Abstract
DNA methylation is widespread in most species, from bacteria to mammals, and is crucial for genomic imprinting, gene expression, and embryogenesis. DNA methylation occurs via two major classes of enzymatic reactions: maintenance-type methylation catalyzed by DNA (cytosine-5-)-methyltransferase (DNMT) 1, and de novo methylation catalyzed by DNMT 3 alpha (DNMT3A) and -beta (DNMT3B). The expression pattern and regulation of DNMT genes in primordial germ cells (PGCs) and germ line cells has not been sufficiently established in birds. Therefore, we employed bioinformatics, RT-PCR, real-time PCR, and in situ hybridization analyses to examine the structural conservation and conserved expression patterns of chicken DNMT family genes. We further examined the regulation of a candidate de novo DNA methyltransferase gene, cDNMT3B by cotransfection of cDNMT3B 3′UTR- and cDNMT3B 3′UTR-specific miRNAs through a dual fluorescence reporter assay. All cDNMT family members were differentially detected during early embryonic development. Of interest, cDNMT3B expression was highly detected in early embryos and in PGCs. During germ line development and sexual maturation, cDNMT3B expression was reestablished in a female germ cell-specific manner. In the dual fluorescence reporter assay, cDNMT3B expression was significantly downregulated by four miRNAs: gga-miR-15c (25.82%), gga-miR-29b (30.01%), gga-miR-383 (30.0%), and gga-miR-222 (31.28%). Our data highlight the structural conservation and conserved expression patterns of chicken DNMTs. The miRNAs investigated in this study may induce downregulation of gene expression in chicken PGCs and germ cells.
Introduction
Methylation represents the addition of a methyl group or replacement of an atom by a methyl group in a substrate. The addition of a methyl group to the 5-position of the cytosine nucleotide in the sequence CpG by catalyzing enzymes is called DNA methylation. DNA methylation is widespread in most species, from bacteria to mammals. In mammals, DNA methylation is crucial for normal development, most likely due to its importance in genomic imprinting, X-chromosome inactivation, chromatin modification, silencing of endogenous retroviruses, gene expression, and embryo development [1], [2]. DNA methylation is primarily negative during zygote formation, and is established through cell division during embryonic development [2]. The occurrence of DNA methylation is closely connected with chromatin organization by histone acetylation and methylation. Histone acetyltransferases (HDACs) mainly catalyze the enzymatic reactions of histone acetylation [3]. DNA methylation occurs via two major classes of enzymatic reaction: maintenance-type methylation and de novo methylation. Maintenance-type methylation activity involves the maintenance of methylation patterns in the daughter strands of every DNA replication cycle. De novo methylation activity involves the recognition and transfer of methyl groups to unmethylated DNA [4].
There are three enzymes in the DNA (cytosine-5-)-methyltransferase (DNMT) family: DNMT1, DNMT 3 alpha (DNMT3A), and DNMT 3 beta (DNMT3B). All catalyze DNA methylation activity. DNMT1 is a member of the maintenance-type methyltransferase family, which is responsible for the maintenance of DNA methylation patterns [5]. DNMT3A and the closely related DNMT3B are de novo methyltransferases, which are responsible for the establishment of new methylation patterns [2], [5]. DNMT1 and DNMT3A expressions are ubiquitous, whereas DNMT3B is expressed at a low level in most tissues except the testis, pancreas, thyroid, and bone marrow. DNA methylation and DNMT family proteins play global functions in vertebrate species. DNMTs act as potential molecular targets in cancer therapy. Overexpression of DNMTs has been shown to influence tumor cell resistance to cytotoxicity of oxidative stress [6]. DNMT1 is associated with the perpetuation of fibroblast activation and fibrogenesis in the kidney [7]. DNMT1 and DNMT3A are required for neuronal synaptic plasticity, learning, and memory [8].
Compared to mammalian species [9], the expression pattern and regulation of DNMT genes during germ line development has not been sufficiently established in birds. In this study, we examined the conservation and functional domains of cDNMT family proteins using bioinformatics analysis, and further examined the conserved expression patterns of cDNMT family genes during early embryonic development, germ line development, and sexual maturation of testis and ovaries using reverse transcription PCR (RT-PCR), quantitative real-time PCR (qRT-PCR), and in situ hybridization analyses. To examine the regulation of the candidate de novo DNA methyltransferase gene cDNMT3B at the post-transcriptional level, we performed cotransfection analysis using cDNMT3B 3′UTR- (3 prime untranslated regions) and cDNMT3B 3′UTR-specific microRNAs (miRNAs). All cDNMT family members were differentially detected during early embryonic development. Of interest, cDNMT3B expression was highly detected in early embryos, primordial germ cells (PGCs), and germ cells at least until embryonic day E14.5. After hatching, cDNMT3B expression was reestablished in a female germ cell-specific manner. In the dual fluorescence reporter assay, cDNMT3B expression was significantly downregulated by all miRNAs examined. The miRNAs investigated in this study may induce downregulation of gene expression in chicken PGCs and germ cells.
Materials and Methods
Experimental animals and animal care
The care and experimental use of White Leghorn chickens were approved by the Institute of Laboratory Animal Resources, Seoul National University (SNU-070823-5), Korea. Chickens were maintained according to a standard management program at the University Animal Farm, Seoul National University. The procedures for animal management, reproduction, and embryo manipulation adhered to the standard operating protocols of our laboratory.
Sex determination
Freshly laid eggs were incubated with intermittent rocking at 37°C under 60–70% relative humidity. Sex was determined on embryonic day E2.5. Approximately 0.2 µL of embryonic blood was collected from the dorsal aorta, diluted in 15 µL of 1× phosphate buffered saline (PBS, pH 7.4), and boiled at 94°C for 10 min to prepare the DNA template for PCR. Each 20-µL PCR reaction contained 2 µL of DNA template, 2 µL of PCR buffer, 1.6 µL of 2.5-mM dNTP mixture, 10 pmol of each forward and reverse primer of chicken W chromosome (F: 5′-CTA TGC CTA CCA CAT TCC TAT TTG C-3′ and R: 5′-AGC TGG ACT TCA GAC CAT CTT CT-3′), and 1 unit of Taq DNA polymerase. The thermal conditions for 35 cycles were 94°C for 30 s, 66°C for 30 s, and 72°C for 30 s. Female sex was identified based on the strong bands detected in the agarose gel after separation of PCR products by gel electrophoresis.
Sample collection
We collected whole embryos at EG&K [10] stage X (freshly laid eggs), H&H [11] stage 3 (12-h incubation), H&H stage 6 (24-h incubation), and H&H stage 12 (48-h incubation). We collected blood PGCs (bPGCs) on E2.5, and gonadal PGCs (gPGCs) and gonadal stromal cells (GSCs) on E6.5 by magnetic-activated cell sorting (MACS) [12]. bPGCs were cultured (hereafter known as cPGCs) up to passage 30, as previously described [13]. Apart from the whole embryos and cell samples, we collected embryonic gonads on E4.5; embryonic brains, kidneys, livers, stomachs, muscles, lungs, and gonads from male and female embryos on E6.5; gonads from male and female embryos on E8.5, E9.5, E10.5, E11.5, E12.5, E13.5, E14.5 and E15.5; and testes and ovaries at 1 day, 12 weeks, and 25 weeks of age. Total RNA was extracted from three batches of the aforementioned samples using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocols. Approximately 1 µg of Oligo(dT)20-primed total RNA from each sample was reverse transcribed with the Superscript III First-Strand Synthesis System (Invitrogen) according to the manufacturer's protocols. All cDNA samples were diluted to 10% and used for RT-PCR, qRT-PCR, and subcloning experiments. Another batch of limited samples were directly used/frozen in liquid nitrogen for the in situ hybridization experiment.
Analysis of structural features and conservation of cDNMT family proteins
The protein sequences of chicken DNMT family members were obtained from a BLAST search of the Chicken Genome Database at the National Center for Biotechnology Information (NCBI). The protein sequences of chicken DNMT1 (NP_996835), DNMT3A (NP_001020003), and DNMT3B (NP_001019999) were used to search for homologous DNMT family members in all vertebrate species using the NCBI BLASTP search engine. We retrieved all DNMT family protein sequences from at least nine other vertebrate species, including human, chimpanzee, cattle, pig, horse, rat, mouse, opossum, and zebrafish (see Table 1 for GenBank accession numbers). The percent identities and conservation of chicken DNMT family protein sequences with other vertebrate DNMT family protein sequences were analyzed using the NCBI BLASTP and CLUSTAL X programs, respectively. The conserved functional domains of chicken DNMT family protein sequences were identified using the Pfam-A family matrices [14].
Table 1. GenBank accession numbers of DNMT family proteins of different vertebrate species obtained by BLASTP searches.
| Species | DNMT1 | DNMT3A | DNMT3B |
| Chicken | NP_996835 | NP_001020003 | NP_001019999 |
| Human | NP_001124295 | NP_783328 | NP_008823 |
| Chimpanzee | XP_001163364 | XP_001148246 | XP_514580 |
| Cattle | NP_872592 | AAP75901 | NP_861529 |
| Pig | NP_001027526 | NP_001090906 | NP_001155876 |
| Horse | XP_001916472 | XP_001503030 | XP_001916549 |
| Rat | NP_445806 | NP_001003958 | NP_001003959 |
| Mouse | NP_034196 | NP_031898 | NP_001003961 |
| Opossum | NP_001028141 | XP_001380132 | XP_001362485 |
| Zebrafish | NP_571264 | NP_001018150 | NP_001020621 |
RT-PCR analysis
We performed RT-PCR analysis to examine the tissue-specific expression of cDNMT1, cDNMT3A, and cDNMT3B during early embryonic development. The cDNA from stages X, 3, 6, and 12 embryos; bPGCs, cPGCs, gPGCs, and GSCs; and the brains, kidneys, livers, stomachs, muscles, lungs, and gonads of male and female embryos at E6.5 were amplified using cDNMT1, cDNMT3A, cDNMT3B, and chicken glyceraldehyde-3-phosphate dehydrogenase (cGAPDH, NM_204305)-specific primers (Table 2). Each 20-µL PCR reaction mix contained 2 µL of cDNA, 2 µL of PCR buffer, 1.6 µL of 2.5 mM dNTP mixture, 10 pmol of each forward and reverse primer, and 1 unit of Taq DNA polymerase. PCR was performed with an initial incubation at 94°C for 5 min, followed by 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The reaction was terminated by a final incubation at 72°C for 7 min.
Table 2. Primers used for cDNMT family members, cGAPDH, cDAZL and cSYCP3.
| Gene | Accession no. | *Primer sequences (5′-3′) | Size (bps) |
| cDNMT1 | NM_206952 | F : CTGAGATGCCCTCCCCCAAGR : GTCCTCCCGTCGTCCTCCAC | 454 |
| F : TGTCCATCTTCGACGCCAACR : CATAGATGGGCTTCACGGCA | 174 | ||
| cDNMT3A | NM_001024832 | F : GCAAGCAGCAGAGCAGGGAAR : CCACCAACAGGTCCACGCA | 577 |
| F : GGGTGAGCGACAAAAGGGACR : TGGAGTTGGAGCGAGTGGTG | 234 | ||
| cDNMT3B | NM_001024828 | F : GAACCCAGCCACCTTCCACCR : AGTGATGTTGCCCTCGTGCC | 547 |
| F : ACCAGCCAAGAGGAGACCCAR : TGGCGAGCGAGAGGTCATTA | 269 | ||
| cGAPDH | NM_204305 | F : CACAGCCACACAGAAGACGGR : CCATCAAGTCCACAACACGG | 443 |
| F : CCGTGTTGTGGACTTGATGGR : GAGGAGTGGGGGAGACAGAA | 175 | ||
| cDAZL | NM_204218 | F: CGTCAACAACCTGCCAAGGAR: TTCTTTGCTCCCCAGGAACC | 540 |
| cSYCP3 | XM_416330 | F: GCAGAAAGCAGAGGAACAGGAGGR: TGGACTGAAGAGACTTGCGAACA | 281 |
*First primer pairs were used for RT-PCR analysis and cRNA probe synthesis, and second primer pairs were used for qRT-PCR analysis. cDAZL primer pairs were used for cRNA probe synthesis only. cSYCP3 primer pairs were used for qRT-PCR only.
qRT-PCR analysis
We performed qRT-PCR analysis to examine the relative quantification of the expression level of cDNMT1, cDNMT3A, and cDNMT3B during early embryonic development and germ line development. The cDNA from stages X, 3, 6, and 12 embryos; bPGCs, cPGCs, gPGCs, and GSCs; male and female gonads at E6.5, E8.5, E10.5, E12.5, and E14.5; and testes and ovaries of chickens at 1 day, 4 weeks, 12 weeks, and 25 weeks of age were amplified with the forward and reverse primers of cDNMT1, cDNMT3A, cDNMT3B, and cGAPDH (Table 2). Quantification was performed using an iCycler Real Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). Each 20-µL PCR reaction mix contained 2 µL of cDNA, 2 µL of PCR buffer, 1.6 µL of 2.5 mM dNTP mixture, 10 pmol of each forward and reverse primer, 1 µL of 20× Eva green (Biotium Inc., Hayward, CA, USA), and 1 unit of Taq DNA polymerase. The reaction was carried out in optical 96-well standard plates (Applied Biosystems Inc., Foster City, CA, USA). PCR was performed with an initial incubation at 94°C for 3 min, followed by 40 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The reaction was terminated by a final incubation at the dissociation temperatures. Furthermore, expression of cDNMT3B during limited time points (E8.5, E10.5, E12.5, E14.5 and 1-day) of meiotic stages were compared with a meiosis specific gene chicken synaptonemal complex protein 3 (cSYCP3, please see table 2 for primers). The relative quantification of cDNMT1, cDNMT3A, cDNMT3B and cSYCP3 expression was calculated using the 2−ΔΔCt method after the threshold cycle (Ct) was normalized with the Ct of cGAPDH.
We performed miRNA qRT-PCR analysis to examine the relative quantification of the expression level of gga-miR-15c, gga-miR-29b, gga-miR-383 and gga-miR-222 during germ line development. First strand cDNA was synthesized from total RNA (1 µg) of male and female gonads on E9.5, E10.5, E11.5, E12.5, E13.5, E14.5 and E15.5 using the miRNA first strand cDNA synthesis kit (Stratagene, Santa Clara, CA, USA). To elongate the miRNAs, total RNAs were first treated with E. coli poly-A polymerase (PAP) to generate a poly-A tail at the 3′-end of each RNA molecule. Following polyadenylation, cDNAs were synthesized using the RT adaptor primer. qRT-PCR analysis for the complete miRNA first strand cDNAs was performed using the High-Specificity miRNA QPCR Core Reagent Kit (Stratagene). Each 25-µL PCR reaction mix contained 4 µL of miRNA cDNA, 2.5 µL of 10× core PCR buffer, 2.75 µL of 50 mM MgCl2, 10 µL of 20 mM dNTPs, 1.25 µL of 20× Eva Green, 1.0 µL of 3.125 µM universal reverse primer, 1.0 µL of 3.125 µM miRNA-specific forward primer and 0.5 µL of High-Specificity polymerase. Each miRNA forward primer was designed according to the guidelines provided by Stratagene (http://www.stratagene.com/miRNAguide). The threshold cycle of miRNA expression were normalized with chicken snoRNA (endogenous control). Please see table 3 for miRNA-specific primers.
Table 3. qRT-PCR primers used for gga-miR-15c, gga-miR-29b, gga-miR-383, gga-miR-222 and SnoRNA.
| miRNA | Forward primer sequences (5′-3′) |
| gga-miR-15c | TAGCAGCACATCATGGTTTG |
| gga-miR-29b | TAGCACCATTTGAAATCAGT |
| gga-miR-383 | AGATCAGAAGGTGATTGTGGCT |
| gga-miR-222 | AGCTACATCTGGCTACTGGGTCTC |
| SnoRNA | GGGATGTAAAAAAATACTTGCTATC |
5-methylcytosine staining
5-methylcytosine staining (5 meC) was performed to examine the DNA methylation pattern in PGCs as previously described [15]. Briefly, MACS-sorted PGCs at E2.5, E4.5, and E6.5 were mounted on glass slides treated with 3-aminopropyltriethoxysilane (APES, Sigma-Aldrich, St. Louis, MO, USA) and then fixed with 3.7% (w/v) paraformaldehyde in 1× PBS for 10 min. The cells were washed three times for 5 min each in 1× PBS and permeabilized with 0.5% (v/v) Triton X-100 and 1% (w/v) bovine serum albumin (BSA) in PBS for 30 min. The cells were then washed with 1× PBS, treated with 4N HCl for 20 min, and blocked with 0.1% Triton X-100 and 1% BSA in 1× PBS for 30 min. The cells were incubated with 5 meC antibody (abCAM, Cambridge, UK) and diluted in blocking buffer (1∶200) at 4°C overnight. After primary antibody incubation, cells were washed in 1× PBS and incubated with Alexa 488 dye-conjugated secondary antibodies (Molecular Probes, Carlsbad, CA, USA) for 1 h at room temperature in the dark. Finally, the cells were mounted with ProLongH Gold antifade reagent with 4′-6-diamidino-2-phenylindole (DAPI, Invitrogen) and imaged with a confocal laser microscope (Carl Zeiss, Oberkochen, Germany).
cRNA probes
To prepare cRNA probes for cDNMT1, cDNMT3A, cDNMT3B, and cDAZL (positive control), the cDNA from gPGCs at E6.5 was amplified with the respective primers (Table 2). PCR products were loaded onto 1% agarose gels and separated by gel electrophoresis at 5 v/cm for 30 min. Size-corrected DNAs were subcloned into a pGEM-T plasmid vector (Promega, Madison, WI, USA) and transformed to Escherichia coli strain DH5α. The sequences of recombinant plasmids containing each gene were verified using the Automated DNA sequencer 3730×l (Applied Biosystems), and then the recombinant plasmids were amplified using T7- and SP6-specific primers (T7: 5′-TGT AAT ACG ACT CAC TAT AGG G-3′ and SP6: 5′-CTA TTT AGG TGA CAC TAT AGA AT-3′) to prepare templates for labeling cRNA probes. Digoxigenin (DIG)-labeled cRNA probes of cDNMT1, cDNMT3A, cDNMT3B, and cDAZL were prepared using a DIG RNA labeling kit (Roche Diagnostics, Indianapolis, IN, USA) by an in vitro transcription method.
In situ hybridization of whole mount and cryosections
The expression levels of cDNMT1, cDNMT3A, and cDNMT3B mRNA during early embryonic development, PGC differentiation, germ line development, and sexual maturation were examined at limited time points from stage X to 25 weeks of age by in situ hybridization. The expression patterns of cDNMT1, cDNMT3A, and cDNMT3B were compared to that of the germ line-specific gene cDAZL at E4.5 and E6.5. Whole embryos (stages X, 6, and 12) collected in Petri dishes, cryosections of gonads, testes, and ovaries (E4.5 to 25 weeks old) mounted on APES-treated glass slides were fixed with 4% (w/v) paraformaldehyde in 1× PBS. The samples were permeabilized with 1% (v/v) Triton X-100 and incubated in a prehybridization mixture containing 50% formamide, 25% 20× standard saline citrate (SSC: 150 mM NaCl and 15 mM sodium citrate; pH 7.0), and 25% diethylpyrocarbonate (DEPC)-treated distilled water. After prehybridization, samples were incubated in a hybridization mixture containing 10% dextran sulfate sodium, 0.02% BSA, 250 µg/mL yeast tRNA, and DIG-labeled cRNA probes in a prehybridization mixture for 18 h at 55°C. The samples were washed for stringency in a series of solutions as previously described [16]. Nonspecific binding was blocked with a 1% (w/v) blocking reagent before the samples were incubated with a sheep anti-DIG-AP antibody (Roche) for 12 h at 4°C. The mRNA signals were visualized as a dark brown color using a substrate solution containing nitroblue tetrazolium (NBT), 5-bromo-4-chloro-3-indolyl phosphate (BCIP), and levamisole. After signal development, the sections were counterstained with 1% (w/v) methyl green (Sigma-Aldrich). Photographs of whole mount embryos were taken with a Stereoscopic Zoom Microscope SMZ1000 (Nikon Corporation, Tokyo, Japan). Photographs of sections were taken with a Zeiss Axiophot light microscope (Carl Zeiss).
Regulation of cDNMT3B using 3′UTR target miRNAs
To examine the regulation of cDNMT3B expression, we selected four miRNAs including gga-miR-15c (5′-UAG CAG CAC AUC AUG GUU UGU A-3′), gga-miR-29b (5′-UAG CAC CAU UUG AAA UCA GUG UU-3′), gga-miR-383 (5′-AGA UCA GAA GGU GAU UGU GGC U-3′), and gga-miR-222 (5′-AGC UAC AUC UGG CUA CUG GGU CUC-3′) specific to the 3′UTR region of cDNMT3B from miRDB, a microRNA target prediction and functional annotation database [17]. cDNMT3B 3′UTR and cDNMT3B 3′UTR mutants were cloned into a pcDNA3 plasmid encoding enhanced green fluorescence protein (eGFP), and cDNMT3B 3′UTR-specific miRNAs were cloned into a pDsRed2-N1 plasmid encoding red fluorescence protein (RFP) under a CMV promoter (Clontech, Palo Alto, CA, USA) for the dual fluorescence reporter assay. Plasmids containing cDNMT3B 3′UTR and miRNA were prepared in a 100-mL LB culture of transformed XL1-Blue E. coli using an EndoFree Plasmid Maxi kit (Qiagen, Valencia, CA, USA) and cotransfected into 293 FT cells using the transfection procedures provided by Invitrogen. After 48 h of cotransfection, eGFP expression was examined under the microscope, and the relative eGFP expression was analyzed by fluorescence-activated cell sorting (FACS) using BD FACSCalibur (BD Biosciences, San Jose, CA, USA). Furthermore, conservation of cDNMT3B 3′UTR region and target miRNAs in chicken, human and mouse were analyzed using the CLUSTAL X programs.
Results
Conserved structural features and sequence analysis of cDNMT family proteins
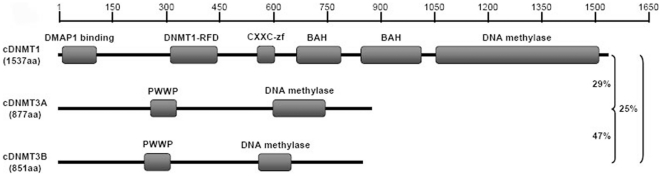
The mRNA and protein sequences of chicken DNMT family genes were obtained from the NCBI Gallus gallus Genome Database. Of these, the mRNA sequence of cDNMT1 contains an open reading frame of 4614 base pairs (bp) encoding a 1537-amino acid protein. The percent identity of cDNMT1 protein with other vertebrate DNMT1 proteins over the entire alignment indicated significant identities: 82% to opossum; 75% to human, cattle, and horse; 73% to chimpanzee; 71% to rat, mouse, and zebrafish; and 70% to pig. cDNMT1 had significant hits to five different functional domains including DMAP1 (DNMT1-associated protein 1) binding domain (at position 8–100 aa, E-value 3e-16), DNMT1-specific replication foci domain (at position 310–445 aa, E-value 4.1e-38), CXXC zinc finger domain (at position 557–603 aa, E-value 1.5e-14), bromo-adjacent homology (BAH) domains (at positions 667–791 aa and 842–1011 aa, E-values 9.1e-22 and 7.1e-18), and C-5 cytosine-specific DNA methylase domain (at position 1054–1508 aa, E-value 3.8e-48), which is also conserved in all investigated vertebrate DNMT1 proteins (Fig. 1 and Fig. S1). The mRNA sequence of the second member, cDNMT3A, contains an open reading frame of 2634 bp encoding an 877-amino acid protein. The percent identity of cDNMT3A protein with other vertebrate DNMT3A proteins was: 93% to pig; 87% to horse and opossum; 86% to human, chimpanzee, cattle, rat, and mouse; and 81% to zebrafish. cDNMT3A had significant hits to two functional domains including the PWWP domain (at position 254–327 aa, E-value 1.4e-23) and C-5 cytosine-specific DNA methylase domain (at position 599–741 aa, E-value 1.7e-12), which is also conserved in other vertebrate DNMT3A proteins (Fig. 1 and Fig. S2). The mRNA and protein length of the third member, cDNMT3B, is comparatively shorter than those of other members of the DNMT family. cDNMT3B mRNA consists of an open reading frame of 2556 bp encoding an 851-amino acid protein. cDNMT3B shares significant identities with other vertebrate DNMT3B proteins for up to 70% to cattle, 67% to opossum, 66% to horse, 64% to human and chimpanzee, 60% to pig, 54% to zebrafish, 45% to rat, and 43% to mouse. Similar to cDNMT3A, cDNMT3B also showed significant hits to the PWWP domain (at position 236–309 aa, E-value 1.9e-20) and C-5 cytosine-specific DNA methylase domain (at position 569–657 aa, E-value 2e-06), which is conserved in all vertebrate DNMT3B proteins (Fig. 1 and Fig. S3). Furthermore, cDNMT1 shares 29% identity with cDNMT3A and 25% identity with cDNMT3B. The identity between cDNMT3A and cDNMT3B is 47% (Fig. 1).
Figure 1. Graphic diagram of the conserved functional domains.
The conserved functional domains of cDNMT1, cDNMT3A, and cDNMT3B protein sequences found using the Pfam-A family matrices with default parameters.
Expression of cDNMT family members during early embryonic development examined by RT-PCR
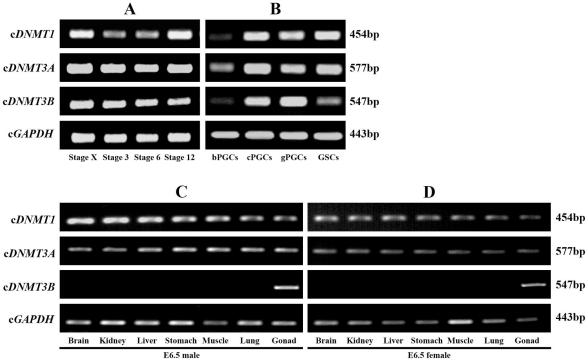
Expression of cDNMT1, cDNMT3A, and cDNMT3B in early embryos at stages X, 3, 6, and 12; bPGCs, cPGCs, gPGCs, and GSCs; and the brains, kidneys, livers, stomachs, muscles, lungs, and gonads of male and female embryos at E6.5 were examined by RT-PCR. cDNMT1 and cDNMT3A showed similar patterns of expression in all tissues/cells examined. During early embryonic development, cDNMT1 and cDNMT3A were detected at a strong level in the embryos at stages X, 3, 6, and 12 (Fig. 2A). Both cDNMT1 and cDNMT3A were detected at low levels in bPGCs compared to cPGCs, gPGCs, and GSCs; however, cDNMT1 expression was much lower in bPGCs (Fig. 2B). cDNMT1 and cDNMT3A expressions were detected at moderate levels in all somatic and gonadal samples of male and female embryos examined at E6.5 (Fig. 2C–D). cDNMT3B expression differed from that of the other members. Expression was strongly detected in all early embryos, cPGCs, gPGCs, and male and female gonads (Fig. 2A–D); weakly detected in bPGCs and GSCs (Fig. 2B); and not detected or barely detected in all somatic tissues of male and female embryos at E6.5 (Fig. 2C–D).
Figure 2. Expression of cDNMT1, cDNMT3A, and cDNMT3B during early embryonic development examined by RT-PCR.
cDNA from EG&K stage X, H&H stage 3, stage 6, and stage 12 (2A); blood PGCs (bPGCs), cultured PGCs (cPGCs), gonadal PGCs (gPGCs), and gonadal stromal cells (GSCs) (2B); and the brains, kidneys, livers, stomachs, muscles, lungs, and gonads of male and female embryos at E6.5 (2C, 2D) were amplified with cDNMT1, cDNMT3A, cDNMT3B, and chicken glyceraldehyde 3 phosphate dehydrogenase (cGAPDH)-specific primers.
Expression of cDNMT family members during early embryonic development, germ-line development, and sexual maturation examined by qRT-PCR
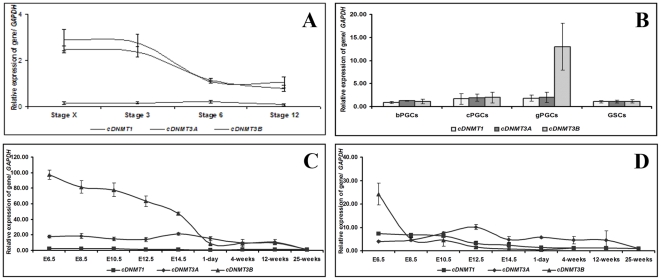
Expression of cDNMT1, cDNMT3A, and cDNMT3B in early embryos at stages X, 3, 6, and 12; bPGCs, cPGCs, gPGCs, and GSCs; male and female gonads at E6.5, E8.5, E10.5, E12.5, and E14.5; and testes and ovaries 1 day, 4 weeks, 12 weeks, and 25 weeks of age were examined by qRT-PCR. Relative quantification of the expression levels of each gene was normalized with cGAPDH. cDNMT1 expression was detected at a low level in stage X to 12 embryos relative to cGAPDH. cDNMT3A and cDNMT3B expressions were initially high at stages X and 3, then decreased to a moderate level at stages 6 and 12 (Fig. 3A). When we examined the expression patterns of cDNMT family members in different PGC and GSC samples, cDNMT1 and cDNMT3A expressions were detected in all PGC and GSC samples; however, cDNMT1 and cDNMT3A expressions were slightly high in bPGCs, cPGCs, and gPGCs compared to that of GSCs. cDNMT3B expression was significantly high in gPGCs when compared to the other members of the cDNMT family. Furthermore, cDNMT3B expression was 11.7-fold, 6.6-fold, and 11.5-fold higher in gPGCs compared to its expression in bPGCs, cPGCs, and GSCs, respectively (Fig. 3B). Regarding the expression patterns of cDNMT family members during germ line development and sexual maturation, cDNMT1 and cDNMT3A expressions were detected in low to moderate levels on E6.5 to 25 weeks of age in males and females. cDNMT3B expression was different during embryonic and post-hatch development. In males, it was significantly detected at high to moderate levels on E6.5 to E14.5. After E14.5, it decreased and was detected at a low level until 25 weeks of age. In females, cDNMT3B expression was significantly high at E6.5. After E8.5, it was detected at moderate to low levels until 25 weeks of age (Fig. 3C–D).
Figure 3. Expression of cDNMT1, cDNMT3A, and cDNMT3B during early embryonic development, germ line development, and sexual maturation examined by qRT-PCR.
cDNA from EG&K stage X, H&H stage 3, stage 6, and stage 12 (3A); blood PGCs (bPGCs), cultured PGCs (cPGCs), gonadal PGCs (gPGCs), and gonadal stromal cells (GSCs) (3B); male (3C) and female (3D) gonads on embryonic days E6.5, E8.5, E10.5, E12.5, and E14.5; and testes and ovaries from 1-day-, 4-week-, 12-week- and 25-week-old chickens were amplified with cDNMT1-, cDNMT3A-, and cDNMT3B-specific primers. The threshold cycle of cDNMT family genes were normalized with chicken glyceraldehyde-3-phosphate dehydrogenase (cGAPDH). Relative gene expression was calculated using the 2−ΔΔCt method.
mRNA localization of cDNMT family members during early embryonic development
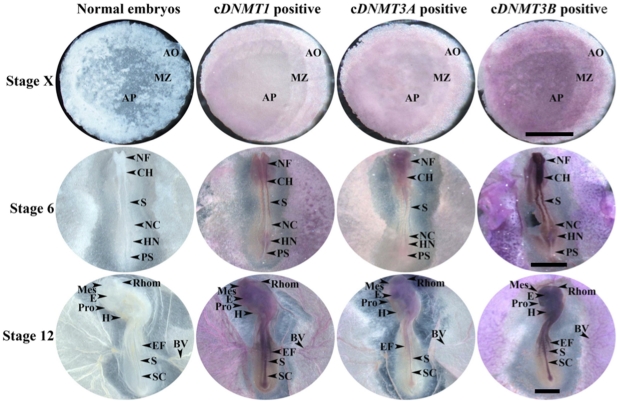
Expression patterns of cDNMT1, cDNMT3A, and cDNMT3B mRNA during early embryonic development at stages X, 6, and 12 were examined by whole mount in situ hybridization. cDNMT1 mRNA expression was weakly detected in the area pellucida at stage X. At stage 6, cDNMT1 mRNA expression was localized in the head fold, neural tube, and primitive streak areas. At stage 12, it was localized in all parts of the embryonic body including the head fold, optic vesicles, neural tube, ventricles, somites, and blood vessels. cDNMT3A mRNA expression was slightly high in the area pellucida at stage X. At stages 6 and 12, it was strongly detected in the head area compared to all other parts of the embryonic body. When compared to other members, cDNMT3B mRNA expression was strongly detected in the area pellucida at stage X and all parts of the developing embryos at stages 6 and 12 (Fig. 4). In addition, all cDNMT family members were not detected or weakly detected in the area opaca at stage X and in the yolk sac at later stages.
Figure 4. Expression patterns of cDNMT1, cDNMT3A, and cDNMT3B mRNA during early embryonic development.
Whole-mount embryos of EG&K stage X, H&H stage 6, and stage 12 were hybridized with antisense cRNA probes against cDNMT1, cDNMT3A, and cDNMT3B. AP = area pellucida, MZ = marginal zone, AO = area opaca, NF = neural fold, CH = chord, S = somite, NC = notochord, HN = Hensen's node, PS = primitive streak, Pro = prosencephalon, Mes = mesencephalon, Rhom = rhombencephalon, E = eye, H = heart, EF = edge of the foregut, SC = spinal cord, BV = blood vessel. Common bar – 5 mm.
DNA methylation pattern in PGCs
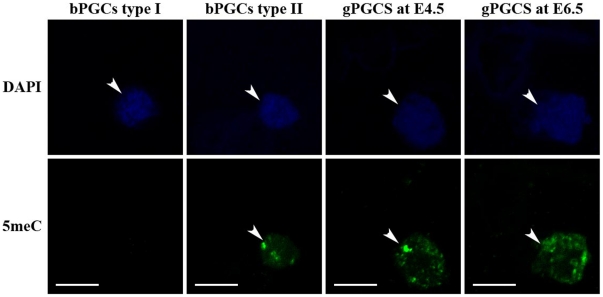
Immunocytochemical staining was performed to confirm the DNA methylation pattern in PGCs. In chicken embryos, PGCs usually occur in the circulation at around E2.0 to E3.0, and they enter differentiating germinal ridges by around E3.5 to E4.0. MACS-sorted bPGCs at E2.5, and gPGCs at E4.5 and E6.5, were subjected to 5 meC antibody staining. 5 meC expression was poorly detected in some bPGCs (designated as type I), suggesting that some bPGCs were undergoing genome-wide DNA demethylation at this stage. However, staining was detectable in many bPGCs (designated as type II), which were undergoing DNA methylation. 5 meC staining was intensive and conserved in almost all PGCs after they entered into the differentiating germinal ridges at E4.5 and settled in the gonads at E6.5 (Fig. 5).
Figure 5. DNA methylation pattern in primordial germ cells (PGCs).
Blood PGCs (bPGCs) at embryonic day E2.5 and gonadal PGCs (gPGCs) at E4.5 and E6.5 were stained with 5-methylcytosine (5 meC) and Alexa 488 dye-conjugated secondary antibody. After staining, cells were mounted with ProLongH Gold antifade reagent with 4′-6-diamidino-2-phenylindole (DAPI) and imaged with a confocal laser microscope. Arrowheads indicate DAPI and 5 meC stained PGCs. Common bar – 10 µm.
mRNA localization of cDNMT family members during PGCs entry into embryonic gonads
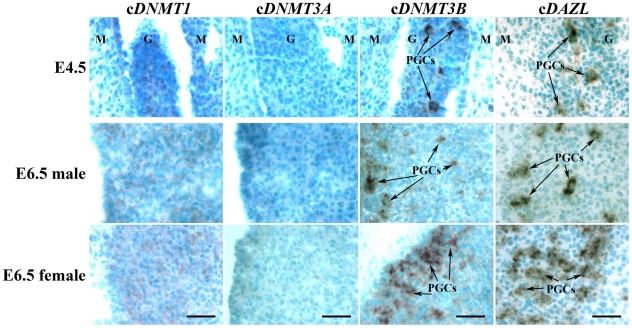
Cryosections of undifferentiated and differentiated (male and female) gonads at E4.5 and E6.5, respectively, were subjected to in situ hybridization to examine the mRNA expression pattern of cDNMT family members during PGC entry into embryonic gonads. Figure 6 shows the mRNA expression pattern of cDNMT family members and cDAZL (positive control) in the left gonads of E4.5 and E6.5 embryos. cDNMT1 mRNA expression was moderately detected in the PGCs and stromal cells at E4.5 and E6.5 gonads. cDNMT3A mRNA expression was detected at low levels in the PGCs, and was almost undetectable in the stromal cells. In contrast, cDNMT3B mRNA expression was strongly detected in the PGCs, which entered undifferentiated gonads/gonadal ridges at E4.5. After gonadal differentiation, cDNMT3B-positive PGCs were widespread in the male gonads. However, cDNMT3B-positive PGCs were detected only in the peripheral area in the female gonads. cDNMT3B mRNA expression was detected at very low levels in the stromal cells from E4.5 to E6.5. cDNMT3B mRNA expression was highly comparable to the expression pattern of cDAZL, a PGC and germ cell marker (Fig. 6).
Figure 6. Expression patterns of cDNMT1, cDNMT3A, and cDNMT3B mRNA during PGCs entry into embryonic gonads compared to cDAZL.
Transverse sections of left gonads at E4.5 and E6.5 (male and female) were hybridized with antisense cRNA probes against cDNMT1, cDNMT3A, cDNMT3B, and cDAZL. M = mesonephros, G = gonads. Arrows indicate PGCs strongly express cDNMT3B and cDAZL mRNA. Common bar – 50 µm.
mRNA localization of cDNMT family members during germ line development and sexual maturation
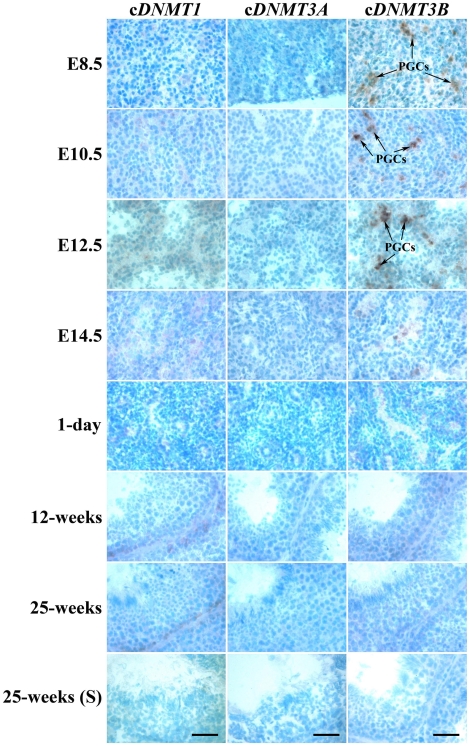
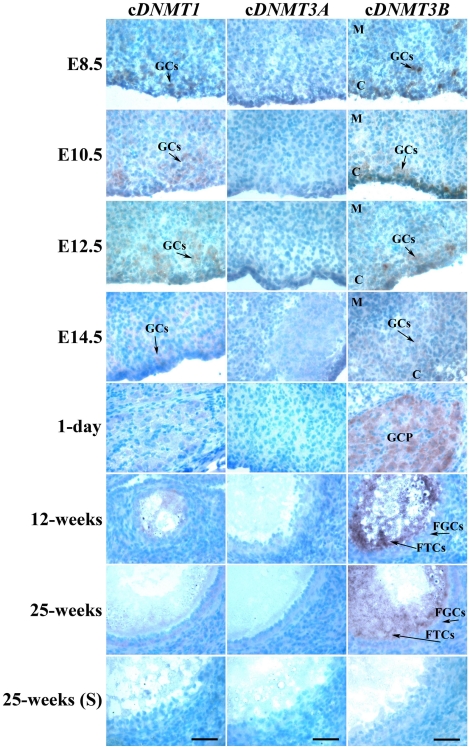
Cryosections of male and female gonads at E8.5, E10.5, E12.5, and E14.5, and testes and ovaries at 1 day, 12 weeks, and 25 weeks of age were subjected to in situ hybridization to examine the mRNA expression patterns of cDNMT family members during germ line development and sexual maturation. In males, cDNMT1 mRNA expression was detected at low levels in the PGCs (until E12.5) and prospermatogonia (on E14.5). cDNMT1 mRNA expression was detected at a basal level in the prospermatogonia in 1-day-old testis. At 12 and 25 weeks of age, cDNMT1 mRNA expression reappeared at a low level in the spermatogonia cells near the basement membrane. cDNMT3A mRNA expression was detected at a low level from E8.5 to E14.5, after which it maintained a basal level until 25 weeks of age. As expected, cDNMT3B mRNA expression was detected at a strong level in the PGCs (until E12.5). After PGC differentiation, it was detected at a low level in the prospermatogonia until 1 day of age and in germ line cells until testes were 25 weeks old (Fig. 7). In females, cDNMT1 mRNA expression was detected at a moderate level in the PGCs and oogonia cells from E8.5 to E12.5. On E14.5 and 1 day of age, it was detected at a low level in the primary oocytes. From 12 weeks of age, it was detected at low levels in the secondary oocytes, maturing ova, and follicular cells. cDNMT3A mRNA expression was detected at a low level from E8.5 to E12.5. After E12.5, it was detected at a basal level in the ovaries. cDNMT3B mRNA expression was similar to cDNMT3A until E14.5. Interestingly, however, it was strongly detected in the primary oocytes deposited in the germ cell/oocyte pool at 1 day of age, and secondary oocytes, maturing ova, and follicular cells in 12-week-old and 25-week-old ovaries (Fig. 8).
Figure 7. Expression patterns of cDNMT1, cDNMT3A, and cDNMT3B during male germ line development and sexual maturation.
Cryosections of male gonads at E8.5, E10.5, E12.5, and E14.5, and testes of 1-day-, 12-week-, and 25-week-old chickens were hybridized with antisense cRNA probes against cDNMT1, cDNMT3A, and cDNMT3B. Testis sections at 25 weeks of age were hybridized with sense (S) cRNA probes of respective genes as negative control. Arrows indicate PGCs strongly express cDNMT3B. Common bar – 50 µm.
Figure 8. Expression patterns of cDNMT1, cDNMT3A, and cDNMT3B during female germ line development and sexual maturation.
Cryosections of female gonads at E8.5, E10.5, E12.5, and E14.5, and ovaries of 1-day-, 12-week-, and 25-week-old chickens were hybridized with antisense cRNA probes against cDNMT1, cDNMT3A, and cDNMT3B. Ovary sections at 25 weeks of age were hybridized with sense (S) cRNA probes of respective genes as a negative control. C = cortex, M = medulla, GCs = germ cells, GCP = germ cell pool, FGCs = follicular granulosa cells, FTCs = follicular theca cells. Common bar – 50 µm.
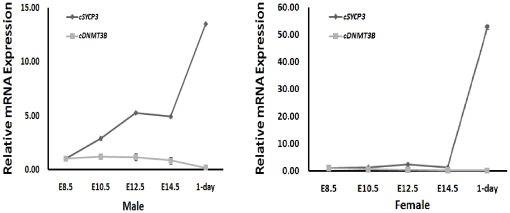
In order to investigate the correlation between cDNMT3B expression and meiotic events, we examined the expression patterns of cDNMT3B with a meiosis specific gene cSYCP3 on limited time points of meiotic stages by qRT-PCR. In males, both cDNMT3B and cSYCP3 expression were detected at a low level on E8.5. After this period, cDNMT3B expression was continuously decreased until 1-day. However, cSYCP3 expression was continuously increased until 1-day. In females, cDNMT3B expression was detected at a low level in all stages. cSYCP3 expression was detected at al low level until E14.5. After E14.5, cSYCP3 expression was sharply increased (Fig. 9).
Figure 9. Expression of cDNMT3B compared with cSYCP3 during limited points of meiotic stages examined by qRT-PCR.
cDNA from male and female gonads on embryonic days E8.5, E10.5, E12.5, E14.5 and 1-day were amplified with cDNMT3B- and cSYCP3-specific primers. The threshold cycle of cDNMT3B and cSYCP3 genes were normalized with chicken glyceraldehyde-3-phosphate dehydrogenase (cGAPDH). Relative gene expression was calculated using the 2−ΔΔCt method.
Expression patterns of miRNA and Regulation of cDNMT3B
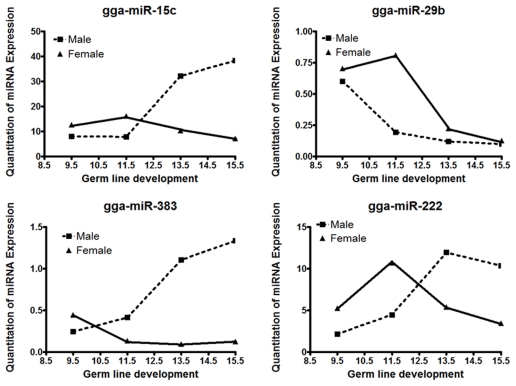
Expression patterns of miRNAs gga-miR-15c, gga-miR-29b, gga-miR-383 and gga-miR-222 were examined during meiotic stages of germ line development by qRT-PCR. During male germ line development, miR-15c, miR-383 and miR-222 were detected at a low level on E9.5 to E11.5. After E11.5, all these miRNAs were detected at a high level. In contrast to other miRNAs, miR-29b expression was detected at a high level on E9.5, and continuously decreased until E15.5. During female germ line development, miR-15c, miR-29b and miR-222 showed similar patterns of expression. All these miRNAs were detected at a moderate level on E9.5, slightly increased until E11.5, and then continuously decreased until E15.5. miR-383 expression was detected at a moderate level on E9.5, after this period, the expression was decreased to a low level in all stages (Fig. 10).
Figure 10. Expression of gga-miR-15c, gga-miR-29b, gga-miR-383 and gga-miR-222 during meiotic stages of germ line development examined by qRT-PCR.
cDNA from male and female gonads on embryonic days E9.5, E10.5, E11.5, E12.5, E13.5, E14.5 and E15.5 were amplified with respective miRNA-specific forward primers and universal reverse primers. The threshold cycle of miRNA expression were normalized with chicken snoRNA (endogenous control).
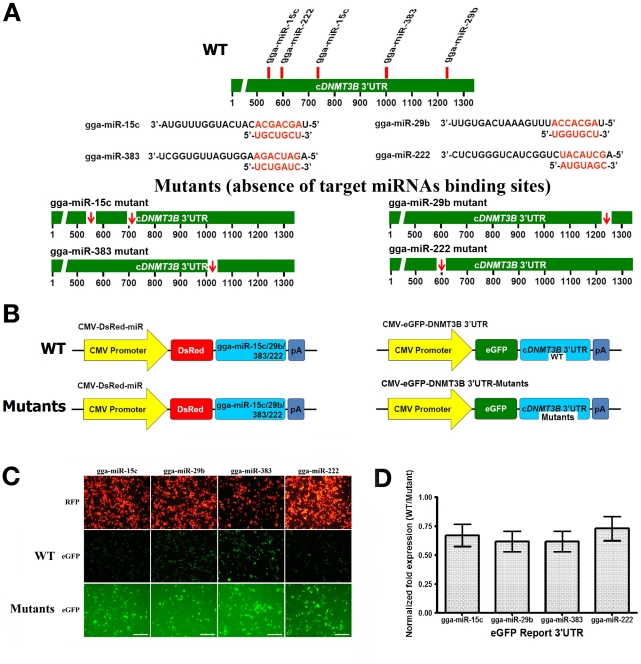
To validate the downregulation of cDNMT3B at the post-transcription levels, the miRNAs, gga-miR-15c (target score: 74, seed location: 544 and 732), gga-miR-29b (target score: 62, seed location: 1223), gga-miR-383 (target score: 53, seed location: 996), and gga-miR-222 (target score: 50, seed location: 591) were selected for the cDNMT3B 3′-UTR based on an online database miRDB (Fig. 11A). Expression vectors were constructed and combined with eGFP or RFP (Fig. 11B). cDNMT3B 3′UTR and cDNMT3B 3′UTR mutants for each miRNA binding sites generated by point mutation cloned into a pcDNA3 plasmid encoding eGFP and cDNMT3B 3′UTR-specific miRNAs including gga-miR-15c, gga-miR-29b, gga-miR-383, and gga-miR-222 cloned into a pDsRed2-N1 plasmid, respectively, encoding RFP were cotransfected into 293 FT cells. After 48 h of cotransfection, cDNMT3B 3′UTR mutants encoding eGFP expression was constant as a control. cDNMT3B 3′UTR intensity significantly decreased in all investigated miRNAs when compared to the eGFP expression of cDNMT3B 3′UTR mutants (Fig. 11C). FACS analysis was performed to further examine the inhibition rate of cDNMT3B 3′UTR encoding eGFP expression from miRNA modulation (Fig. 11D). Compared to each mutants, cDNMT3B 3′UTR eGFP expression was slightly decreased by gga-miR-15c (25.82%), gga-miR-29b (30.01%), gga-miR-383 (30.0%), and gga-miR-222 (31.28%). In addition, chicken DNMT3B 3′UTR region is highly conserved with human and mouse DNMT3B 3′UTR regions. However, the cDNMT3B 3′UTR specific miRNAs binding sites are not conserved in human and mouse (Fig. S4).
Figure 11. Regulation of cDNMT3B.
(11A) The miRNA binding sites and mutants of each miRNA for cDNMT3B 3′UTR. (11B) Schematic diagram of constructed expression vectors for the dual fluorescent reporter assay. In the dual fluorescent reporter assay, cDNMT3B 3′UTR and mutants of cDNMT3B 3′UTR were cloned into a pcDNA3 plasmid encoding enhanced green fluorescence protein (eGFP) and cDNMT3B 3′UTR-specific miRNAs including gga-miR-15c, gga-miR-29b, gga-miR-383, and gga-miR-222 cloned into a pDsRed2-N1 plasmid encoding red fluorescence protein (RFP) were cotransfected into 293 FT cells. After 48 h of cotransfection, eGFP expression was examined under a confocal laser microscope (11C, Common bar – 20 µm). The inhibition rate of eGFP expression from miRNA modulation was calculated by fluorescence-activated cell sorting (FACS, 11D). The y-axis represents the normalized fold expression (WT vs Mutants). Error bars indicate the standard error of triplicate analysis.
Discussion
The C-terminus of cDNMT1, cDNMT3A, and cDNMT3B contains the highly conserved catalytic domain DNA methylase. DNA methylase enzymes specifically methylate the C-5 carbon of cytosines to produce C5-methylcytosine. Cytosine-specific methyltransferases transfer methyl groups from S-adenosylmethionine to cytosines in CpG dinucleotides, which modulate gene expression and cell differentiation during embryogenesis [18]. The noncatalytic N-terminus of cDNMT1 is an independent domain structure that interacts with different regulatory proteins and DNA, and significantly differs from the N-terminus of cDNMT3A and cDNMT3B. The noncatalytic N-terminus of cDNMT1 consists of a DMAP1 binding domain, DNMT1-specific replication foci domain, CXXC zinc finger domain, and BAH domains, which are also conserved in mammalian DNMT1 proteins. DMAP1 is a transcriptional corepressor that binds to the N-terminal amino acids of DNMT1. DNMT1-specific replication foci domain function noncatalytically to target the proteins toward replication foci, and allow DNMT1 protein to methylate the correct residues [18]. The CXXC zinc finger domain contains eight conserved cysteine residues that bind to nonmethyl-CpG dinucleotides. The CXXC domain is found in a variety of chromatin-associated proteins [19], [20]. The BAH domain appears to act as a protein–protein interaction module specialized in gene silencing. The BAH module might play an important role by linking DNA methylation, replication, and transcriptional regulation [21]. The noncatalytic N-terminus of cDNMT3A and cDNMT3B consists of a unique domain, the Proline-Tryptophan-Tryptophan-Proline motif (PWWP domain), which binds to histone-4 methylated at lysine-20 (H4K20me). The methylation of H4K20 is involved in organizing higher-order chromatin, maintaining genome stability, and regulating cell cycle progression [22].
In mammals, many researchers have reported that DNMTs are expressed at high levels in undifferentiated embryonic stem cells (ESCs), embryonic carcinoma cells, germ cells, and gonads [4], [23], [24]. Our results in this investigation suggest that cDNMT1, cDNMT3A, and cDNMT3B are highly active in early embryos for the maintenance of methylation patterns and de novo methylation. During midembryonic development, cDNMT3B mRNA expression was strongly detected in PGCs when compared to its expression in germ cells, suggesting that cDNMT3B is more highly active than other members of the cDNMT family to ensure de novo methylation in PGCs. During late embryonic development and germ line development, cDNMT family members were weakly detected in both sexes. However, cDNMT3B expression was reestablished in female germ cells after hatching. DNA methylation occurs in a nonrandom manner within the genome, and the generated methylation pattern is gene- and tissue-specific. The generation of the methylation pattern requires de novo methylation during embryogenesis [25]. De novo methylation is largely suppressed in differentiated somatic cells; however, it is upregulated in germ cells and is believed to play a critical role in the establishment of genomic imprinting in the gametes [24]. cDNMT3B expression in follicular theca cells remains unclear. Our experiments suggest that cDNMT3B expression in PGCs and germ cells is higher than that of other members. On the other hand, cDNMT1 expression in PGCs and germ cells is higher than that of cDNMT3A, which suggests that cDNMT1 might be significantly involved in de novo methylation and interaction with cDNMT3B. Although its preference for hemimethylated DNA is unique among DNMTs, DNMT1 also has a significant capacity for de novo methylation [23], [26]. In the mouse, Dnmt1 expression is strong in PGCs, growing oocytes, and proliferating male germ cells, but is downregulated during late embryonic development, which supports our findings that genome-wide DNA methylation occurs after germ cell proliferation is arrested, when the DNMT1 expression is downregulated [27].
miRNAs are short, noncoding RNAs that usually bind to their complementary sequences in the 3′UTR of target mRNA, resulting in gene silencing or downregulation at the post-transcriptional level [28]. Many miRNAs cloned from chicken embryos were previously reported to be ESC-specific in the mouse and human, indicating their contribution to basic cellular functions and maintenance of pluripotency [29]. Regulation of cDNMT3B using the cDNMT3B 3′UTR target miRNAs gga-miR-15c, gga-miR-29b, gga-miR-383, and gga-miR-222 is of great interest in this study. We found that cDNMT3B encoding eGFP expression was significantly decreased by all investigated miRNAs when compared to the control eGFP expression. To our knowledge, the expression and functions of miR-15c have not been well studied. miR-383 expression is abundant in meiotic prophase cells and primary spermatocytes in mammals [30]. miR-383 significantly interacts with growth arrest and the DNA damage-inducible gamma (GADD45G) gene, which inhibits cell growth and induces apoptosis. The silencing of GADD45G could be reversed by genetic double knockout of DNMT1 and DNMT3B, indicating a direct epigenetic mechanism [31]. The expression and functions of miR-29b and miR-222 have been extensively characterized in mammals.
miR-29b directly targets DNMT3A and DNMT3B, and indirectly targets DNMT1, thereby leading to downregulation of genes, reduction of global DNA methylation, and re-expression of the DNA hypermethylated and silenced tumor suppressor genes [32]. miR-29b significantly regulates many collagen genes, matrix metalloproteinase 2 (MMP2), integrin beta1 (ITGB1), progranulin (PGRN), podoplanin (PDPN), and other genes related to the extracellular matrix [33], [34], [35]. Furthermore, it plays an important role during osteoblast differentiation and gonadogenesis [36], [37]. It is expressed in mouse PGCs, and its expression is upregulated in a female-specific manner when male-specific de novo methylation of the PGC genome occurs [37]. miR-222 induces cell growth and cell cycle progression via direct targeting of cyclin-dependent kinase inhibitors 1B and 1C (p27 and p57). miR-222 significantly interacts with several target genes including: p27 and p57 [38], [39], [40]; phosphatase and tensin homolog (PTEN) [41]; estrogen receptor alpha (ERalpha) [42]; pro-apoptotic gene PUMA [43]; protein phosphatase 2A subunit B (PPP2R2A) [44]; and β1-syntrophin [45]. Functionally, it is involved in inflammation-mediated vascular remodeling [46], is modulated during myogenesis, and plays a role both in the progression from myoblasts to myocytes and in the achievement of the fully differentiated phenotype [47]. miR-222 also participates in ESC differentiation by regulating ESCs terminally withdrawing from the cell cycle [48].
Downregulation of DNMT3B causes hypomethylation in germ line cells and somatic cells [49]. Because DNMTs are associated with genomic imprinting, gene expression, and embryonic development, cDNMT3B downregulation might cause hypomethylation and downregulation of genes that are normally germ cell-specific or somatic cell-specific. We examined the expression of gga-miR-15c, gga-miR-29b, gga-miR-383 and gga-miR-222 during meiotic stages of germ line development by qRT-PCR. In correlation with cDNMT3B expression, gga-miR-15c, gga-miR-383 and gga-miR-222 expressions were increased after E11.5 in males. On the other hand, all four miRNAs were detected at a low level during meiotic stages in females. Earlier publications reported the downregulation of DNMT3B by miR-29b or miR-383 particularly in tumor cells [31], [32]. Our studies in chickens reinforce that miR-15c, miR-29b, miR-383 and miR-222 may downregulate DNMT3B in PGCs from female embryos after they enter meiosis. We also suggest that miR-15c, miR-383 and miR-222 may downregulate DNMT3B in PGCs from male embryos.
Our study highlights the bioinformatics analysis of sequence conservation and functional domains of cDNMT family proteins, and the conserved expression patterns of cDNMT family genes during early embryonic development, germ line development, and sexual maturation of testis and ovaries. All cDNMT family members were differentially expressed during early embryonic development. Of interest, expression of the de novo DNA methyltransferase gene cDNMT3B was highly detected in the early embryos and PGCs. During late germ line development and sexual maturation, cDNMT3B expression was reestablished in a female germ cell-specific manner. Correlation between cDNMT3B and miRNAs expressions during meiotic stages of germ line development suggests that gga-miR-15c, gga-miR-383 and gga-miR-222 may downregulate cDNMT3B in vivo in both male and female chickens. Gga-miR-29b is believed to downregulate cDNMT3B in a sex specific manner. Our dual fluorescent reporter assay suggests that gga-miR-29b, gga-miR-383 and gga-miR-222 may cause maximum (30.01–31.28%) downregulation of cDNMT3B in vitro.
Supporting Information
Sequence comparison of vertebrate DNMT1 proteins. The protein sequences of DNMT1 from chicken, human, chimpanzee, pig, cattle, horse, rat, mouse, opossum, and zebrafish were aligned using the CLUSTAL X program and edited with the BioEdit program. Dark/light gray shaded sequences indicate amino acids identical/similar to those in chicken DNMT1, and dashes represent gaps in the sequence. Arrows indicate the positions of the DMAP1 (DNMT1-associated protein 1) binding domain, DNMT1-specific replication foci domain, CXXC zinc finger domain, BAH (bromo-adjacent homology) domains, and DNA methylase domain in the chicken sequence.
(PDF)
Sequence comparison of vertebrate DNMT3A proteins. The protein sequences of DNMT3A from chicken, human, chimpanzee, pig, cattle, horse, rat, mouse, opossum, and zebrafish were aligned using the CLUSTAL X program and edited with the BioEdit program. Dark/light gray shaded sequences indicate amino acids identical/similar to those in chicken DNMT3A, and dashes represent gaps in the sequence. Arrows indicate the positions of the PWWP domain and DNA methylase domain in the chicken sequence.
(PDF)
Sequence comparison of vertebrate DNMT3B proteins. The protein sequences of DNMT3B from chicken, human, chimpanzee, pig, cattle, horse, rat, mouse, opossum, and zebrafish were aligned using the CLUSTAL X program and edited with the BioEdit program. Dark/light gray shaded sequences indicate amino acids identical/similar to those in chicken DNMT3B, and dashes represent gaps in the sequence. Arrows indicate the positions of the PWWP domain and DNA methylase domain in the chicken sequence.
(PDF)
Comparison of chicken DNMT3B 3′UTR and DNMT3B 3′UTR specific miRNA binding sites with human and mouse DNMT3B 3′UTR using the CLUSTAL X program. miR-15c, miR-29b, miR-383 and miR-222 binding sites in chicken and corresponding human and mouse sequences are shown in red colour.
(PDF)
Acknowledgments
We thank Dr. Gwonhwa Song, Ph.D. (WCU Biomodulation Major, Department of Agricultural Biotechnology, Seoul National University, Korea) for suggestions and critical review of this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the BioGreen 21 Program (20070401034010) and by the World Class University (WCU) program (R31-10056) through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Li E, Beard C, Jaenisch R. Role for DNA Methylation in Genomic Imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 2.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 3.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. Journal of Biological Chemistry. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 4.Tajima S, Tsuda H, Wakabayashi N, Asano A, Mizuno S, et al. Isolation and Expression of a Chicken DNA Methyltransferase Cdna. Journal of Biochemistry. 1995;117:1050–1057. doi: 10.1093/oxfordjournals.jbchem.a124805. [DOI] [PubMed] [Google Scholar]
- 5.Yen RWC, Vertino PM, Nelkin BD, Yu JJ, Eldeiry W, et al. Isolation and Characterization of the Cdna-Encoding Human DNA Methyltransferase. Nucleic Acids Research. 1992;20:2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra MV, Bisht KS, Sun L, Muldoon-Jacobs K, Awwad R, et al. DNMT1 as a molecular target in a multimodality-resistant phenotype in tumor cells. Molecular Cancer Research. 2008;6:243–249. doi: 10.1158/1541-7786.MCR-07-0373. [DOI] [PubMed] [Google Scholar]
- 7.Bechtel W, McGoohan S, Zeisberg EM, Muller GA, Kalbacher H, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nature Medicine. 2010;16:544–U575. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng J, Zhou Y, Campbell SL, Le T, Li E, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nature Neuroscience. 2010;13:423–U437. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Salle S, Trasler JM. Dynamic expression of DNMT3a and DNMT3b isoforms during male germ cell development in the mouse. Developmental Biology. 2006;296:71–82. doi: 10.1016/j.ydbio.2006.04.436. [DOI] [PubMed] [Google Scholar]
- 10.Eyal-Giladi H, Kochav S. From cleavage to primitive streak formation: a complementary normal table and a new look at the first stages of the development of the chick. I. General morphology. Dev Biol. 1976;49:321–370. doi: 10.1016/0012-1606(76)90178-0. [DOI] [PubMed] [Google Scholar]
- 11.Hamburger V, Hamilton HL. A Series of Normal Stages in the Development of the Chick-Embryo, (Reprinted from Journal of Morphology, Vol 88, 1951). Developmental Dynamics. 1992;195:231. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 12.Han JY, Park TS, Kim JN, Kim MA, Lim D, et al. Gene expression profiling of chicken primordial germ cell ESTs. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JW, Kim S, Kim TM, Kim YM, Seo HW, et al. Basic Fibroblast Growth Factor Activates MEK/ERK Cell Signaling Pathway and Stimulates the Proliferation of Chicken Primordial Germ Cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, et al. Pfam: clans, web tools and services. Nucleic Acids Research. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–U876. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rengaraj D, Gao F, Liang XH, Yang ZM. Expression and regulation of type II integral membrane protein family members in mouse male reproductive tissues. Endocrine. 2007;31:193–201. doi: 10.1007/s12020-007-0027-6. [DOI] [PubMed] [Google Scholar]
- 17.Wang XW. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. Rna-a Publication of the Rna Society. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 19.Bestor TH. Activation of Mammalian DNA Methyltransferase by Cleavage of a Zn Binding Regulatory Domain. Embo Journal. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cross SH, Meehan RR, Nan XS, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nature Genetics. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 21.Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. Febs Letters. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Reddy B, Thompson J, Wang HB, Noma K, et al. Regulation of Set9-Mediated H4K20 Methylation by a PWWP Domain Protein. Molecular Cell. 2009;33:428–437. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okano M, Xie SP, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genetics. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 24.Chen TP, Ueda Y, Xie SP, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. Journal of Biological Chemistry. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 25.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. Embo Journal. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PDS, et al. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Developmental Biology. 2009;334:213–223. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakai Y, Suetake I, Itoh K, Mizugaki M, Tajima S, et al. Expression of DNA methyltransferase (Dnmt1) in testicular germ cells during development of mouse embryo. Cell Structure and Function. 2001;26:685–691. doi: 10.1247/csf.26.685. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao P, Zhou H, Xiao ZD, He JH, Huang MB, et al. Identification of novel chicken microRNAs and analysis of their genomic organization. Gene. 2008;418:34–40. doi: 10.1016/j.gene.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Lian J, Zhang XS, Tian H, Liang N, Wang Y, et al. Altered microRNA expression in patients with non-obstructive azoospermia. Reproductive Biology and Endocrinology. 2009;7 doi: 10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying JM, Srivastava G, Hsieh WS, Gao ZF, Murray P, et al. The stress-responsive gene GADD45G is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clinical Cancer Research. 2005;11:6442–6449. doi: 10.1158/1078-0432.CCR-05-0267. [DOI] [PubMed] [Google Scholar]
- 32.Garzon R, Liu SJ, Fabbri M, Liu ZF, Heaphy CEA, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Taylor NE, Lu LM, Usa K, Cowley AW, et al. Renal Medullary MicroRNAs in Dahl Salt-Sensitive Rats miR-29b Regulates Several Collagens and Related Genes. Hypertension. 2010;55:974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao J, Herl LD, Farese RV, Gao FB. MicroRNA-29b Regulates the Expression Level of Human Progranulin, a Secreted Glycoprotein Implicated in Frontotemporal Dementia. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortez MA, Nicoloso MS, Shimizu M, Rossi S, Gopisetty G, et al. miR-29b and miR-125a Regulate Podoplanin and Suppress Invasion in Glioblastoma. Genes Chromosomes & Cancer. 2010;49:981–990. doi: 10.1002/gcc.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li ZY, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, et al. Biological Functions of miR-29b Contribute to Positive Regulation of Osteoblast Differentiation. Journal of Biological Chemistry. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takada S, Berezikov E, Choi YL, Yamashita Y, Mano H. Potential role of miR-29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos. Rna-a Publication of the Rna Society. 2009;15:1507–1514. doi: 10.1261/rna.1418309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27(Kip1)*. Journal of Biological Chemistry. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 39.Wurz K, Garcia RL, Goff BA, Mitchell PS, Lee JH, et al. MiR-221 and MiR-222 Alterations in Sporadic Ovarian Carcinoma: Relationship to CDKN1B, CDKN1C and Overall Survival. Genes Chromosomes & Cancer. 2010;49:577–584. doi: 10.1002/gcc.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frenquelli M, Muzio M, Scielzo C, Fazi C, Scarfo L, et al. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010;115:3949–3959. doi: 10.1182/blood-2009-11-254656. [DOI] [PubMed] [Google Scholar]
- 41.Zhang CZ, Han L, Zhang AL, Fu YC, Yue XA, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. Bmc Cancer. 2010;10 doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, et al. MicroRNA Cluster 221–222 and Estrogen Receptor alpha Interactions in Breast Cancer. Journal of the National Cancer Institute. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Molecular Cancer. 2010;9 doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong QWL, Ching AKK, Chan AWH, Choy KW, To KF, et al. MiR-222 Overexpression Confers Cell Migratory Advantages in Hepatocellular Carcinoma through Enhancing AKT Signaling. Clinical Cancer Research. 2010;16:867–875. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 45.De Arcangelis V, Serra F, Cogoni C, Vivarelli E, Monaco L, et al. beta1-syntrophin modulation by miR-222 in mdx mice. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dentelli P, Rosso A, Orso F, Olgasi C, Taverna D, et al. microRNA-222 Controls Neovascularization by Regulating Signal Transducer and Activator of Transcription 5A Expression. Arteriosclerosis Thrombosis and Vascular Biology. 2010;30:1562–U1125. doi: 10.1161/ATVBAHA.110.206201. [DOI] [PubMed] [Google Scholar]
- 47.Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, et al. Microrna-221 and Microrna-222 Modulate Differentiation and Maturation of Skeletal Muscle Cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian K, Hu LL, Chen H, Li HX, Liu N, et al. Hsa-miR-222 Is Involved in Differentiation of Endometrial Stromal Cells in Vitro. Endocrinology. 2009;150:4734–4743. doi: 10.1210/en.2008-1629. [DOI] [PubMed] [Google Scholar]
- 49.Velasco G, Hube F, Rollin J, Neuillet D, Philippe C, et al. Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9281–9286. doi: 10.1073/pnas.1000473107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence comparison of vertebrate DNMT1 proteins. The protein sequences of DNMT1 from chicken, human, chimpanzee, pig, cattle, horse, rat, mouse, opossum, and zebrafish were aligned using the CLUSTAL X program and edited with the BioEdit program. Dark/light gray shaded sequences indicate amino acids identical/similar to those in chicken DNMT1, and dashes represent gaps in the sequence. Arrows indicate the positions of the DMAP1 (DNMT1-associated protein 1) binding domain, DNMT1-specific replication foci domain, CXXC zinc finger domain, BAH (bromo-adjacent homology) domains, and DNA methylase domain in the chicken sequence.
(PDF)
Sequence comparison of vertebrate DNMT3A proteins. The protein sequences of DNMT3A from chicken, human, chimpanzee, pig, cattle, horse, rat, mouse, opossum, and zebrafish were aligned using the CLUSTAL X program and edited with the BioEdit program. Dark/light gray shaded sequences indicate amino acids identical/similar to those in chicken DNMT3A, and dashes represent gaps in the sequence. Arrows indicate the positions of the PWWP domain and DNA methylase domain in the chicken sequence.
(PDF)
Sequence comparison of vertebrate DNMT3B proteins. The protein sequences of DNMT3B from chicken, human, chimpanzee, pig, cattle, horse, rat, mouse, opossum, and zebrafish were aligned using the CLUSTAL X program and edited with the BioEdit program. Dark/light gray shaded sequences indicate amino acids identical/similar to those in chicken DNMT3B, and dashes represent gaps in the sequence. Arrows indicate the positions of the PWWP domain and DNA methylase domain in the chicken sequence.
(PDF)
Comparison of chicken DNMT3B 3′UTR and DNMT3B 3′UTR specific miRNA binding sites with human and mouse DNMT3B 3′UTR using the CLUSTAL X program. miR-15c, miR-29b, miR-383 and miR-222 binding sites in chicken and corresponding human and mouse sequences are shown in red colour.
(PDF)