Abstract
Background
Green fluorescent protein (GFP) and its fusion proteins have been used extensively to monitor and analyze a wide range of biological processes. However, proteolytic cleavage often removes GFP from its fusion proteins, not only causing a poor signal-to-noise ratio of the fluorescent images but also leading to wrong interpretations.
Methodology/Principal Findings
Here, we report that the M153R mutation in a ratiometric pH-sensitive GFP, pHluorin, significantly stabilizes its fusion products while the mutant protein still retaining a marked pH dependence of 410/470 nm excitation ratio of fluorescence intensity. The M153R mutation increases the brightness in vivo but does not affect the 410/470-nm excitation ratios at various pH values.
Conclusions/Significance
Since the pHluorin(M153R) probe can be directly fused to the target proteins, we suggest that it will be a potentially powerful tool for the measurement of local pH in living cells as well as for the analysis of subcellular localization of target proteins.
Introduction
Green fluorescent protein (GFP) and related fluorescent proteins have been utilized to monitor and analyze a wide range of biological processes such as gene expression, protein localization and cell motility. These fluorescent proteins can also be used as the indicator of Ca2+ or ATP concentrations, or pH because they provide a high sensitivity in detection and are not toxic to living cells [1]–[3]. A GFP-derivative, pHluorin, is a ratiometric pH indicator with excitation wavelength at 410 and 470 nm and emission at 508 nm [3]. The relative emission intensity of pHluorin at the two excitation wavelengths show a remarkable pH dependence, thereby pH can be measured by the 410/470 nm excitation ratio, R410/470. The R410/470 ratio changes in less than 0.5 ms when pH is changed, indicating that the ratiometric measurement by pHluorin can detect a rapid pH change [4]. Since ratiometric methods, in which dual-wavelength measurements detect changes in the fluorescence absorption or emission spectra upon ion-binding, are independent of the concentration of the indicator, a precise and quantitative pH measurement of living cells can be easily carried out using pHluorin.
Since local pH is one of the most important parameters for probing the activities of live cells, pH imaging is becoming an efficient and useful method in various fields of biological sciences. Unlike GFP itself, however, GFP fusion proteins are fairly susceptible to proteolytic cleavage and so GFP is often released from target proteins, resulting in the poor signal-to-noise ratio of the fluorescent images. We therefore tried to improve the stability of fusion proteins in the experimental system that we study.
The flagellar motor of Salmonella enterica is powered by the electrochemical proton gradient across the cytoplasmic membrane. Two integral membrane proteins, MotA and MotB, form a proton channel to couple proton flow to torque generation. An interaction of MotA with a rotor protein FliG is required for torque generation [5]. The rotation-dependent proton influx has been estimated to be about 1,200 protons per revolution [6]. Since a decrease in intracellular pH significantly reduces flagellar motor rotation [7], [8], the proton release from the proton channel to the cytoplasm plays an important role in the torque generation process, and local pH near the motor must be tightly controlled. We therefore tried to measure local pH of the cytoplasmic side the motor by expressing pHluorin fusion proteins to flagellar motor proteins, such as pHluorin-FliG and pHluorin-MotB. However, the fusion proteins were susceptible to proteolytic digestion in the cell, making the high sensitivity pH imaging difficult.
In this study, we show that the M153R mutation in pHluorin markedly improves the stability of its fusion proteins while the mutant protein still retaining the pH dependence of 410/470 nm excitation ratio of fluorescence intensity to be useful as a pH sensor.
Results
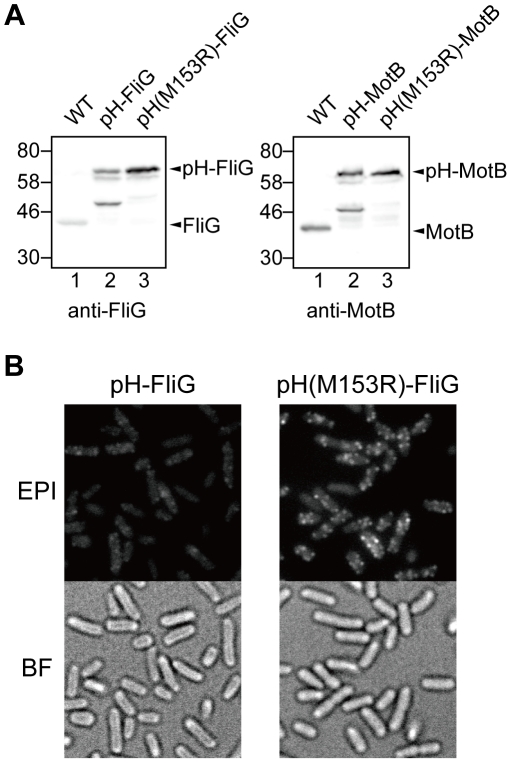
To investigate local pH near the bacterial flagellar motor, the ratiometric pHluorin probe must be localized to the motor. Since it has been reported that the GFP-FliG and GFP-MotB fusion proteins are partially functional [9], [10], we fused pHluorin to the N-termini of FliG and MotB to produce Salmonella pHluorin-fliG and pHluorin-motB strains, respectively. However, these fusion proteins were unstable, and about a half of them were cleaved into a 47 kDa fragment as shown on the immunoblots (Figure 1A, lane 2 in both panels). Neither intact FliG nor MotB was observed, suggesting that the cleavage must occur within pHluorin. To identify the cleavage sites, a His8 tag was attached to pHluorin-MotB at its C terminus to facilitate protein purification. The cleavage product of pHluorin-MotB-His8 was purified by Ni-NTA affinity chromatography, and its molecular mass was measured by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. The molecular mass was around 43.2–44.8 kDa. In search of possible proteolytic fragments having these masses, we identified possible cleavage sites to be at Met-153, Ala-154, Lys-162 or Ala-163 of pHluorin. Therefore, we carried out site-directed mutagenesis of these residues to see if any of such mutations have a stabilizing effect on the fusion proteins. The M153R mutation markedly stabilized both pHluorin-FliG and pHluorin-MotB (Figure 1A, lane 3 in both panels) while the other mutations showed no improvement (data not shown).
Figure 1. Effects of the M153R mutation in pHluorin on the protein stability of its fusion products.
(A) Immunoblotting, using polyclonal anti-FliG (left panel) or anti-MotB antibody (right panel), of whole cell proteins prepared from SJW1103 (WT), YVM1002 (pHluorin-FliG, indicated as pH-FliG), YVM1004 (pHluorin(M153R)-FliG, indicated as pH(M153R)-FliG), YVM1001 (pHluorin-MotB, indicated as pH-MotB) and YVM1003 (pHluorin(M153R)-MotB, indicated as pH(M153R)-MotB). The positions of molecular mass markers (kDa) are shown on the left. (B) Fluorescence images (EPI) and bright field images (BF) of YVM1002 and YVM1004. The cells were grown overnight in LB at 30°C and observed by fluorescence microscopy.
We next investigated whether the M153R mutation increases the signal to noise (S/N) ratio. Since the turnover of GFP-FliG between the cytoplasmic pool and functional motors does not occur [11], we analyzed the subcellular localization of pHluorin-FliG and pHluorin(M153R)-FliG by epi-illumination fluorescence microscopy. The M153R mutation substantially increased the number of fluorescent spots of pHluorin-FliG (Figure 1B). The fluorescence intensities of the pHluorin-FliG and pHluorin(M153R)-FliG spots were 1,065±278 A.U. (n = 109) and 3,560±1322 A.U. (n = 126), respectively, indicating that the M153R mutation resulted in a remarkable improvement in the S/N ratio of the fluorescent images.
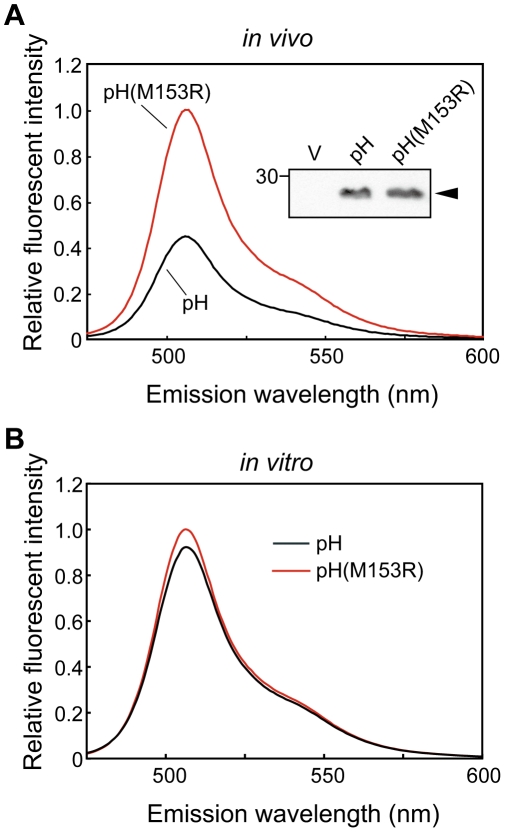
To test whether the M153R mutation affects the brightness of pHluorin alone in vivo, we transformed a Salmonella wild-type strain, SJW1103, with a plasmid encoding pHluorin(M153R) on pKK223-3 and analyzed the fluorescent intensity with a spectrophotometer (Figure 2A). We used SJW1103 expressing pHluorin as a control. Immunoblotting with polyclonal GFP antibody revealed that the expression level of pHluorin(M153R) was the same as that of pHluorin (Figure 2A, inset) and wild-type GFP (data not shown), indicating that the M153R mutation does not alter the stability of pHluorin itself. Interestingly, the fluorescent intensity of pHluorin(M153R) was approximately 2.5-fold brighter than that of pHluorin. When the fluorescence intensities of purified pHluorin and pHluorin(M153R) were measured at the same protein concentration, there was no difference in the fluorescence intensity (Figure 2B). This result indicates that the M153R mutation does not increase the intrinsic brightness of properly matured pHluorin molecules. Therefore, we conclude that the M153R mutation improves the folding efficiency of pHluorin in vivo.
Figure 2. Effect of the M153R mutation on Ratio410/470.
(A) Fluorescent intensities of SJW1103/pYC001 (pHluorin) and SJW1103/pYVM001 (pHluorin(M153R)) cells grown in T-broth at 30°C. Emission spectra with 395 nm excitation were measured by a fluorescence spectrophotometer. The measurements were done at 23°C. Inset: Immunoblotting, using polyclonal anti-GFP antibody, of whole cell proteins. (B) Fluorescent intensities of purified pHluorin and pHluorin(M153R).
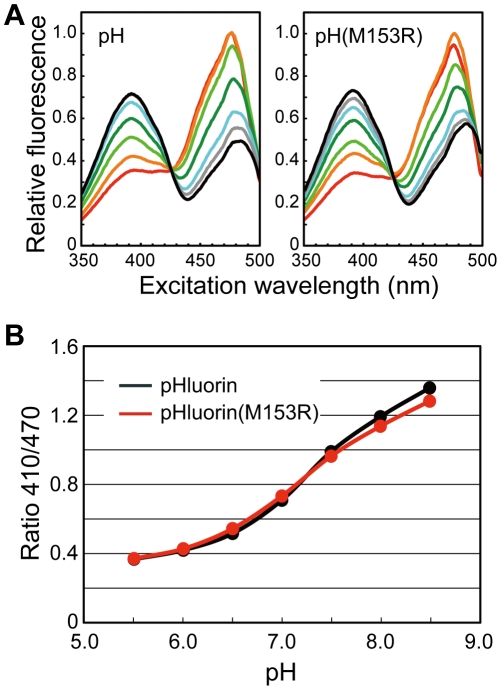
It has been reported that the M153A mutation shifts the excitation wavelength of GFP(S65T) to a longer wavelength [12]. We therefore measured the excitation spectra of purified pHluorin and pHluorin(M153R) (Figure 3). The M153R mutation changed neither the excitation wavelengths nor the emission intensity ratios at the excitation wavelengths of 410 and 470 nm over a pH range from 5.5 to 8.5.
Figure 3. pH dependence of fluorescence excitation spectra and Ratio410/470 of pHIuorin and pHluorin(M153R).
(A) Fluorescence excitation spectra. The different colored lines refer to different pH values. red, pH 5.5; orange, pH 6.0; light green, pH 6.5; green, pH 7.0; cyan, pH 7.5; grey, pH 8.0; black, pH 8.5. (B) Ratio410/470. The fluorescence excitation spectra of purified proteins were recorded on a fluorescence spectrophotometer. The measurements were done at 23°C.
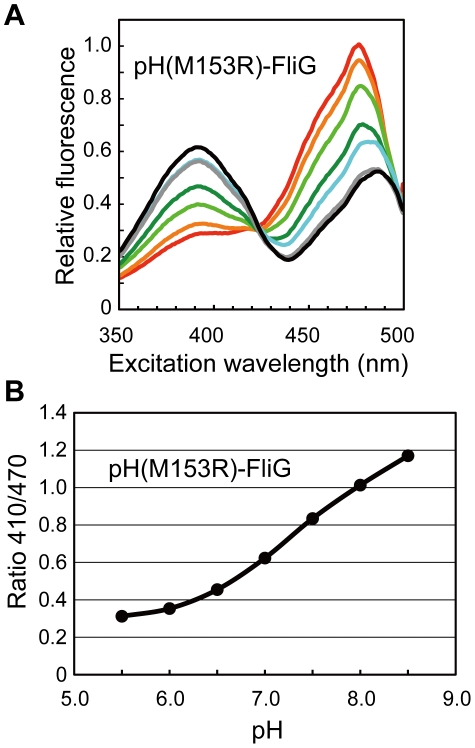
We next examined whether pHluorin(M153R)-FliG can be used as a ratiometric pH sensor. The R410/470 ratio of purified pHluorin(M153R)-FliG-His6 showed a pH-dependence from 0.3 at pH 5.5 to 1.2 at pH 8.5, which is as large as that of pHluorin(M153R) (Figure 4), indicating that pHluorin(M153R)-FliG can be a useful tool to measure local pH near the flagellar motor.
Figure 4. pH dependence of fluorescence excitation spectra and Ratio410/470 of pHluorin(M153R)-FliG-His6.
(A) Fluorescence excitation spectra. The different colored lines refer to different pH values. red, pH 5.5; orange, pH 6.0; light green, pH 6.5; green, pH 7.0; cyan, pH 7.5; grey, pH 8.0; black, pH 8.5. (B) Ratio410/470. The fluorescence excitation spectra of the purified protein were recorded on a fluorescence spectrophotometer. The measurements were done at 23°C.
Discussion
GFP has been used to determine subcellular protein localization. A peptide linker between GFP and the target protein is required for the stability and function of fusion proteins. However, GFP is often removed from fusion proteins by proteolytic cleavage. Here, we directly fused the ratiometric pHluorin probe to the N-termini of Salmonella FliG and MotB and found that these fusion proteins are also unstable in vivo (Figure 1A). As pHluorin itself is stable (Figure 2, inset), the fusion to target proteins presumably induces a conformational change in pHluorin, resulting in proteolytic cleavage of the fusion proteins. Site-directed mutagenesis revealed that the M153R mutation in pHluorin considerably improved the stability of its fusion proteins (Figure 1A). The M153R mutation also increased not only the number of fluorescent spots of pHluorin-FliG in Salmonella cells but also their fluorescence intensity, improving the S/N ratio of the images (Figure 1B). The M153R mutation did not change the 410/470-nm excitation ratios of pHluorin (Figure 3), indicating that pHluorin(M153R) can be used as a pH sensor. The 410/470-nm excitation ratios of pHluorin(M153R)-FliG-His6 also showed a similar pH-dependence (Figure 4). Since the pHluorin(M153R) probe can be directly fused to the target proteins, we believe that the pHluorin(M153R) probe can be a potentially powerful tool not only for the analysis of subcellular localization of target proteins but also for local pH measurement in living cells. We are currently developing a high-resolution pH imaging system using pHIuorin(M153R) as a probe.
Materials and Methods
Bacteria, plasmids, DNA manipulations and media
Bacterial strains and plasmids used in this study are listed in Table 1. Procedures for DNA manipulation were carried out as described previously [13]. L-broth, T-broth and motility medium were prepared as described [7]. TcS plates were prepared as described by Maloy and Nunn [14]. Ampicillin and tetracycline were added to LB at a final concentration of 100 µg/ml and 15 µg/ml, respectively.
Table 1. Strains and Plasmids used in this study.
| Strains and Plasmids | Relevant characteristics | Source or reference |
| E. coli | ||
| BL21(DE3) pLysS | T7 expression host | Novagen |
| Salmonella | ||
| SJW1103 | Wild type for motility and chemotaxis | [19] |
| YVM1001 | pHluorin-motB | This study |
| YVM1002 | pHluorin-fliG | This study |
| YVM1003 | pHluorin(M153R)-motB | This study |
| YVM1004 | pHluorin(M153R)-fliG | This study |
| YVMT001 | motB::tetRA | This study |
| YVMT002 | fliG::tetRA | This study |
| Plasmids | ||
| pGST-pHluorin | pGEX2T/GST-pHluorin | [3] |
| pYC001 | pKK223-3/pHluorin | [8] |
| pNSK22pH | pTrc99A/MotA+pHluorin-MotB-His8 * | This study |
| pNSK22pH(M153R) | pTrc99A/MotA+pHluorin(M153R)-MotB-His8 * | This study |
| pYVM001 | pKK223-3/pHluorin(M153R) | This study |
| pYVM007 | pGEX2T/GST-pHluorin(M153R) | This study |
| pYVM013 | pTrc99A/pHluorin(M153R)-FliG-His6 | This study |
*In this pHluorin-MotB-His8 fusion construct, N-terminal 28 residues of MotB (Met1- Lys28) are attached to the N-terminus of pHluorin as described before [9].
Construction of Salmonella strains expressing pHluorin-fusion proteins
To construct Salmonella pHluorin-fliG, and pHluorin-motB strains, the fliG or motB gene on the chromosome was replaced by the gfp-fliG or gfp-motB allele, respectively, by using the λRed homologous recombination system developed by Datsenko and Wanner [15]. First, the tetRA genes were inserted into the 5′-end of fliG or motB to create fliG::tetRA or motB::tetRA, respectively. Then, to replace the tetRA genes by pHluorin (GenBank accession No. AF058694), the pHluorin or pHluorin(M153R) gene was amplified by PCR using pYC001 or pYVM001 as a template and primers shown in Table 2. The PCR products were purified using a QIAquick PCR purification kit (QIAGEN). The fliG::tetRA or motB::tetRA strain transformed pKD46 [15], which has a temperature-sensitive replicon, was grown in 5-ml L-broth containing ampicillin and 0.2% L-arabinose at 30°C until OD600 had reached 0.6. The cells were washed three times with ice-cold H2O and suspended in 50 µl of ice-cold H2O. 50 µl of cells were electroporated with 100 to 200 ng of purified PCR products using 0.1-cm cuvettes at 1.8 kV. Shocked cells were incubated in 1 ml L-broth for 1 h at 37°C. Then one-half were spread onto TcS plates because tetracycline-resistant cells cannot grow in TcS plates [15] and incubated overnight at 42°C to remove pKD46. The constructs were confirmed by DNA sequencing. The pHluorin-fliG, pHluorin(M153R)-fliG, pHluorin-motB, and pHluorin(M153R)-motB alleles are placed under the control of their native promoters.
Table 2. Primers used for construction of pHluorin-fliG and pHluorin-motB strains.
| Name | Sequence |
| pH-motB_Fw | 5′-gcagtgagaaacccaaaccagcagcagacgactgaggaagcatgagtaaaggagaagaacttttc-3′ |
| pH-motB_Rv | 5′-ctgcggcgttttacgacgacaatgggatgagcctgattttttttgtatagttcatccatgccatg-3′ |
| pH-fliG_Fw | 5′-gcgcgtggtggcgctggtcattcgccagtggatgagtaacgatcatgagtaaaggagaagaacttttc-3′ |
| pH-fliG_Rv | 5′-ggtcatcaacaaaatgacgcttttatcggtaccgctaagattacttttgtatagttcatccatgccatg-3′ |
Preparation of whole cell proteins and immunoblotting
Salmonella cells were grown overnight at 30°C in T-broth with shaking. Cell pellets were suspended in a SDS-loading buffer and normalized by cell density to give a constant amount of cells. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunoblotting with polyclonal anti-FliG, anti-MotB and anti-GFP antibodies was carried out as described previously [16].
Purification of proteolytic products of pHluorin-MotB-His8
Membranes containing overproduced pHluorin-MotB-His8 fusion proteins were solubilized by 0.2% (w/v) of dodecylphosphocholine (Anatrace) and their proteolytic cleavage products were purified by Ni-NTA affinity chromatography as described [17]. Molecular mass of the cleavage products was analyzed by a mass spectrometer (Voyager DE/PRO, Applied Biosystems) as described [13].
Site-directed mutagenesis
Site-directed mutagenesis was carried out using QuickChange site-directed mutagenesis method as described in the manufacturer's instructions (Stratagene). The mutations were confirmed by DNA sequencing.
Fluorescence microscopy
Epi-fluorescence of pHluorin fusion proteins was observed by an inverted fluorescence microscope (IX-71, Olympus) with a 150× oil immersion objective lens (UApo150XOTIRFM, NA 1.45, Olympus) and an Electron-Multiplying Charge-Coupled Device (EMCCD) camera (C9100-02, Hamamatsu Photonics) as described before [10].
Purification and Spectroscopy of pHluorin, pHluorin(M153R) and pHluorin(M153R)-FliG-His6
pHluorin and pHluorin(M153R) were purified as described before [8]. pHluorin-FliG-His6 was purified by Ni-NTA affinity chromatography as described [17]. Fluorescence excitation spectra of purified pHluorin, pHluorin(M153R), and pHluorin-FliG-His6 in buffers of defined pH were recorded on a fluorescence spectrophotometer (RF-5300PC, Shimadzu), and the 410/470-nm excitation ratios of pHluorin fluorescence intensities were determined at different pH values to generate a calibration curve as described previously [18].
For measurements of intracellular pH, wild-type cells carrying pYC001 or pYVM001 were grown with shaking in T-broth at 30°C until the cell density had reached an OD600 of 1.0. The cells were washed twice with motility buffer and resuspended in motility buffer. The cells were diluted 1∶100 into motility buffer, and the fluorescence excitation spectra of the cells were recorded on a fluorescence spectrophotometer. The 410/470-nm excitation ratios were calculated and converted to pH values based on the calibration curve previously generated.
Acknowledgments
We thank M. K. Macnab for critically reading the manuscript, helpful comments and continuous support and encouragement, J. Rothman for a gift of the pHluorin probe, and J. Armitage and J. Chandler for suggestions and advice on constructing a pHluorin-MotB fusion protein.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: YVM is a research fellow of the Japan Society for the Promotion of Science. TM is a PRESTO researcher of Japan Science and Technology Agency. This work has been supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to YVM, SK, and KN) and by the Takeda Science Foundation (to TM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc Natl Acad Sci USA. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imamura H, Nhat KP, Togawa H, Saito K, Iino R, et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci USA. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miesenböck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 4.Hess ST, Heikal AA, Webb WW. Fluorescence Photoconversion Kinetics in Novel Green Fluorescent Protein pH Sensors (pHluorins). J Phys Chem B. 2004;108:10138–10148. [Google Scholar]
- 5.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 6.Meister M, Lowe G, Berg HC. The proton flux through the bacterial flagellar motor. Cell. 1987;49:643–650. doi: 10.1016/0092-8674(87)90540-x. [DOI] [PubMed] [Google Scholar]
- 7.Minamino T, Imae Y, Oosawa F, Kobayashi Y, Oosawa K. Effect of intracellular pH on the rotational speed of bacterial flagellar motors. J Bacteriol. 2003;185:1190–1194. doi: 10.1128/JB.185.4.1190-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura S, Kami-ike N, Yokota PJ, Kudo S, Minamino T, et al. Effect of intracellular pH on the torque-speed relationship of bacterial proton-driven flagellar motor. J Mol Biol. 2009;386:332–338. doi: 10.1016/j.jmb.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 9.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 10.Morimoto YV, Nakamura S, Kami-ike N, Namba K, Minamino T. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol Microbiol. 2010;78:1117–1129. doi: 10.1111/j.1365-2958.2010.07391.x. [DOI] [PubMed] [Google Scholar]
- 11.Fukuoka H, Inoue Y, Terasawa S, Takahashi H, Ishijima A. Exchange of rotor components in functioning bacterial flagellar motor. Biochem Biophys Res Commun. 2010;394:130–135. doi: 10.1016/j.bbrc.2010.02.129. [DOI] [PubMed] [Google Scholar]
- 12.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 13.Saijo-Hamano Y, Minamino T, Macnab RM, Namba K. Structural and functional analysis of the C-terminal cytoplasmic domain of FlhA, an integral membrane component of the type III flagellar protein export apparatus in Salmonella. J Mol Biol. 2004;343:457–466. doi: 10.1016/j.jmb.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 14.Maloy SR, Nunn WD. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minamino T, Macnab RM. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minamino T, Macnab RM. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol. 2000;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 18.Morimoto YV, Che Y-S, Minamino T, Namba K. Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS lett. 2010;584:1268–1272. doi: 10.1016/j.febslet.2010.02.051. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J Gen Microbiol. 1984;130:255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]