methylation at time points following the removal of 5-azaCdR at the ESR1 locus as described in the legend to Figure 1 D. DNA methylation was analyzed by bisulfite sequencing at the indicated time points. Each line represents a single colony isolate (9–12 isolated per sample). Open circles, unmethylated CpG; filled circles, methylated CpG. pp, passages post 5-azaCdR treatment. E. Histone modifications and RNA Pol II occupancy were determined by ChIP as described in the legend to Figure 2. Plotted is the mean (±standard deviation) of the fold change in enrichment relative to untreated MDA-MB231 cells from a single time course experiment assayed in triplicate. Similar results were obtained from a second independent time course (Supplemental Figure 3).
We also examined alterations in the histone profiles at the CDH1 and ESR1 CpG islands throughout the 5-azaCdR time course. As observed at TMS1, 5-azaCdR-induced reactivation of CDH1and ESR1was associated with the accrual of H3ac, H3K4me2, the loss of H3K9me2 and the re-establishment of Pol II occupancy (Figures 4E and 5E). H4K20me3 was not present at significant levels at either gene in untreated MDA-MB231 cells (data not shown) (11)and was not further analyzed. Although 5-azaCdR treatment induced a similar set of epigenetic changes at all three genes, there were differences in the ability to maintain the induced histone profiles after drug removal. At CDH1, low levels of H3K4me2 and H3ac were maintained throughout the time course, while these marks were completely depleted from ESR1 by 27 passages after 5-azaCdR removal (Figures 4E and 5E). Somewhat surprisingly, Pol II was still present at both CDH1 and ESR1 6 passages after drug removal, despite the lack of detectable gene expression. Ultimately, however, both CDH1 and ESR1exhibited a complete loss of Pol II occupancy between 6 and 27 passages after drug removal (Figures 4E, 5E). These differences in maintained histone profile may reflect the subtle differences in the extent of remethylation of individual alleles observed in genomic bisulfite sequencing (Figures 4D, 5D). Similar results were observed in a second independent time course (Supplemental Figures 2, 3). Thus, although a similar subset of epigenetic alterations are induced by treatment with 5-azaCdR at TMS1, CDH1 and ESR1, complete DNA remethylation and the return to stable gene repression correlated with an inability to maintain Pol II occupancy after drug removal.
Discussion
Here we establish that the degree and long-term stability of tumor suppressor gene reactivation induced by 5-azaCdR is locus-specific and correlates with the ability to attain and maintain Pol II promoter occupancy. Detailed analysis of the TMS1/ASC locus showed that transient exposure to 5-azaCdR induces DNA demethylation, depletion of H3K9me2, and the re-acquisition of H3ac, and H3K4me2. This allows for the re-engagement of Pol II on the TMS1 promoter and gene re-activation. Using a single molecule approach, we show that these acquired active marks (H3ac, H3K4me2, and Pol II) preferentially associate with demethylated alleles whereas H3K9me2 was selectively enriched on those alleles that remain methylated. H4K20me3, a mark typically associated with heterochromatin, was unaffected by 5-azaCdR treatment and was retained on both unmethylated and methylated alleles. After 3 months in the absence of drug, a subpopulation of unmethylated alleles persists, and remains associated with the active marks and with Pol II, whereas TMS1 alleles that re-methylated lost both H3K4me2 and Pol II occupancy while maintaining H3ac. Thus, following the removal of 5-azaCdR, the TMS1 CpG island is maintained in a unique epigenetic state, consisting of two distinct subpopulations of alleles neither of which fully resembles the repressed state in untreated MDA-MB231 cells, or that induced immediately after 5-azaCdR treatment.
Consistent with previous studies (8, 11, 26), we found that H3K9me2, H3K4me2 and DNA methylation status are tightly linked at the TMS1 locus (as well as at CDH1and ESR1)(18–20). Our single molecule approach confirms that H3K9me2 is selectively depleted from the unmethylated alleles (i.e. it remains selectively associated with methylated alleles), suggesting that DNA methylation is necessary to maintain H3K9me2. Recent work demonstrates that the histone methyltransferase G9a, thought to catalyze most H3K9me2 in euchromatin, interacts with components of the DNMT1 complex (31–33), suggesting a model in which DNA methylation and H3K9me2 may be coordinately maintained during DNA replication. Given that the incorporation of 5-azaCdR into DNA precipitates the degradation of DNMT1 (34), it is possible that the depletion of H3K9me2 is an indirect consequence of loss of DNMT1. After removal of 5-azaCdR, H3K9me2 remained predominantly associated with methylated allele s. Although we cannot distinguish between alleles that remethylated after drug removal from those that were never demethylated, these data together with the finding that H3K9me2 levels recover over time in parallel with DNA methylation (Figure 2) suggest that alleles that remethylate also recover H3K9me2.
After the removal of 5-azaCdR, we find H3ac to be associated with both methylated and unmethylated TMS1 alleles. Studies on the hTERT gene similarly found H3ac to associate with both methylated and unmethylated alleles (26). Our data suggest that drug-induced DNA demethylation is necessary to re-establish H3ac at TMS1, but once established, the presence of H3ac does not prevent the remethylation of DNA. Nor does DNA remethylation drive the de-acetylation of H3. Thus, although the combination of DNMT and HDAC inhibitors results in the synergistic re-activation of epigenetically silenced genes (35), they may have little impact on DNA remethylation and ultimately gene re-silencing after the cessation of treatment. Indeed, treatment with an HDAC inhibitor does not prevent the remethylation of the p16 gene after 5-azaCdR-induced DNA demethylation (36).
Unlike many densely methylated genes reactivated by 5-azaCdR treatment which have a propensity to undergo remethylation and resilencing after drug removal (18, 36), we found that a subset (~20%) of molecules are maintained in an unmethylated state at the TMS1 locus for more than 80 days in culture in the absence of drug. These alleles were selectively occupied by RNA Pol II and marked by H3K4me2, and likely account for the residual low-level gene expression observed. In contrast, ESR1and CDH1were completely resilenced and all alleles remethylated over the same time frame. This raises the question of whether certain genes or genomic regions are differentially affected by 5-azaCdR treatment. Studies on the MLH1gene in colon cancer cells have similarly shown the maintenance of a stable subpopulation of unmethylated alleles (~7%) for at least 44 days after 5-aza-CdR-induced reactivation (19). This study showed that 5-aza-CdR induced DNA demethylation was associated with the eviction of a single nucleosome near the transcription start site of MLH1and that the maintenance of this pattern of nucleosome depletion was both heritable and selective for the stably unmethylated subset of alleles (19). Although we did not examine nucleosome occupancy directly in this study, we have previously shown that the TMS1 CpG island is spanned by positioned nucleosomes flanking a single nucleosome gap at the transcription start site in its unmethylated and actively transcribed state (11). We propose that this 5-azaCdR-induced nucleosome eviction allows for the re-engagement of the Pol II complex with the demethylated promoter, perhaps filling the gap left behind by the evicted nucleosome, and it may be the maintenance of Pol II occupancy and/or the associated H3K4me2/3 deposition that prevents DNA re-methylation. Consistent with this idea, recent studies examining histone methylation at 5-azaCdR-induced genes across the genome showed that reactivation of methylated genes was associated with accumulation of H3K4me2 distributed to either side of a characteristic “dip” centered over transcription start (28). CDH1and ESR1, which exhibited total allelic remethylation, also failed to maintain significant Pol II occupancy after 5-azaCdR removal. The idea that Pol II occupancy may somehow protect CpG islands from de novo methylation is further supported by recent work showing that resistance of CpG islands to aberrant DNA methylation in cancer cells is better correlated with Pol II occupancy than gene expression on a genome-wide scale (37).
The fact that many genes appear to re-silence to some extent after the removal of 5-azaCdR may be a direct result of the underlying chromatin structure. Whereas some repressive marks like H3K9me2 are reversed by 5-azaCdR-induced demethylation, others (H3K9me3, H3K27me3) are not affected, and can be maintained in the absence of DNA methylation (20, 36). The persistence of such chromatin-mediated repression mechanisms may put the region at risk of re-methylation after drug removal. We show here that H4K20me3 is similarly unaffected by alterations in DNA methylation and remains associated with both unmethylated and methylated TMS1 alleles after the initial treatment with 5-azaCdR and throughout the course of DNA remethylation. These data suggest that there is a mechanism to maintain H4K20me3 at TMS1 that is independent of DNA methylation. At present, the mechanisms targeting H4K20me3 to specific loci are unclear. Current models suggest that H4K20 methylation proceeds in a step-wise fashion with PR-SET7 catalyzing mono-methylation at this position, which is then acted on by SUV4-20H to catalyze di-and tri-methylation (38–41). Consistent with the idea that SUV4-20H is actively targeted to the TMS1 locus in MDA MB231 cells, we find that whereas an initial co-treatment with 5-azaCdR and transient knockdown of SUV4-20H leads to depletion of DNA methylation and H4K20me3, and synergistic reactivation of TMS1 gene expression, it does not change the kinetics of TMS1 re-silencing or remethylation after drug removal compared to 5-aza-CdR alone (data not shown). Indeed, recent work from our lab suggests that whereas 5-azaCdR-mediated DNA demethylation allows for re-association of Pol II with the locus, the residual presence of H4K20me3 inhibits Pol II elongation, resulting in the accumulation of initiated Pol II in the promoter proximal region and down regulation of the full length transcript, (P. Kapoor-Vazirani, J. Kagey, and P. Vertino, submitted for publication). Whether this ‘enforcement’ of RNA Pol II promoter-proximal pausing puts the gene at risk of subsequent re-methylation remains to be determined.
The above considerations raise the question of whether the continued long-term inhibition of histone methyltransferases like SUV4-20H might facilitate the maintenance of tumor suppressor gene activity after demethylation-induced reactivation. Thus far, our attempts to address this question have been hampered by the inability to create stable cell populations knocked down for SUV4-20H and the lack of inhibitors specific for this mark. Histone methyltransferase inhibitors have not yet been widely explored in cancer therapy, but several agents are beginning to emerge. Treatment of cancer cells with BIX01294, a small molecule inhibitor with specificity for the G9a and GLP H3K9 methyltransferases, leads to a global and gene-specific depletion of H3K9me2 and re-activation of at least some genes (42). 3-deazaneplanocin A(DzNEP) was originally reported to have selective effects on H3K27me3, which appear to be mediated through the destabilization of EZH2 and other PRC2 complex components (43). As an inhibitor of S-adenosyl homocyteine hydrolase, DzNEP might be expected to have indiscriminant effects on many S-adenosyl-L-methionine dependent methyltransferases, and subsequent studies have shown that this agent has broad effects on the global levels of numerous histone methylation marks, including both ‘active’ (eg. H3K4me3, H3K79me3) and repressive (eg. H3K27me3, H3K9me2, H3R2me2) modifications (44). Nevertheless, recent preclinical studies suggest that this agent may be particularly active in cancer types with a dependency on EZH2 (45). As such agents continue to be developed, future combination therapies and dosing schedules that incorporate both DNA methylation inhibitors and histone methyltransferase inhibitors will undoubtedly follow.
The DNA methyltransferase inhibitors are now in widespread use for the treatment of MDS, and are currently in clinical trials for AML and other solid tumor types (15). Molecular analyses of bone marrow biopsies from patients treated with these agents have demonstrated that global and gene-specific DNA demethylation is achievable in vivo(46, 47). The degree of demethylation varies between patients, and whether this is an important indicator of clinical response remains controversial and may depend upon the compartment being analyzed (i.e. repetitive element methylation versus methylated tumor suppressor genes) or the surrogate marker measured (DNA methylation versus gene expression) (14, 48). In most cases, a gradual return to pre-treatment methylation levels has been observed within a few weeks or by the start of the next treatment cycle (46, 48, 49). We show here that, at least in cell culture, the kinetics of DNA remethylation and gene silencing do not necessarily parallel each other, and vary at different loci. Whereas CDH1and ESR1were resilenced within a week after drug removal and before the complete remethylation of H3K9me2 and DNA, a subset of TMS1 alleles remained stably unmethylated and occupied by Pol II for more than 3 months. The remethylation potential of a particular gene may be determined in part by the histone code present at that gene prior to demethylation, and the persistence of certain histone methylation marks (eg. H4K20me3). A thorough understanding of the underlying causes of these differences would be valuable in both developing new combined approaches incorporating inhibitors of other histone modifying enzymes and improving existing epigenetic therapy regimens.
Supplementary Material
MDA-MB231 cells were left untreated or treated with 0.5uM 5-azaCdR for six days. Chromatin was isolated immediately after treatment (5-azaCdR) or at the indicated time after drug removal (pp, passages post 5-azaCdR). Histone modifications and RNA Pol II occupancy were analyzed by ChIP followed by qPCR. Percent enrichment was determined by comparison of immunoprecipitated DNA relative to input DNA at each time point using primers specific to the CDH1 (Supplementary Table I). Plotted is the mean (± standard deviation) of the fold change in enrichment relative to untreated MDA-MB231 cells from a second time course experiment assayed in triplicate.
MDA-MB231 cells were left untreated or treated with 0.5uM 5-azaCdR for six days. Chromatin was isolated immediately after treatment (5-azaCdR) or at the indicated time after drug removal (pp, passages post 5-azaCdR). Histone modifications and RNA Pol II occupancy were analyzed by ChIP followed by qPCR. Percent enrichment was determined by comparison of immunoprecipitated DNA relative to input DNA at each time point using primer set 3 of the TMS1 locus (Kapoor-Vazirani et al., 2008). Plotted is the mean (± standard deviation) of the fold change in enrichment relative to untreated MDA-MB231 cells from a second time course experiment assayed in triplicate.
MDA-MB231 cells were left untreated or treated with 0.5uM 5-azaCdR for six days. Chromatin was isolated immediately after treatment (5-azaCdR) or at the indicated time after drug removal (pp, passages post 5-azaCdR). Histone modifications and RNA Pol II occupancy were analyzed by ChIP followed by qPCR. Percent enrichment was determined by comparison of immunoprecipitated DNA relative to input DNA at each time point using primers specific to the ESR1 locus (Supplemental Table I). Plotted is the mean (± standard deviation) of the fold change in enrichment relative to untreated MDA-MB231 cells from a second time course experiment assayed in triplicate.
Acknowledgments
The authors wish to thank Pritty Patel, Harold Saavedra, and Arsene Adon for technical assistance and helpful discussions.
Footnotes
This work was supported by NIH grants 2RO1 CA077337 (to PMV), a Department of Defense CDMRP Breast Cancer Program predoctoral fellowship W81XWH-08-1-0362 (to JDK) and an American Cancer Society postdoctoral fellowship PF-07-130-01-MGO (to MTM)
References
- 1.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 2.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009;15:3927–37. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2 (Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 7.McCabe MT, Lee EK, Vertino PM. A multifactorial signature of DNA sequence and polycomb binding predicts aberrant CpG island methylation. Cancer Res. 2009;69:282–91. doi: 10.1158/0008-5472.CAN-08-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–61. [PubMed] [Google Scholar]
- 9.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–15. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–8. [PubMed] [Google Scholar]
- 11.Kapoor-Vazirani P, Kagey JD, Powell DR, Vertino PM. Role of hMOF-dependent histone H4 lysine 16 acetylation in the maintenance of TMS1/ASC gene activity. Cancer Res. 2008;68:6810–21. doi: 10.1158/0008-5472.CAN-08-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman JG, Baylin SB. Gene silencing in cancer inassociation with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 13.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 14.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15:3938–46. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oki Y, Aoki E, Issa JP. Decitabine--bedside to bench. Crit Rev Oncol Hematol. 2007;61:140–52. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–40. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 17.Matei DE, Nephew KP. Epigenetic therapies for chemoresensitization of epithelial ovarian cancer. Gynecol Oncol. 2010;116:195–201. doi: 10.1016/j.ygyno.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coffee B, Zhang F, Ceman S, Warren ST, Reines D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am J Hum Genet. 2002;71:923–32. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–44. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–9. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 21.McConnell BB, Vertino PM. TMS1/ASC: the cancer connection. Apoptosis. 2004;9:5–18. doi: 10.1023/B:APPT.0000012117.32430.0c. [DOI] [PubMed] [Google Scholar]
- 22.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–4. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine JJ, Stimson-Crider KM, Vertino PM. Effects of methylation on expression of TMS1/ASC in human breast cancer cells. Oncogene. 2003;22:3475–88. doi: 10.1038/sj.onc.1206430. [DOI] [PubMed] [Google Scholar]
- 25.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–8. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 26.Zinn RL, Pruitt K, Eguchi S, Baylin SB, Herman JG. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
- 27.Stimson KM, Vertino PM. Methylation-mediated silencing of TMS1/ASC is accompanied by histone hypoacetylation and CpG island-localized changes in chromatin architecture. J Biol Chem. 2002;277:4951–8. doi: 10.1074/jbc.M109809200. [DOI] [PubMed] [Google Scholar]
- 28.McGarvey KM, VanNeste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–9. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 30.Nass SJ, Herman JG, Gabrielson E, Iversen PW, Parl FF, Davidson NE, et al. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res. 2000;60:4346–8. [PubMed] [Google Scholar]
- 31.Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–41. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 36.Egger G, Aparicio AM, Escobar SG, Jones PA. Inhibition of histone deacetylation does not block resilencing of p16 after 5-aza-2′-deoxycytidine treatment. Cancer Res. 2007;67:346–53. doi: 10.1158/0008-5472.CAN-06-2845. [DOI] [PubMed] [Google Scholar]
- 37.Takeshima H, Yamashita S, Shimazu T, Niwa T, Ushijima T. The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res. 2009 doi: 10.1101/gr.093310.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Mizzen CA. The multiple facets of histone H4-lysine 20 methylation. Biochem Cell Biol. 2009;87:151–61. doi: 10.1139/O08-131. [DOI] [PubMed] [Google Scholar]
- 39.Pannetier M, Julien E, Schotta G, Tardat M, Sardet C, Jenuwein T, et al. PR-SET7 and SUV4–20H regulate H4 lysine-20 methylation at imprinting control regions in the mouse. EMBO Rep. 2008;9:998–1005. doi: 10.1038/embor.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–95. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–61. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferro MT, Steegman JL, Escribano L, Heiurichs B, Parada L, Garcia-Sagredo JM, et al. Ph-positive chronic myeloid leukemia with t(8;21)(q22;q22) in blastic crisis. Cancer Genet Cytogenet. 1992;58:96–9. doi: 10.1016/0165-4608(92)90143-v. [DOI] [PubMed] [Google Scholar]
- 43.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–88. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puppe J, Drost R, Liu X, Joosse SA, Evers B, Cornelissen-Steijger P, et al. BRCA1-deficient mammary tumor cells are dependent on EZH2 expression and sensitive to Polycomb Repressive Complex 2-inhibitor 3-deazaneplanocin A. Breast Cancer Res. 2009;11:R63. doi: 10.1186/bcr2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mund C, Hackanson B, Stresemann C, Lubbert M, Lyko F. Characterization of DNA demethylation effects induced by 5-Aza-2′-deoxycytidine in patients with myelodysplastic syndrome. Cancer Res. 2005;65:7086–90. doi: 10.1158/0008-5472.CAN-05-0695. [DOI] [PubMed] [Google Scholar]
- 47.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–9. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 48.Fandy TE, Herman JG, Kerns P, Jiemjit A, Sugar EA, Choi SH, et al. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood. 2009;114:2764–73. doi: 10.1182/blood-2009-02-203547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braiteh F, Soriano AO, Garcia-Manero G, Hong D, Johnson MM, Silva Lde P, et al. Phase I study of epigenetic modulation with 5-azacytidine and valproic acid in patients with advanced cancers. Clin Cancer Res. 2008;14:6296–301. doi: 10.1158/1078-0432.CCR-08-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MDA-MB231 cells were left untreated or treated with 0.5uM 5-azaCdR for six days. Chromatin was isolated immediately after treatment (5-azaCdR) or at the indicated time after drug removal (pp, passages post 5-azaCdR). Histone modifications and RNA Pol II occupancy were analyzed by ChIP followed by qPCR. Percent enrichment was determined by comparison of immunoprecipitated DNA relative to input DNA at each time point using primers specific to the CDH1 (Supplementary Table I). Plotted is the mean (± standard deviation) of the fold change in enrichment relative to untreated MDA-MB231 cells from a second time course experiment assayed in triplicate.
MDA-MB231 cells were left untreated or treated with 0.5uM 5-azaCdR for six days. Chromatin was isolated immediately after treatment (5-azaCdR) or at the indicated time after drug removal (pp, passages post 5-azaCdR). Histone modifications and RNA Pol II occupancy were analyzed by ChIP followed by qPCR. Percent enrichment was determined by comparison of immunoprecipitated DNA relative to input DNA at each time point using primer set 3 of the TMS1 locus (Kapoor-Vazirani et al., 2008). Plotted is the mean (± standard deviation) of the fold change in enrichment relative to untreated MDA-MB231 cells from a second time course experiment assayed in triplicate.
MDA-MB231 cells were left untreated or treated with 0.5uM 5-azaCdR for six days. Chromatin was isolated immediately after treatment (5-azaCdR) or at the indicated time after drug removal (pp, passages post 5-azaCdR). Histone modifications and RNA Pol II occupancy were analyzed by ChIP followed by qPCR. Percent enrichment was determined by comparison of immunoprecipitated DNA relative to input DNA at each time point using primers specific to the ESR1 locus (Supplemental Table I). Plotted is the mean (± standard deviation) of the fold change in enrichment relative to untreated MDA-MB231 cells from a second time course experiment assayed in triplicate.