Abstract
Purpose
TrkB has been involved in poor cancer outcome. TrkB mutations have been reported in non-small cell lung cancer. In this study, we aimed at characterizing the role of 3 potentially sensitizing TrkB mutations previously reported in lung cancer.
Experimental design
We characterized three activation loop mutants of TrkB (M713I, R715G and R734C) in terms of pathway activation/phosphorylation, migration, anchorage independent growth and sensitivity to a Trk inhibitor, using NIH3T3 cells and Baf3 cells. We also sequenced the tyrosine kinase domain of TrkB in a large number of lung cancer samples of East-Asian origin and cell lines.
Results
None of the mutants were constitutively active in NIH3T3 transformation and migration assays. M713I and R734C mutants showed low levels of autophosphorylation in comparison with wild-type TrkB. Although R715G showed similar level of autophosphorylation to wild-type TrkB upon brain-derived neurotrophic factor stimulation, the mutant was not as competent as wild-type TrkB in supporting IL-3 independent growth of Baf3 cells. In addition, the Trk inhibitor AZD6918 inhibited wild-type TrkB induced cell migration and cell growth, whereas the mutants were relatively resistant to the Trk inhibitor compared to wild-type TrkB. We could not confirm the presence of non-synonymous mutation in 78 lung cancer samples and 29 cell lines.
Conclusions
Wild-type but not mutant TrkB enhances cell migration and transformation. Our study suggests that TrkB mutations should not be used for selection of patients with lung cancer treated with Trk inhibitors. High expression of wild-type TrkB might be beneficial for studies of Trk inhibitors.
Introduction
Receptor tyrosine kinases (RTK) regulate critical cellular processes, such as cell proliferation, metabolism, and migration. Deregulation of RTKs has been reported in various types of cancer, and RTK-targeted therapies, such as inhibition of EGFR in NSCLC, have been successfully developed (1).
NTRK1-3 gene family encodes tropomyosin receptor kinases (Trk A, B and C), which are activated by neurotrophins. Nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF) and neurotrophin3 bind to TrkA, TrkB and Trk C, respectively. Upon ligand binding, the tyrosine kinase and its downstream signaling are activated. Members of the Trk family are highly expressed in cells of neural origin, and are involved in neural maintenance and development (2).
Although functions related to neural cells have been extensively examined, the Trk receptor was originally described as an oncogene. Oncogenic Trk was reported as a fusion gene between the 5’ region of the tropomyosin and the tyrosine kinase domain of Trk A derived from Inv (1q) inversion. The fused protein resulted in constitutive activation of the tyrosine kinase. This type of constitutively active TrkA fusions was reported in a subset of papillary thyroid cancers and colon cancers (3, 4).
Overexpression of TrkB has been reported in several malignancies, such as neuroblastoma (5), prostate cancer, pancreatic ductal adenocarcinoma (6), multiple myeloma (7) and lung cancer (8). High levels of TrkB correlate with poor outcome (9). In vitro, TrkB has been shown to be involved in cancer cell proliferation, anoikis, cell migration/invasion and epithelial-mesenchymal transition (10, 11). These results suggest that TrkB may have a significant impact on the malignant phenotype of tumors in vivo. Accumulating evidence suggests that TrkB is a potential target for cancer therapy. Several small molecules that inhibit Trk signaling have been developed recently and are being tested in Phase I and II trials (12)
Besides fusion genes, activating mutations in the ATP binding site of tyrosine kinase domains have been shown to result in constitutively activate tyrosine kinases (13, 14). Two reports described TrkB mutations in non-small cell lung cancer (NSCLC) (15, 16). Some of these reported mutations are located in the tyrosine kinase domain. However the significance of these mutations has not been elucidated. In this study, we characterized the function of three reported mutations, which by their localization might be activating mutations. We also have sequenced the TrkB tyrosine kinase domain in a large number of lung cancers and cell lines. .
Materials and methods
Cell lines and reagents
We used 29 lung cancer cell lines in this study (A549, NCI-H1355, NCI-H1373, NCI-H1466, NCI-H1944, NCI-H2077, NCI-H2087, NCI-H2122, NCI-H23, NCI-H2347, NCI-H3122, NCI-H854, NCI-H322, NCI-H322, NCI-H358, NCI-H820, NCI-H720, NCI-H1299, NCI-H1725, NCI-H460, NCI-H1173, NCI-H128, NCI-H211, NCI-H592, NCI-H620, NCI-H678, NCI-H69, NCI-H82, NCI-H1717, Corl23). NIH3T3 cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS). Baf3 cells were grown in RPMI supplemented with 10% FBS and 10% WEHI3B conditioned media. Baf3 and WEHI3B cells were a kind gift from Dr Meyerson (MGH, Boston). The Trk inhibitor AZD6918 was obtained from Astrazeneca (Boston, MA). Antibodies for phospho-TrkB(pY516), pErk, pAkt and alpha-tubulin were purchased from Cell signaling technology (Danvers, MA). Anti-pan-Trk (C14) was purchased from Santa-Cruz Biotechnology (Santa Cruz, CA). Anti-phosphotyrosine (4G10) antibody was purchased from Millipore (Billerica, MA). Brain derived neurotrophic factor (BDNF) was from Sigma-Aldrich (St. Louis, MO).
Tumor samples and DNA sequencing
Tumor samples were obtained from Aichi cancer center, Kyushu University hospital and Vrij Universiteit Medical Center in Amsterdam, through protocols approved by the institutional review boards of their facility (17). A list of the tumor pathological diagnosis is reported in table 1. Diagnosis of large cell neuroendocrine carcinoma was based on both morphological appearance and immunohistochemical demonstration of at least one of the neuroendocrine specific marker such as Chromogranin-A, synaptophysin and CD56. DNA was extracted from paraffin embedded slides. Sections containing more than 70% tumor cells were dissected and processed with DNeasy tissue and blood kit (Qiagen, Valencia, CA). Exons 17 to 21 including tyrosine kinase and PLC-g binding domains of TrkB was sequenced by PCR-direct sequencing. Primers are described in supplemental Table 1.
Table 1.
Summary of TrkB mutations in lung cancer
| Histology | Pt.No. | mutations | country | |
|---|---|---|---|---|
| This study | ADC | 17 | ND | Asia(Japan) |
| LCNEC | 28 | ND | ||
| SCLC | 10 | ND | ||
| ADC | 12 | ND | Europe(Netherland) | |
| SCC | 6 | ND | ||
| LCC | 5 | ND | ||
| Cell lines | ||||
| ADC | 17 | ND | ||
| SCC | 1 | ND | ||
| LCC | 3 | ND | ||
| SCLC | 8 | ND | ||
| TC | 1 | ND | ||
| Marchetti et al. | ADC | 228 | ND | Europe(Italy) |
| SCC | 184 | ND | ||
| LCC | 31 | ND | ||
| LCNEC | 29 | 4(13.8%) | ||
| SCLC | 39 | ND | ||
| TC | 17 | ND | ||
| AC | 10 | ND | ||
| Ding et al. | ADC | 188 | 6(3.2%) | USA |
| total | NSCLC(without cell lines) | 728 | 10(1.39%) | |
| SCLC(without cell lines) | 49 | ND | ||
ADC=adenocarcinoma; LCC=large cell carcinoma; LCNEC=large cell neuroendocrine carcinoma
SCC=squamous cell carcinoma; SCLC=small cell carcinoma; TC=typical carcinoid; AC=atypical carcinoid
ND=not detected; NSCLC=non-small cell lung cancer
Plasmid construction and retroviral infection
Full-length TrkB cDNA was obtained by PCR using a pBabepuro/TrkB plasmid as a template. The TrkB plasmid was a kind gift from Dr. Peeper (NKI, Amsterdam). The TrkB ORF was subcloned into pQCXIN retrovirus vector (Clontech, Mountain View, CA). TrkB mutants were generated using QuickChangeII site-directed mutagenesis kit (Stratagene, La Jolla, CA) with wild type pQCXIN/TrkB vector as a template according to the manufacturer’s instruction. Primers for mutagenesis are shown in supplemental table 1. The mutations were confirmed by sequencing. NIH3T3 and Baf3 cells were infected with retrovirus according to standard protocols as described previously (18). Briefly, TrkB expressing retroviruses were produced by cotransfection of GP2-293 cells with pVSV-G (Clontech) using nanofect transfection reagent (Qiagen). Cells were infected with TrkB retroviruses in the presence of 4 µg/ml polybrene (Sigma). Stable populations were obtained by selection in medium containing 800 µg/ml of G418 (Invitrogen, Carlsbad, CA).
Wound healing assay
NIH3T3 cells were plated in 12-well tissue plates and maintained in DMEM medium. At 80–90% confluency, the tip of a micropipette was used to create a linear scratch. The cells were then washed with phosphate-buffered saline to remove floating cellular debris and fed for an additional 8 h with DMEM medium without FBS. Cell migration was judged by photographs taken immediately after scratching and at designated times after scratching using a digital camera. Using Image J software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA), the wound area was measured, and the wound closure area was calculated as follows: the wound closure area= area of wound at time 0 h – area of wound at time 8 h.
Cell proliferation assay
Anchorage independent cell growth/Anoikis assay: A total of 5000 NIH3T3 cells were plated onto 24-well low-attachment tissue plate (Corning, New York, NY), and maintained for 2 weeks. Pictures were taken using a digital camera. Cell growth assay for baf3 cells: A total of 3000 cells were plated onto 96-well plate with/without IL3 or BDNF stimulation. Cells were collected and counted at the indicated time points by Cellometer Auto T4 (Nexcelom Bioscience, Lawrence, MA). MTS assay: Changes in Baf3 cells proliferation were examined by the addition of AZD6918 at various concentrations. The number of surviving Baf3 cells was determined after 72 hours of treatment by measuring the dissolved formazan products after the addition of MTS as described by the manufacturer (Promega, Madison, WI).
Results
Mutation analysis
We sequenced the tyrosine kinase domain of TrkB in 29 lung cancer cell lines, 78 NSCLC samples (28 large cell neuroendocrine carcinomas (LCNEC), 29 adenocarcinomas, 6 squamous cell carcinomas and 5 large cell carcinomas) and 10 small cell lung cancer (SCLC) samples. Although TrkB mutations have been reported in NSCLC by others (Fig. 1) (15, 16), we could not detect mutations in our samples. The results of our study and previous studies are summarized in Table 1.
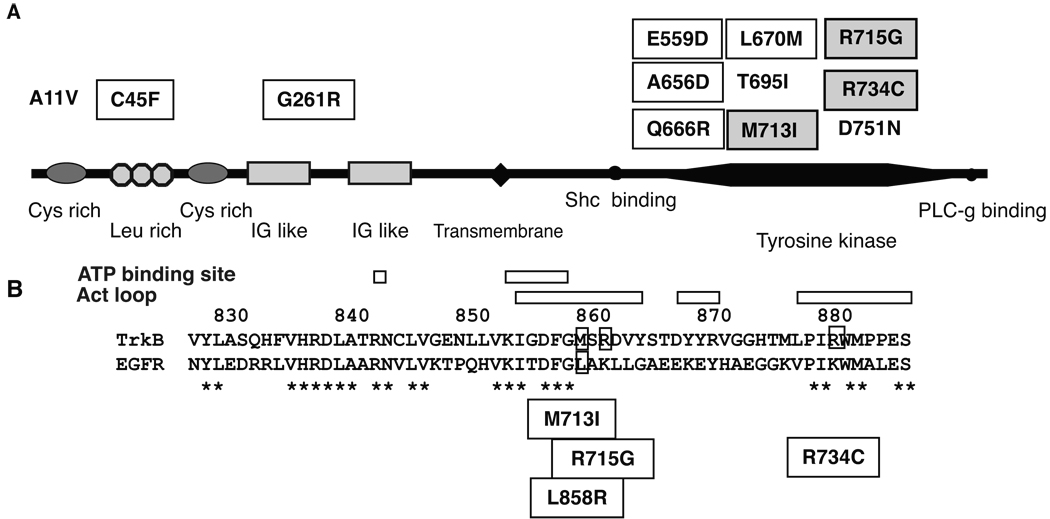
Figure 1.
(A) Schematic representation of TrkB domain, Cys rich, cycteine rich region; Leu rich, leucine rich region; IG like, immunoglobulin like-domain. Reported non-synonymous mutations are indicated. Mutations with square are reported in lung adenocarcinoma (15). Mutations with shaded square are reported in LCNEC (16). A11V was found in glioblastoma (36). T696I and D751N are found in colon cancer (37). (B) Protein alignment of TrkB and EGFR. * indicates matched amino acid sequence. Positions of activation loop and ATP binding site are indicated. Positions of EGFR mutation L858R and TrkB mutations, M713I, R715G and R734G, are also indicated.
Characterization of mutated TrkB
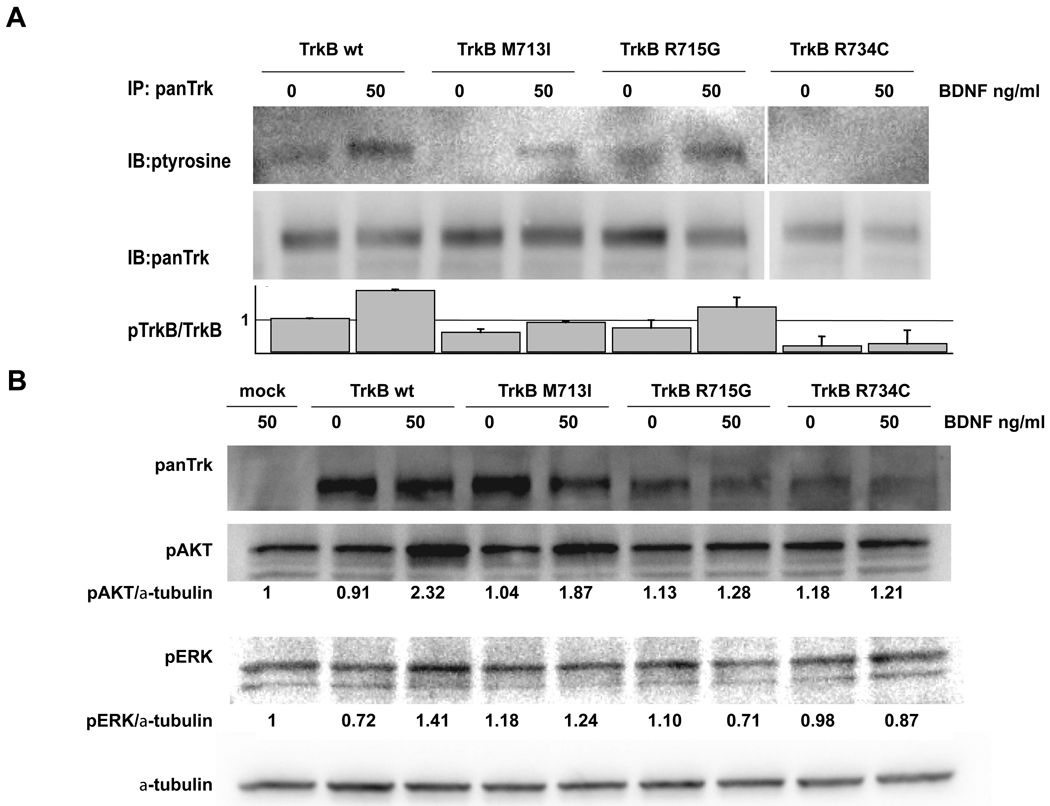
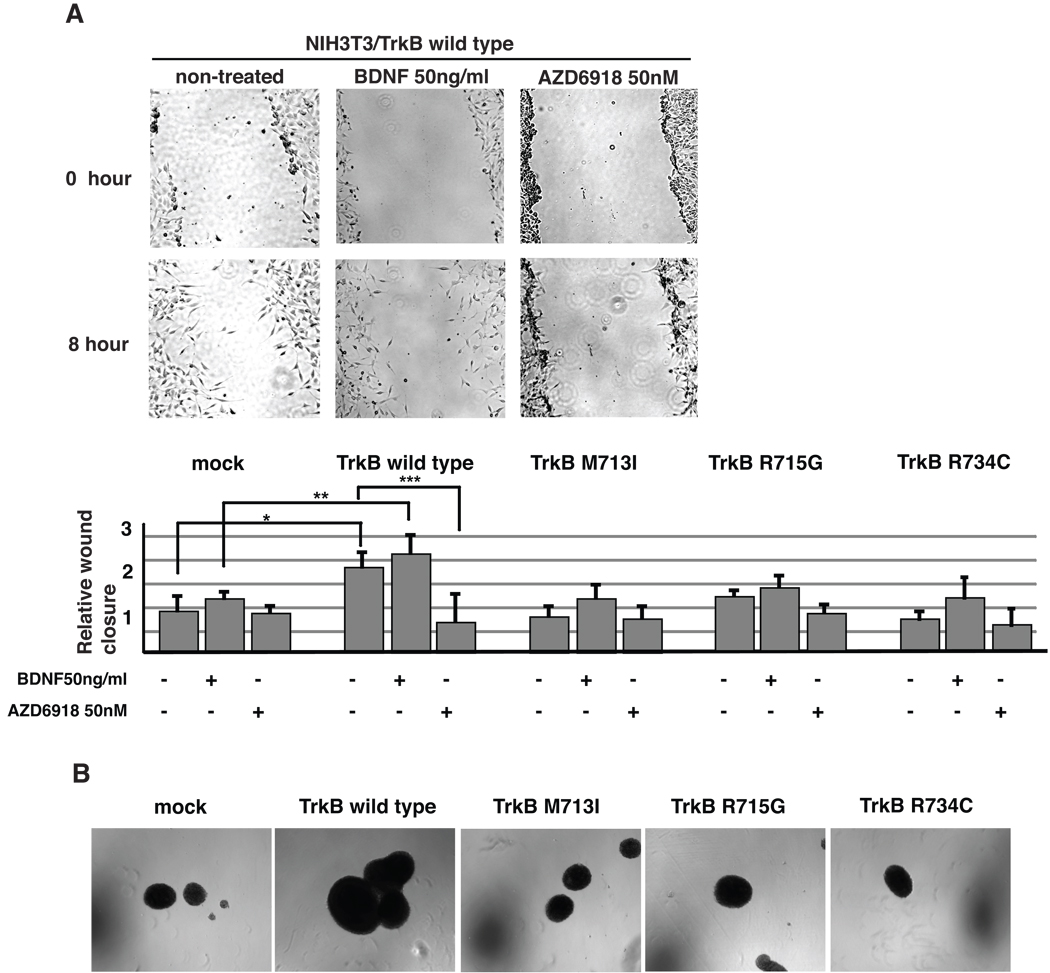
M713I, R715G and R734C are TrkB mutations recently reported in LCNECs (19). Protein alignment of TrkB and EGFR revealed that the three mutations were located in the activation loop of TrkB flanking the position corresponding to the EGFR mutation L858R (Fig. 1B). In order to study the functional significance of these three mutations, we stably introduced wild type and mutant (M713I, R715G and R734C) TrkBs into TrkB-negative NIH3T3 cells respectively. Western blot analysis revealed that TrkB autophosphorylation was detectable in the wild type and R715G TrkB expressing cells, but not in the M713I and R734G TrkB cells (Figure 2A). Upon BDNF stimulation, the level of phosphorylation was similar between wild type and R715G cells, while it was lower in M713I and R734C than in wild type cells. Downstream signaling, AKT and ERK phosphorylation were preferably observed in wild type TrkB transfected cells (Fig.2B). Similar TrkB phosphorylation and down stream signaling were observed in H322 and H2122 lung cancer cell lines (data not shown). In addition, wild type TrkB expressing 3T3 cells shows greater migration ability than their mock transfected counterpart. On the other hand, there were no significant differences between mutated TrkB- and mock-transfected cells (Fig 3A). A Trk inhibitor, AZD6918, is potent in vitro inhibition of pan-Trk kinases and in vivo anti-tumor growth in a neuroblastoma model (20). The drug inhibited wild type TrkB induced migration (Fig 3A). Furthermore wild type but not mutated TrkB transfected NIH3T3 cells showed anchorage independent growth (Fig 3B).
Figure 2.
(A) Autophosphorylation of TrkB. NIH3T3 cells stably transfected with wild type and mutated TrkB were treated with/without 50ng of BDNF for one hour. Immunoprecipitaiton was performed with anti-panTrk antibody, and immunoblots were performed with the indicated antibodies. Densitometrical analysis was performed with GeneTools (Syngene, Frederick, MD). The experiments were repeated three times, and representative results are shown. Relative pTrk/TrkB ratios were indicated, non-treated NIH3T3/wild type TrkB as 1. Bars, standard deviation. (B) Activation of TrkB downstream signaling by BDNF. TrkB was stimulated as described above. Western blots were performed with the indicated antibodies. Relative expression levels of pAkt and pERK were indicated.
Figure 3.
TrkB in cell migration and anchorage independent growth. (A) Cell migration ability was determined with wound healing assay. Wild type and mutated TrkBs expressing NIH3T3 cells were treated with 50ng/ml of BDNF or 50nM of AZD6918. After 8 hours incubation, wound closure area was determined as described in Materials and Methods. Experiments were repeated at least three times. Representative pictures are shown. Results are expressed as relative wound closure area, non-treated mock as 1; bars, standard deviation; *, P=0.001; **, P=0.011; ***, P=0.007. Statistical significance (defined as p < 0.05) was determined by Student’s t test. (B) Anchorage independent cell growth. The cells were seeded onto low-attachment plate, and maintain 2 weeks. Using 4x objective lenses, photographs were taken.
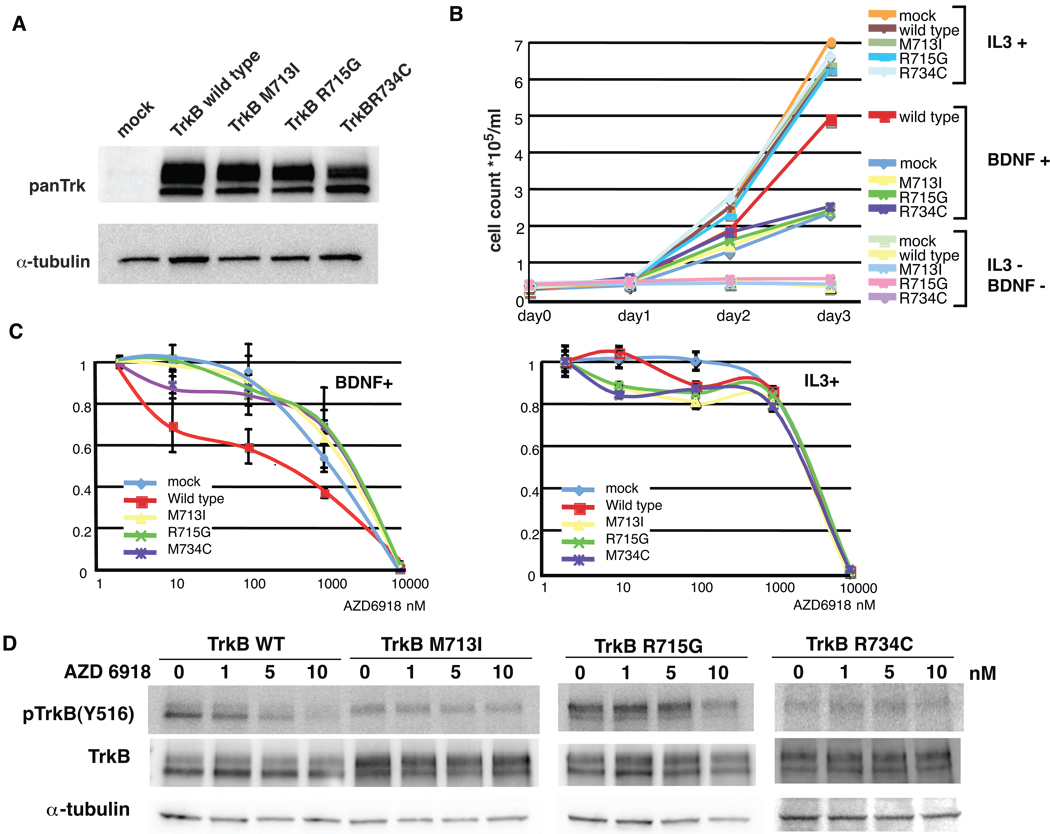
To evaluate the transformation capacity of TrkB (M713I, R715G, and R734C) mutants, we introduced wild type and mutant TrkBs into Baf3 cells (21) (Fig.4A). Baf3 cells are IL3 dependent, and withdrawal of IL3 results in cell death. TrkB expressing Baf3 cells could not grow in the absence of IL3 and BDNF stimulation, suggesting that wild type and mutant TrkBs are neither constitutively active nor capable of overcoming IL3 dependency. (Fig. 4A and B). Interestingly, in line with a previous finding in NIH3T3 transformation assay (22), wild type TrkB transfected Baf3 cells showed accelerated growth rate compared with mock transfectants upon BDNF stimulation, whereas no difference in growth was observed between the mutant TrkB- and mock-transfected cells (Fig. 4B), suggesting that the mutant TrkBs are BDNF non-responsive mutants. It is interesting to note that BDNF could rescue Baf3 cells from IL-3 withdrawal regardless of ectopic TrkB expression, suggesting that BDNF could activate cell survival/proliferation signaling through TrkB independent pathway. Taken together, these results indicate that the observed outgrowth of wild type TrkB transfected Baf3 cells over mock- and mutant TrkB-transfected cells in the presence of BDNF may be attributed to BDNF-stimulated TrkB signaling. AZD6918 inhibited BDNF/TrkB induced cell proliferation at as low as 10nM with wild-type TrkB being the most sensitive, whereas there was no difference between TrkB expressing cells and mock cells IL3 induced cell growth upon exposure to the Trk inhibitor (Fig. 4C). Inhibitory effects of the drug were confirmed by Western blotting (Fig. 4D). At 5 nM, AZD6918 inhibited TrkB Y516 autophosphorylation, and the three mutants seemed to be relatively resistant to the drug. In addition, we evaluated endogenous TrkB expression in 17 NSCLC cell lines, and tested sensitivity to AZD6918 in 4 cell lines (supplementary Fig. 1). Although two cell lines expressed detectable level of TrkB, IC50s of AZD6918 were above 1µM in all tested cell lines.
Figure 4.
Functional characterization of TrkB in Baf3 cells.
(A) Wild type and mutated TrkBs expressing Baf3 cells. Expression of TrkB was confirmed by Western blotting (B) Cell growth assay. The Baf3 cells were seeded onto 96-well plate with IL3 stimulation (WEHI3B conditioned media was added at day 0; final concentration was 10%), BDNF stimulation (BDNF was added daily; final concentration was 100ng/ml) or without treatment. (C) Dose response curves in the Baf3 cells treated with AZD6918. Left panel; the Baf3 cells were stimulated with BDNF. Right panel; The Baf3 cells were stimulated with IL3. The conditions of stimulation with BDNF and IL3 were same as in described above. Bars, standard deviation. (D) Inhibition of phosphoTrkB with AZD6918. The wild type and Mutated TrkB constructs were transiently transfected into MCF7 cells. The cells were treated with various concentration of AZD6918 for 1 hour. Western blotting was performed with indicated antibodies.
Discussion
Overexpression of TrkB in cancers has been described, and is indicative of aggressive tumor behavior. Several mechanisms underlying overexpression of TrkB have been reported. In neuroblastoma, TrkB is often overexpressed in association with amplification of MYCN locus (23, 24). Overexpression of TrkB through an autocrine loop was reported in prostate cancer and malignant myeloma (7, 25). TrkA and TrkC oncogenic fusion proteins have been reported, but not TrkB (3, 4). Although the mechanism for gain of oncogenic function is mainly due to activation of the tyrosine kinase, TrkB has a kinase independent function. The kinase domain deficient splicing variant TrkB-T1 may enhance metastasis of pancreatic adenocarcinoma cells through activation of Rho-Rock pathway (26). Mutations in the tyrosine kinase domain are a possible mechanism of activation of TrkB. TrkB mutations have been described in NSCLC (Table 1). Ding et al reported 6 mutations out of 188 lung adenocarcinomas. Marchetti et al reported 4 mutations out of 29 LCNECs, however they failed to find mutations in 443 NSCLCs, including 228 adenocarcinomas, 184 squamous cell carcinoma and 31 large cell carcinoma without neuroendocrine features. In this study, we could not detect non-synonymous mutations in 78 lung cancer samples and 29 cell lines. In addition, another group have sequenced the entire TrkB gene in 51 lung cancer samples and 32 lung cancer cell lines and found no mutations (J. Brown, Astrazeneca, personal communication). Possible causes of the discrepancy in this series are as follows: (1) Ethnic difference: our samples mainly (60/78) consist of East-Asian (Japanese) patients, and all of LCNEC samples were from Japanese patients, whereas all patients in the previous reports were recruited in Western countries. Striking differences in the frequency of EGFR mutations between Caucasians and East Asians are well known (27, 28). (2) False negative: Marchetti et al. performed single-strand conformation polymorphism (SSCP) as an initial screening, and only SSCP positive tumors were sequenced. It may be possible that SSCP may not be sensitive enough to resolve existing mutations. (3) False positive: it has been documented that DNA extracted from paraffin embedded tissue easily led to PCR artifact (19). Taken together with the previous reports, the frequency of Trk mutation in NSCLC was quite low (10 out of 722, or 1.39%; Table 1). In this study, though no non-synonymous mutation was detected, we found 5 known silent SNPs (I616I; rs2289657) out of 78 tumor samples (data not shown). A synonymous polymorphism in ERCC1 has reported to be correlated with ERCC1 expression, different prognosis and response to chemotherapy (29–31). Meaning of the polymorphism in TrkB remains to be explored.
We selected three mutations reported before for further characterization. However, the mutations that we tested in this study were not activating mutations. Intriguingly, although R715G showed comparable levels of phospho TrkB to wild type TrkB, the mutant TrkB failed to show transforming ability (Fig. 3 and Fig. 4). Our results suggest that mutations of TrkB cannot be used as a positive marker for patient selection. To date, small molecule inhibitors targeting kinase domain have been most successful in case where the targeted kinases contain activating mutation such as in EGFR. It is unclear whether the Trk inhibitor will be effective as a single agent. Indeed, we tested sensitivity to AZD6918 in 4 lung cancer cell lines, all of their IC50 were as high as 1 to 10 µM (supplementary figure 1). However, our results suggested that the Trk inhibitor might inhibit wild type TrkB-induced cell migration and proliferation in the presence of BDNF. Some studies reported high levels of BDNF and TrkB co-expression in lung cancers (8, 32, 33). The Trk inhibitor may be potentially effective in those cancer patients based on our data (Fig. 4C). Using the Trk inhibitor in combination with cytotoxic agents is another possible option. Several in vitro studies reported that inhibiting Trk signaling might enhances efficacy of cytotoxic agents (34, 35).
In conclusion, although mutations in the TrkB gene may not be used for patient selection, overexpression of wild type TrkB may represent a potential target for treatment.
Translational Relevance
Improvement of sequencing technology has provided a large amount of information for gene mutations in cancer. Some mutations drive oncogenic transformation and are thought to be targets for therapeutic intervention. In this study we demonstrate that the frequency of the TrkB mutations is quite low, and that three previously reported mutations in the activation loop are not activation mutations. These results suggest that TrkB mutations should not be used to select patients with lung cancer. However, overexpressing wild type TrkB enhanced cell proliferation and migration, inhibited by a Trk inhibitor, suggesting TrkB may be a viable target for cancer therapy.
Supplementary Material
Acknowledgement
We thank Dr Meyerson (MGH, Boston) for the Baf3 and WEHI3B cells and Dr. Peeper (NKI, Amsterdam) for the TrkB ORF carrying plasmid. This study and TH were supported by NIH intramural fund.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- 3.Beimfohr C, Klugbauer S, Demidchik EP, Lengfelder E, Rabes HM. NTRK1 re-arrangement in papillary thyroid carcinomas of children after the Chernobyl reactor accident. Int J Cancer. 1999;80:842–847. doi: 10.1002/(sici)1097-0215(19990315)80:6<842::aid-ijc7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 4.Martin-Zanca D, Hughes SH, Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. 1986;319:743–748. doi: 10.1038/319743a0. [DOI] [PubMed] [Google Scholar]
- 5.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 6.Sclabas GM, Fujioka S, Schmidt C, et al. Overexpression of tropomysin-related kinase B in metastatic human pancreatic cancer cells. Clin Cancer Res. 2005;11:440–449. [PubMed] [Google Scholar]
- 7.Pearse RN, Swendeman SL, Li Y, Rafii D, Hempstead BL. A neurotrophin axis in myeloma: TrkB and BDNF promote tumor-cell survival. Blood. 2005;105:4429–4436. doi: 10.1182/blood-2004-08-3096. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S, Guo D, Luo W, et al. TrkB is highly expressed in NSCLC and mediates BDNF-induced the activation of Pyk2 signaling and the invasion of A549 cells. BMC Cancer. 2010;10:43. doi: 10.1186/1471-2407-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desmet CJ, Peeper DS. The neurotrophic receptor TrkB: a drug target in anti-cancer therapy? Cell Mol Life Sci. 2006;63:755–759. doi: 10.1007/s00018-005-5490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douma S, Van Laar T, Zevenhoven J, Meuwissen R, Van Garderen E, Peeper DS. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430:1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 11.Kupferman ME, Jiffar T, El-Naggar A, et al. TrkB induces EMT and has a key role in invasion of head and neck squamous cell carcinoma. Oncogene. 2010;29:2047–2059. doi: 10.1038/onc.2009.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos FP, Kantarjian HM, Jain N, et al. Phase 2 study of CEP-701, an orally available JAK2 inhibitor, in patients with primary or post-polycythemia vera/essential thrombocythemia myelofibrosis. Blood. 2010;115:1131–1136. doi: 10.1182/blood-2009-10-246363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giaccone G. Targeting HER1/EGFR in cancer therapy: experience with erlotinib. Future Oncol. 2005;1:449–460. doi: 10.2217/14796694.1.4.449. [DOI] [PubMed] [Google Scholar]
- 14.Marrari A, Trent JC, George S. Personalized cancer therapy for gastrointestinal stromal tumor: synergizing tumor genotyping with imatinib plasma levels. Curr Opin Oncol. 2010;22:336–341. doi: 10.1097/CCO.0b013e32833a6b8e. [DOI] [PubMed] [Google Scholar]
- 15.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchetti A, Felicioni L, Pelosi G, et al. Frequent mutations in the neurotrophic tyrosine receptor kinase gene family in large cell neuroendocrine carcinoma of the lung. Hum Mutat. 2008;29:609–616. doi: 10.1002/humu.20707. [DOI] [PubMed] [Google Scholar]
- 17.Voortman J, Lee JH, Killian JK, et al. Array comparative genomic hybridization-based characterization of genetic alterations in pulmonary neuroendocrine tumors. Proc Natl Acad Sci U S A. 2010;107:13040–13045. doi: 10.1073/pnas.1008132107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchetti A, Felicioni L, Buttitta F. Assessing EGFR mutations. N Engl J Med. 2006;354:526–568. doi: 10.1056/NEJMc052564. author reply -8. [DOI] [PubMed] [Google Scholar]
- 20.Somaiah N, Simon GR. Molecular targeted therapy in non-small cell lung cancer: an overview of available agents. J Thorac Oncol. 2009;4:S1046–S1056. doi: 10.1097/01.JTO.0000361747.56424.f7. [DOI] [PubMed] [Google Scholar]
- 21.Daley GQ, Baltimore D. Transformation of an interleukin 3-dependent hematopoietic cell line by the chronic myelogenous leukemia-specific P210bcr/abl protein. Proc Natl Acad Sci U S A. 1988;85:9312–9316. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein R, Nanduri V, Jing SA, et al. The trkB tyrosine protein kinase is a receptor for brain-derived neurotrophic factor and neurotrophin-3. Cell. 1991;66:395–403. doi: 10.1016/0092-8674(91)90628-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aoyama M, Asai K, Shishikura T, et al. Human neuroblastomas with unfavorable biologies express high levels of brain-derived neurotrophic factor mRNA and a variety of its variants. Cancer Lett. 2001;164:51–60. doi: 10.1016/s0304-3835(00)00715-1. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawara A, Azar CG, Scavarda NJ, Brodeur GM. Expression and function of TRK-B and BDNF in human neuroblastomas. Mol Cell Biol. 1994;14:759–767. doi: 10.1128/mcb.14.1.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bronzetti E, Artico M, Forte F, et al. A possible role of BDNF in prostate cancer detection. Oncol Rep. 2008;19:969–974. doi: 10.3892/or.19.4.969. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Chang Z, Chiao LJ, et al. TrkBT1 induces liver metastasis of pancreatic cancer cells by sequestering Rho GDP dissociation inhibitor and promoting RhoA activation. Cancer Res. 2009;69:7851–7859. doi: 10.1158/0008-5472.CAN-08-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 28.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 29.Chang PM, Tzeng CH, Chen PM, et al. ERCC1 codon 118 C-->T polymorphism associated with ERCC1 expression and outcome of FOLFOX-4 treatment in Asian patients with metastatic colorectal carcinoma. Cancer Sci. 2008 doi: 10.1111/j.1349-7006.2008.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintela-Fandino M, Hitt R, Medina PP, et al. DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J Clin Oncol. 2006;24:4333–4339. doi: 10.1200/JCO.2006.05.8768. [DOI] [PubMed] [Google Scholar]
- 31.Suk R, Gurubhagavatula S, Park S, et al. Polymorphisms in ERCC1 and grade 3 or 4 toxicity in non-small cell lung cancer patients. Clin Cancer Res. 2005;11:1534–1538. doi: 10.1158/1078-0432.CCR-04-1953. [DOI] [PubMed] [Google Scholar]
- 32.Ricci A, Greco S, Mariotta S, et al. Neurotrophins and neurotrophin receptors in human lung cancer. Am J Respir Cell Mol Biol. 2001;25:439–446. doi: 10.1165/ajrcmb.25.4.4470. [DOI] [PubMed] [Google Scholar]
- 33.Ricci A, Mariotta S, Pompili E, et al. Neurotrophin system activation in pleural effusions. Growth Factors. 2010;28:221–231. doi: 10.3109/08977191003677402. [DOI] [PubMed] [Google Scholar]
- 34.Ho R, Eggert A, Hishiki T, et al. Resistance to chemotherapy mediated by TrkB in neuroblastomas. Cancer Res. 2002;62:6462–6466. [PubMed] [Google Scholar]
- 35.Jaboin J, Kim CJ, Kaplan DR, Thiele CJ. Brain-derived neurotrophic factor activation of TrkB protects neuroblastoma cells from chemotherapy-induced apoptosis via phosphatidylinositol 3'-kinase pathway. Cancer Res. 2002;62:6756–6763. [PubMed] [Google Scholar]
- 36.Cancer_genome_atlas_research_network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bardelli A, Parsons DW, Silliman N, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.