Abstract
Purpose
We have evaluated the eukaryotic translation initiation factor 4E (eIF4E) as a potential biomarker and therapeutic target in breast cancer. eIF4E facilitates nuclear export and translation of specific, growth-stimulatory mRNAs and is frequently overexpressed in cancer.
Experimental design
Breast cancer cells were treated with ribavirin, an inhibitor of eIF4E, and effects on cell proliferation and on known mRNA targets of eIF4E were determined. eIF4E expression was assessed, at the mRNA and protein level, in breast cancer cell lines and in skin biopsies from patients with metastatic disease. Additionally, pooled microarray data from 621 adjuvant untreated, node negative breast cancers were analyzed for eIF4E expression levels and correlation with Distant Metastasis Free Survival (DMFS), overall and within each intrinsic breast cancer subtype.
Results
At clinically relevant concentrations, ribavirin reduced cell proliferation and suppressed clonogenic potential, correlating with reduced mRNA export and protein expression of important eIF4E targets. This effect was suppressed by knockdown of eIF4E. Although eIF4E expression is elevated in all breast cancer cell lines, variability in ribavirin responsiveness was observed, indicating that other factors contribute to an eIF4E-dependent phenotype. Assessment of the prognostic value of high eIF4E mRNA in patient tumors found that significant discrimination between good and poor outcome groups was observed only in luminal B cases, suggesting that a specific molecular profile may predict response to eIF4E-targeted therapy.
Conclusions
Inhibition of eIF4E is a potential breast cancer therapeutic strategy that may be especially promising against specific molecular subtypes and in metastatic as well as primary tumors.
Keywords: eIF4E, protein translation, ribavirin, intrinsic breast cancer subtypes, prognostic markers
Introduction
Eukaryotic translation initiation factor 4E (eIF4E) is an oncogene that is overexpressed in a wide range of cancers (1). It has been reported that greater than 50% of breast cancers express elevated levels of the eIF4E protein, and that high levels correlate with increased angiogenesis, clinical progression and poor prognosis (2–4). The eIF4E protein is present both in the nucleus, where it acts to facilitate export of a subset of specific, growth promoting mRNAs (5), and in the cytoplasm, where it recruits mRNAs with highly structured 5′UTRs to the ribosome for initiation of translation (6, 7). This way, eIF4E mediates a tight regulation of expression of many proteins that are critical to cell division, cell growth and angiogenesis, including cyclin D1, survivin, c-myc, ornithine decarboxylase (ODC), and vascular endothelial growth factor (VEGF) (6). eIF4E activity is modulated by a large number of proteins including the eIF4E binding proteins (4EBP), PML, maskin, CUP and various homeodomain proteins (8–10). Notably, the mammalian target of rapamycin (mTOR) prevents sequestration of eIF4E by 4EBP through phosphorylation of 4EBP, thereby activating eIF4E (11). mTOR is a validated clinical target, as evidenced by the clinical approval of selective TOR complex 1 (TORC1) inhibitors (rapalogs) (12) which, unfortunately, have shown only limited clinical acitivity in phase II breast cancer trials (13, 14). Some clinical limitations of the rapalogs include: i) lack of validated tumor marker(s) predictive of responsiveness, ii) lack of effect on the TORC2 complex, which activates AKT, iii) release of p70S6 kinase mediated negative feedback, resulting in secondary tumor activation of PI3K/AKT, and iv) transcriptional upregulation of eIF4E (15–17). The latter suggests that even complete TORC1 inhibition cannot prevent eIF4E driven breast tumorigenesis, so targeting eIF4E directly may potentially be a more successful therapeutic strategy. Toward this end, novel approaches including antisense and RNAi-mediated downregulation of eIF4E have shown promising preclinical activity in different cancer models and are now under clinical development (18–20).
We have recently shown that ribavirin, an antiviral drug, inhibits eIF4E and has antitumor activity in tumor cells characterized by elevated levels of eIF4E (21, 22) and in patients with specific subtypes of acute myeloid leukemia (AML) (23). Ribavirin binds to eIF4E, thereby inhibiting its association with the m7G cap and blocking translation and/or mRNA export of specific, eIF4E dependent transcripts without affecting translation or export of housekeeping mRNAs (21, 22). In particular, the anti-tumor activity of ribavirin has been associated with decreased protein levels of several eIF4E targets, including cyclin D1, NBS1 and active (phosphorylated) Akt (21, 22). Ribavirin appears to specifically inhibit the proliferation of tumor cells overexpressing eIF4E (22), and this selectivity probably underlies the lack of toxicity observed in our recent clinical trial of ribavirin mono-therapy in patients with poor prognosis, eIF4E overexpressing AML. In this study population, ribavirin triggered dramatic clinical improvements, which correlated with relocalization of eIF4E from the nucleus to the cytoplasm, as well as a decrease in total eIF4E levels (23). Direct evidence of decreased mRNA export of the eIF4E target NBS1, together with reduced protein levels of cyclin D1 and NBS1 were also shown, as was repression of AKT activity.
Not all breast cancers overexpress eIF4E, suggesting that the efficacy of ribavirin in targeting eIF4E is likely to be specific to some tumor subtypes (24). As breast cancer is a highly heterogeneous disease with respect to both biological and clinical behavior, it is imperative to understand the interplay between intrinsic breast cancer characteristics, eIF4E expression levels, and sensitivity to an eIF4E-targeted therapeutic such as ribavirin. Intrinsic breast cancer subtypes are now commonly defined by gene expression profiling as luminal A, luminal B, basal-like, HER2-overexpressing and normal breast-like tumors (25–27). Luminal breast cancers are predominantly steroid receptor (ER, PR) positive, while basal-like breast cancers are “triple-negative” tumors that express neither ER, PR nor HER2 receptors (27). This molecular subtyping of breast cancers has been demonstrated to add prognostic information to standard clinical parameters and to predict likelihood of treatment response (27, 28). In particular, among all ER and/or PR overexpressing breast cancers, luminal A tumors are associated with good prognosis and responsiveness to endocrine therapy, while luminal B tumors are associated with high proliferation rates, worse outcome and resistance to endocrine therapy (27, 28). While targeted therapies exist and are quite effective against HER2-positive and luminal A breast cancers, for other subtypes like luminal B and basal-like breast cancers there is still a great need for targeted therapeutics as well as companion prognostic and predictive biomarkers.
In the current study, we have evaluated eIF4E as a potential biomarker and therapeutic target in breast cancer. We assessed the ability of clinically relevant concentrations of ribavirin to suppress growth of breast cancer cell lines that overexpress eIF4E, and correlated this with the ability of ribavirin to suppress downstream eIF4E targets including cyclin D1, NBS1, VEGF and phospho-AKT. Notably, we showed that ribavirin suppresses proliferation of breast cancer cells in an eIF4E-dependent manner. We further interrogated pooled microarray data from 621 adjuvant naive, node-negative breast cancer cases with respect to their intrinsic molecular subtypes, eIF4E transcript levels and metastatic outcome (distant metastasis-free survival, DMFS). From this analysis, we observed that while eIF4E overexpression commonly occurs in different breast cancer subtypes, its prognostic value is restricted to luminal B cases.
Materials and methods
Cells and Reagents
Cells were maintained in DMEM (MCF-7, MDA-MB-468, MDA-MB-321, ZR75.1) or RPMI 1640 (BT474, SkBr3) with 10% FBS and antibiotics (50 IU/ml penicillin, 50 μg/ml streptomycin). MCF10A were maintained in DMEM supplemented with 5% horse serum (Gibco), 10 μg/ml insulin (Sigma), 20 ng/ml EGF (Peprotech), 100 ng/ml choleratoxin (List Biochemical Laboratories Inc.), 0.5 μg/ml hydrocortisone (Sigma) and antibiotics. During experiments, MCF10A cells were grown in the absence of insulin. Cell lines were obtained from the ATCC prior to 2005, and have not been authenticated since. All culture media, FBS and antibiotics were purchased from Wisent (Quebec, Canada). Lyophilized Ribavirin (Kemprotec Ltd, UK) was dissolved in H20 at a stock concentration of 50 mM and sterile-filtered. Aliquots were kept at −80 °C and thawed only once. Normal breast cells were obtained after ethical approval, with informed consent, from patients undergoing prophylactic mastectomy, and were cultured in MEGM® (Clonetics).
Sulphorhodamine B (SRB) Assay
Cells were seeded in 96-well plates and cultured in the absence or presence of ribavirin. The media was changed and the cells re-treated on day 3 and 5. Cell number was assessed as described (29).
Clonogenic Assay
Cells were seeded at low density in 6-well plates and kept in the presence or absence of Ribavirin for 14 days, with the media changed and the cells re-treated every 3 days. Cells were then fixed in 10% TCA and stained with SRB, as above. All visible colonies were manually counted.
Western blotting
Whole cell extracts were prepared by lysis of cells in protein lysis buffer (1% Triton X-100, 150 mM NaCl and 50 mM Tris-HCl, pH 8.0) supplemented with protease and phosphatase inhibitors. 20–50 micrograms of protein were used for Western blotting to detect total eIF4E (BD Biosciences), phospho-4E-BP1 (Thr37/46), total 4E-BP1, phospho-Akt (Ser473) total Akt and NBS1 (all from Cell Signaling), as well as Cyclin D1 and VEGF (Santa Cruz). An antibody to β-actin (Sigma) was used to confirm equal protein loading.
RNA interference
Cells were transfected with siRNAs at a final concentration of 16nM using Lipofectamine RNAiMax (Invitrogen) following the manufacturer’s instructions for reverse transfection. Transfections were performed in 6-well plates for assessment of knockdown by Western blot and in 96-well plates for assessment of cell proliferation by SRB assay. The sequences of the eIF4E siRNA pair were as follows: 5′-AGA GUG GAC UGC AUU UAA AUU UGdA dT-3′ and 5′-AUC AAA UUU AAA UGC AGU CCA CUC UGC-3′. As non-silencing control, we used AllStars Negative Control siRNA (Qiagen).
Confocal immunofluorescence
Exponentially growing cells were fixed with methanol, permeabilized with 0.5% Triton-X 100 (in PBS), then blocked with 10% fetal bovine serum (in PBS). An eIF-4E-FITC antibody (BD Biosciences) was used to show the cellular distribution of eIF4E, and the VECTASHIELD® Mounting Medium with DAPI was used to visualize the nuclei. Micrographs were collected on a laser scanning confocal microscope (LSM510 Carl Zeiss, Inc) using a 100X objective with a numerical aperture of 1.4, with further two times digital zoom at room temperature.
Cell fractionation and quantitative PCR
Cellular fractionation was performed as previously described (30). Relative quantification of transcripts was performed using the StepOnePlus™ Real-Time PCR System with SYBR Green based detection (Applied Biosystems) using primers described in Table S1. All calculations were done using the relative standard curve method described in Applied Biosystems User Bulletin #2. To confirm purity of the fractions, levels of tRNAlys (cytoplasmic) and U6 snRNA (nuclear) were assessed by PCR, as described previously (5).
Immunoprecipitation (IP)
Cells were incubated with 0.5μM 3H-ribavirin over night, fixed with 1.2% formaldehyde (15 minutes at room temperature), followed by 0.15 M glycine for 5 minutes. After two washes in PBS, cells were lysed in IP buffer (50 mM Tris pH 7.5, 5mM EDTA, 150 mM NaCl, 5 mM MgCl2, 0.5% NP40) followed by sonication. Lysates were pre-cleared with protein G-sepharose (30 minutes at 4 °C) and IP was performed over night with anti-eIF4E or IgG conjugated agarose beads (Santa Cruz). Beads were washed 6 times with IP buffer and resuspended in 2xSDS elution buffer (15 minutes at 98 °C). Eluted proteins were subjected to scintillation counting and western blot.
Patient sample collection and immunohistochemistry (IHC)
3 mm punch skin biopsies from normal skin tissue and skin metastases were obtained after approval by an institutional review board and with informed consent from patients with breast cancer that had progressed on prior anthracycline and taxane-containing regimens. Biopsies were sectioned, stained with anti-eIF4E antibody (BD Bioscience) and analyzed by a board certified pathologist as part of the Histology Platform service at the Institute for Research in Immunology and Cancer (IRIC), Université de Montréal.
Analysis of breast cancer microarray data
Expression data from four independent studies (GSE3521, GSE15852, GSE18672 and GSE10780) were obtained from the Gene Expression Omnibus (GEO) to assess whether eIF4E mRNA is elevated in primary breast cancers and metastatic lesions over normal breast epithelium. eIF4E gene expression levels within each tissue type were visualized using box plots and compared using t-test. In addition, to assess the prognostic value of eIF4E mRNA expression, 683 adjuvant untreated, node negative breast cancer cases annotated for distant-metastasis free survival (DMFS) were pooled from four sources (GSE2034, GSE5327, GSE7390, NKI295(31)), Each sample was assigned to one of five subtypes: luminal A (LumA), luminal B (LumB), HER2, basal, or normal-like (normal). Survival analysis was restricted to a subset of 621 cases with ≤15 year follow-up to avoid curve instability at extremely long follow-up times. Tumors were dichotomized based on median eIF4E mRNA level and association with DMFS was assessed by Kaplan Meier analysis in all 621 cases, as well as within each intrinsic breast cancer subtype. For comparison, the prognostic value of mTOR or the ratio of eIF4E with its binding proteins (4EBP1, 4EBP2 and PML) mRNA expression levels was similarly assessed. As well, eIF4E expression levels within each intrinsic subtype were represented by box plots and compared by t-test. Details on expression data processing and subtype assignment, as well as relevant references for individual GEO datasets, are described in supplemental methods S1.
Results
Ribavirin suppresses proliferation and clonogenic potential of breast cancer cell lines
In patients with AML receiving ribavirin therapy, plasma levels of up to 88 μM have been measured (unpublished data). We treated six breast cancer cell lines, plus the non-tumorigenic mammary epithelial cell line MCF10A (32), with up to 100 μM ribavirin and assessed effects on cell proliferation and clonogenic potential. 7 days of ribavirin treatment revealed that all cell lines tested were growth inhibited with IC50 values that are within the clinically achievable range (Figure 1A). Two cell lines were less sensitive than the others, namely MCF10A and ZR75.1. Additionally, assessment of the growth inhibitory activity of ribavirin in normal primary breast cells (n=3) showed no response at concentrations up to 120 μM. Assessment of anchorage-dependent clonogenic potential in the absence or presence of ribavirin demonstrated significant inhibition at concentration ranging from 1–50 μM (Figure 1B). Between 20 and 100 μM ribavirin were required to completely suppress the ability of the tumor cell lines to form colonies, while MCF10A cells were 50% inhibited at 100 μM. We note that ZR75.1 cells were less sensitive to ribavirin than all other cell lines with comparably elevated eIF4E.
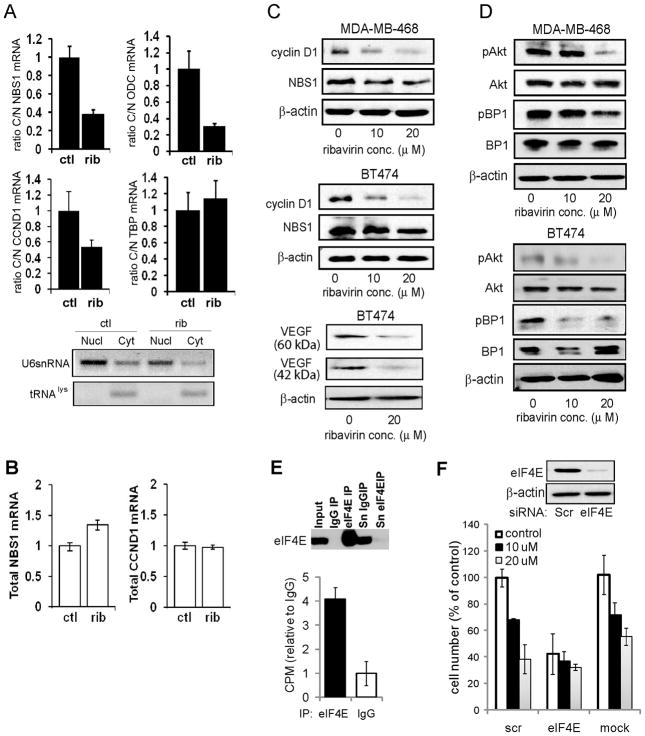
Figure 1. Ribavirin reduces proliferation and clonogenic survival of breast cancer cells.
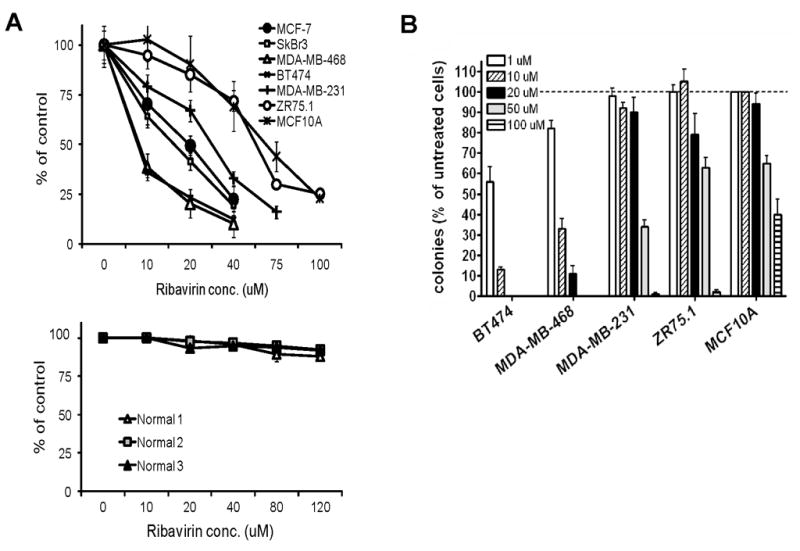
(A) Breast cancer cell lines (top) and primary normal breast cells (bottom) were cultured in the absence or presence of ribavirin at the indicated concentrations for 7 days, and the number of cells was determined as described in Materials and Methods. (B) Anchorage-dependent clonogenic potential was assessed in cells cultured in the absence or presence of ribavirin for 14 days.
Ribavirin growth inhibition is associated with downregulation of known eIF4E targets and is suppressed byknockdown of eIF4E
We used several strategies to confirm that ribavirin targets eIF4E in breast cancer cells. Because the anti-tumor effects of ribavirin have been associated with decreased mRNA export as well as translation of eIF4E sensitive transcripts (22), we first assessed changes in the levels of nuclear and cytoplasmic mRNA of known eIF4E targets. As expected, we observed a reduced ratio of cytoplasmic to nuclear NBS1, ODC and cyclin D1 mRNA in cells treated with ribavirin for 72 hours, indicating inhibition of nuclear mRNA export (Figure 2A). In contrast, the ratio of cytoplasmic to nuclear TBP mRNA was unaffected by ribavirin, consistent with it not being an eIF4E target (Figure 2A). Total mRNA levels of cyclin D1 or NBS1 were not affected by ribavirin treatment (Figure 2B), but a reduction in the protein levels of these eIF4E targets was evident (Figure 2C). Earlier studies in NIH3T3 cells indicated that ribavirin treatment led to reduction in polysomal loading of VEGF with no alteration in mRNA export (22) and, consistent with this, we observed a decrease in the protein level of VEGF (Figure 2C). These data indicate that ribavirin modulates both the nuclear and the cytoplasmic function of eIF4E in breast cancer cells, in accordance with data from other cell types. Ribavirin has previously been demonstrated to suppress the activity of AKT, via a reduction in NBS1 (21, 33). Consistently, we observed a decrease in active, phosphorylated AKT (Figure 2D). We also assessed phosphorylation of 4EBP1, which is a target of mTOR downstream of AKT and observed a dose-dependent reduction in the hyperphosphorylated form of this protein (Figure 2D), confirming suppression of AKT signaling. To demonstrate that ribavirin binds eIF4E in these cells, we treated cells with 3H-labeled ribavirin for 24 hours, and performed immunoprecipitation (IP) of eIF4E followed by scintillation counting. These studies showed a four-fold increase in 3H-ribavirin co-immunoprecipitated with eIF4E, relative to the IgG control (Figure 2E). Finally, we used siRNA to knock down eIF4E in BT474 cells, which are highly responsive to ribavirin, and assessed the effect on cell proliferation in the absence or presence of the drug. As expected, knockdown of eIF4E caused a robust decrease in cell proliferation; moreover, there was little additional suppression of proliferation by ribavirin (Figure 2F).
Figure 2. Ribavirin inhibits mRNA export and protein expression of eIF4E targets, and downregulation of eIF4E suppresses ribavirin growth inhibition.
(A) BT474 cells were treated with 20 μM ribavirin for 3 days and NBS1, ODC, cyclin D1 and TBP mRNA quantified in cellular fractions prepared as described in Materials and Methods. For each fraction and gene, untreated cells were used as calibrator (RQ=1). Bottom: semi-quantitative PCR of tRNAlys and U6 snRNA demonstrate the purity of the fractions. (B) Total mRNA levels were assessed in BT474 cells, treated as in A. (C) Cyclin D1, NBS1 and VEGF protein levels were assessed by western blot in MDA-MB-468 and BT474 cells, treated with the indicated concentrations of ribavirin for 3 days. (D) AKT activity was assessed as the level of phosphorylation of AKT and its downstream target 4E-BP1 in MDA-MB-468 and BT474 cells, treated as in C. (E) BT474 cells were incubated with 3H-ribavirin for 24h, and subjected to immunoprecipitation using anti-eIF4E or IgG as described in Materials and Methods, Western blot analysis (top) and scintillation counting (bottom) shows specific pulldown of 3H-ribavirin with eIF4E. Sn=supernatant collected after the last wash. (F) BT474 cells were transfected with siRNA targeting eIF4E or a scrambled control (scr). Knockdown was assessed by western blot (top) on day 4 post-transfection, and cell number was determined after 5 days incubation −/+ ribavirin at the indicated concentrations. Data from one representative experiment performed in triplicate are shown as means −/+ SD.
eIF4E is highly expressed in the cytoplasm and nucleus of all breast cancer cell lines
eIF4E mRNA and protein levels in exponentially growing cells were assessed by qPCR and Western blot analyses. Consistent with published microarray data, eIF4E was found to be similarly expressed at the mRNA level in all cell lines (34) (data not shown). Western blot analysis further confirmed that all breast cancer cell lines express high protein levels of eIF4E (Figure 3A), similar to the known overexpressing cell line FaDu (35) (supplemental Figure S1). In contrast, lower levels of eIF4E were detected in non-malignant MCF10A cells (Figure 3A). Immunofluorescence and confocal microscopy were used to study eIF4E localization in the breast cancer cell lines and showed that eIF4E is present in the nucleus as well as in the cytoplasm in all cells (Figure 3B and data not shown), which is typical of most cell lines and primary tissues (36).
Figure 3. All breast cancer cell lines express high levels of eIF4E both in the nucleus and the cytoplasm.
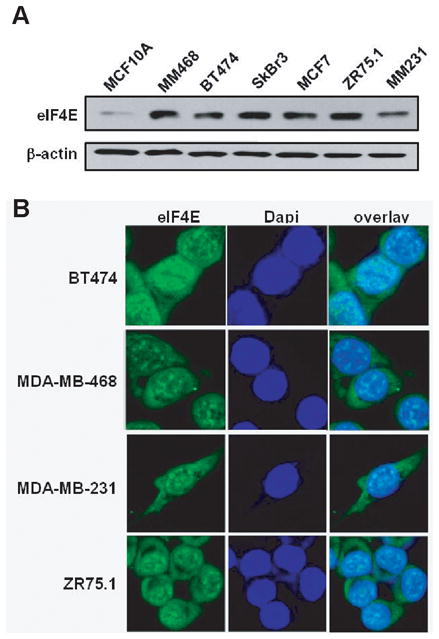
(A) eIF4E protein levels in whole cell extracts from exponentially growing cells were assessed by western blot, which showed elevated levels in all tumor cell lines. (B) Immunofluorescence staining of eIF4E (green) in BT474, MDA-MB-468, MDA-MB-231 and ZR75.1 cells shows that eIF4E is present in the nucleus as well as the cytoplasm.
Metastatic breast cancers show concordant overexpression of eIF4E mRNA and protein
To investigate whether eIF4E mRNA is elevated in patient tumors, and whether metastatic lesions also display high eIF4E expression, we first examined microarray data from three publically available datasets (GSE15852, GSE18672 and GSE10780), and found that in each of the datasets, average eIF4E mRNA levels were significantly higher in primary breast tumors than in normal breast (Figure 4A). Further, analysis of one dataset (GSE3521), containing samples from primary breast cancers as well as metastases and normal breast, showed significantly elevated levels in both primary tumors and metastases (Figure 4B). We also obtained biopsy material from three patients with advanced breast cancer that had metastasized to the skin. All patients had undergone chemotherapy and were experiencing progressive disease at the time of biopsy. Q-PCR analysis showed 2.5–3.5 fold increased levels of eIF4E mRNA compared to normal skin (Figure 4C), although this is likely to be an underestimate due to contamination of the tumor biopsies with normal cells. Immunohistochemistry also demonstrated strong positive staining for eIF4E in malignant skin, while normal skin showed much weaker staining, with the exception of the epidermis (Figure 4D), indicating a concordant increase in eIF4E expression at the mRNA and protein level.
Figure 4. eIF4E is overexpressed in primary and metastatic lesions of patients with breast cancer.
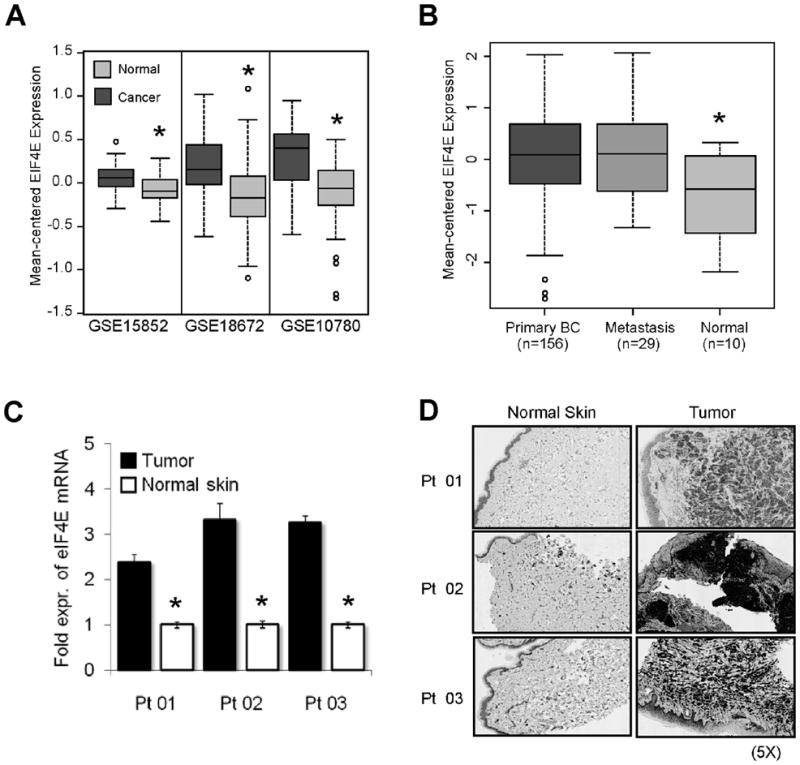
(A–B) Box plots showing elevated eIF4E mRNA levels in (A) primary breast tumors and (B) primary breast tumors and breast cancer metastasis over normal breast tissue. (*) denotes significance by t-test (p<0.05). (C) mRNA levels in malignant and normal skin were assessed by qPCR and results were normalized to RPL13a. For each patient, normal skin was used as a calibrator (RQ=1). (D) Immunohistochemistry shows strong eIF4E staining in malignant skin.
High eIF4E mRNA expression is a prognostic factor in patients with luminal B breast cancer
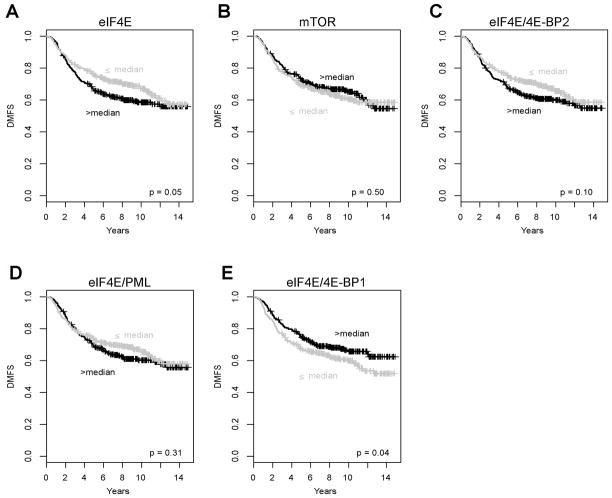
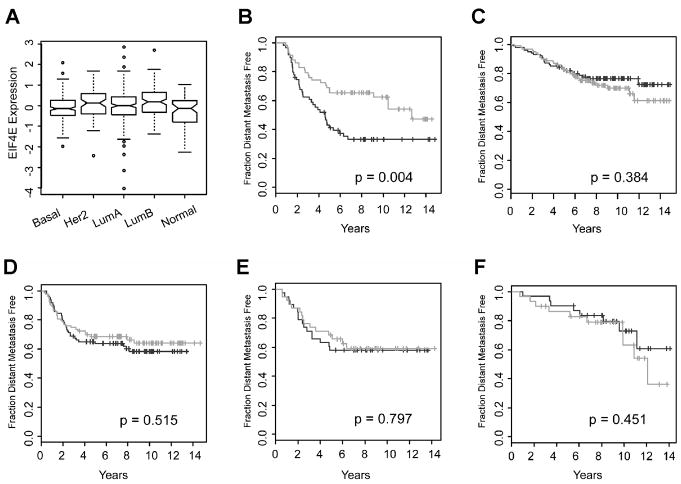
To determine the prognostic value of eIF4E mRNA, we analyzed pooled microarray data from 621 adjuvant untreated, node negative breast cancers for eIF4E expression levels and correlation with Distant Metastasis Free Survival (DMFS). Tumors were dichotomized into High vs. Low expressors at the median, and Kaplan Meier analysis revealed that high eIF4E expressing breast cancers have reduced DMFS when compared to low eIF4E expressors (Figure 5A). In comparison, mTOR mRNA expression did not produce significant curve separation between High vs. Low expressing groups (Figure 5B). eIF4E availability/activity is regulated by many proteins, including 4EBPs, PML and others; thus we also examined the prognostic value of the ratios of eIF4E/BP1, eIF4E/4EBP2 and eIF4E/PML mRNA. Neither eIF4E/4EBP2 nor eIF4E/PML mRNA expression produced significant curve separation between High vs. Low expressing groups (Figure 5C–D). Interestingly, high eIF4E/4EBP1 ratio appears to be associated with better prognosis (Figure 5E), although this is likely due to low 4EBP1 expression (and hence high eIF4E/4EBP1 ratios) in the good prognosis luminal A subtype (Figure S2) When the cohort was subset into intrinsic breast cancer subtypes, comparison of eIF4E mRNA expression levels revealed that luminal B breast cancers have the highest average eIF4E mRNA levels (Figure 6A). These differences are significant for comparisons with basal, luminal A, and normal-like cases (t-test p=1.03E-04; 2.17E-03; 4.85E-07 respectively), but not with the HER2 subtype (t-test p=0.29). When the prognostic value of eIF4E was assessed within each intrinsic subtype, significant discrimination between good and poor outcome groups was observed in luminal B cases (Figure 6B), and not in any of the other subtypes (Figure 6C–F), suggesting that the overall prognostic significance of eIF4E in the pooled dataset may be attributed mainly to its discriminating power in the luminal B subset of breast cancer cases. Interestingly, the prognostic value of the ratio of eIF4E/4E-BP2 and eIF4E/PML show the same intrinsic subtype specificity, where significant curve separations were observed only in luminal B cases (Figure S3 and S4). In contrast, mTOR and the eIF4E/4EBP1 mRNA ratios did not show prognostic significance in any individual intrinsic subtypes (data not shown).
Figure 5. Comparative prognostic performance of eIF4E, mTOR, and eIF4E/4E-BP1, eIF4E/BP2 and eIF4E/PML mRNA.
(A–E) Kaplan Meier plots of distant metastasis events in a cohort of 621 node negative adjuvant naïve breast cancers dichotomized at the median of (A) eIF4E, (B) mTOR, (C) eIF4E/4E-BP2, (D) eIF4E/PML, and (E) eIF4E/4E-BP1 mRNA expression levels. Black: high, light grey: low
Figure 6. High eIF4E mRNA expression is prognostic in luminal B breast cancer.
(A) Box plot depiction of eIF4E mRNA expression levels within each of five intrinsic breast cancer subtypes. (B–F) Kaplan Meier plots of distant metastasis events within individual intrinsic subtypes, dichotomized at median eIF4E expression levels. (B) Luminal B, (C) Luminal A, (D) Basal, (E) Her2, (F) Normal. Black: high eIF4E, light grey: low eIF4E.
Discussion
Overexpression of eIF4E, an oncogene that post-transcriptionally regulates the expression of many genes critical to cell division, proliferation and angiogenesis, is associated with oncogenic transformation in cell culture and with tumor formation, metastatic disease, and increased tumor invasion in mice (37, 38). From a number of small to medium size studies in breast cancer, it has been reported that overexpression of eIF4E at the protein level is a common event, which correlates with aggressive disease and poorer patient prognosis (2–4, 39). In agreement with these reports, we found that eIF4E mRNA is elevated in primary as well as metastatic breast cancer relative to normal breast epithelium. In a limited study of three patients with metastatic skin lesions, we observed elevated levels of eIF4E compared to normal skin from the same patient, both at the mRNA and protein level. Altogether, these findings suggest that transcriptional upregulation of eIF4E occurs early during breast tumorigenesis, although enhanced eIF4E transcript stability may also play a role. It is not well understood how eIF4E transcription is regulated, however the eIF4E promoter contains a c-myc responsive element, an AP-1 binding site, as well as Rel, Myb, NF-κB, SP-1, NF1, STAT, AP-4, ATB and CREB consensus motifs (40). eIF4E mRNA is stabilized by HuR (41), a protein that is commonly overexpressed in cancer, and high levels of cytoplasmic HuR have in fact been associated with poor prognosis in ductal breast carcinoma (42). In addition, amplification of the eIF4E gene has been demonstrated in a few breast cancer samples (43).
This study is the first to demonstrate that clinically relevant concentrations of ribavirin, a confirmed inhibitor of eIF4E (22), suppress proliferation and clonogenecity of a number of breast cancer cell lines (Figure 1). All breast cancer cell lines tested express levels of eIF4E that are substantially higher than that of the non-malignant mammary cell line MCF10A (Figure 3), and all are sensitive to ribavirin within the therapeutic range. In cell proliferation assays, IC50 values varied from 7–50 μM ribavirin; and all cells with elevated eIF4E levels were also sensitive to high levels of ribavirin in colony forming assays with no colonies formed at 100 μM ribavirin, in contrast to the non-malignant MCF10A line where colonies formed even in the presence of this high ribavirin concentration. These results suggest that within the therapeutic concentration range evaluated, all breast cancer cells are responsive to ribavirin, with their observed variation in ribavirin sensitivity likely caused by individual genetic and/or epigenetic features that potentially impact the eIF4E pathway. For instance, not only the expression level but also the activity of eIF4E is a critical determinant of cell growth, and its activity regulation is known to be redundant and multi-factorial (8, 9, 44, 45). Variations in the levels of positive and negative eIF4E regulatory proteins, including 4EBPs, 4E-T, maskin, CUP, PML and others, as well as the activity of the AKT/mTOR pathway or the eIF4E kinase Mnk, may modulate the effectiveness of ribavirin.
As expected, growth inhibition by ribavirin was accompanied by decreased cytoplasmic to nuclear mRNA ratios and protein levels of known eIF4E export targets, as well as the cytoplasmic target VEGF (Figure 2A–C). We also showed for the first time that siRNA mediated knockdown of eIF4E protein in BT474 cells prevents further antiproliferative activity of ribavirin, supporting the essential role of eIF4E in mediating ribavirin’s anti-tumor activity. Given these data, we conclude that eIF4E overexpression is essential for ribavirin responsiveness, and that additional influences may modulate ribavirin sensitivity.
Importantly, and consistent with previous studies in the Head and Neck Squamous Cell Carcinoma (HNSCC) cell line FaDu (21), in fibroblasts and in AML patients undergoing ribavirin treatment, we observed that suppression of cell growth by ribavirin was associated with a decrease in AKT signaling, as shown by decreased phospho-AKT and phospho-BP1 (Figure 2D). This suppression works as a negative feedback towards eIF4E activity, by allowing hypo-phosphorylated BP1 to sequester eIF4E, and thus provides a double anti-proliferative signal in tumor cells that in addition to overexpressing eIF4E may have mutations activating the PI3K/AKT/mTOR pathway. Of note, basal-like breast cancers are associated with frequent PTEN loss and PI3K activation (46, 47) and the sensitivity of MDA-MB-468, a basal cell line that lacks PTEN and has constitutively activated PI3K/AKT signaling, to ribavirin induced growth inhibition implicates potential therapeutic value in this clinically problematic subset of breast cancers. The ability of ribavirin to suppress AKT clearly distinguishes it from clinically available mTOR inhibitors, which have the opposite effect, and suggests that it may indeed be more beneficial to directly target eIF4E, which acts both upstream and downstream of PI3K/AKT/mTOR.
Complementary to the evaluation of eIF4E as a therapeutic target in breast cancer cells, we have performed the first large survey assessing the prognostic value of total eIF4E mRNA expression in a cohort of over 600 node negative adjuvant naïve breast cancers, sufficiently powered to evaluate expression levels within specific intrinsic subtypes in relation to metastatic recurrence. Surprisingly, despite previous links between elevated eIF4E protein expression and poor breast cancer prognosis (2–4, 39), the reduction in DMFS observed in breast cancers with elevated eIF4E mRNA levels were only barely significant (p=0.05) (Figure 5A). This disparity may in part be accounted for by the high degree of heterogeneity among breast cancers and, indeed, when each intrinsic subtype was analyzed separately, high eIF4E mRNA was correlated with poor prognosis specifically in luminal B breast cancers, and this correlation was highly significant. This prognostic association suggests that eIF4E overexpression in luminal B breast cancers not only contributes to their greater clinical aggressiveness and endocrine resistance relative to luminal A breast cancers, but it also points to a possible mechanistic difference in the tumorigenic pathways driving these two types of ER-positive breast cancer. Luminal B breast cancers are potentially more dependent on the downstream translational products induced by overexpressed eIF4E and therefore more susceptible to eIF4E-targeted therapeutics. Of note, high eIF4E/BP2 and eIF4E/PML ratios were also associated with poor prognosis in the luminal B cancers; however eIF4E alone gives the best prognostic performance. This is in contrast to observations at the protein level where parameters which include 4EBP levels improve prognostic performance over eIF4E levels alone (48). Among the breast cancer cell lines we tested for ribavirin responsiveness one of the most sensitive lines was BT474, which is ER-positive and HER2 amplified and potentially considered to be luminal B (34). It is difficult to assign cell lines to intrinsic subtypes, as the subtyping of cell lines performed by Neve et al. did not distinguish between luminal A and luminal B categories (34), but HER2 amplification along with ER and/or PR positivity is commonly employed as an immunohistochemical surrogate of the luminal B subtype (49). However, we note that other sensitive cells lines, such as MDA-MB-468, are of the basal-like subtype (34). It is important to concede that a prognostic marker may not be predictive of response, even to a drug targeting the marker.
Our findings provide strong rationale for the ongoing phase I/II clinical study evaluating ribavirin in the setting of advanced breast cancer (NCT01056757), and correlative studies linked to this trial will help determine if luminal B and possibly some basal-like breast cancers are more sensitive to eIF4E-targeted therapy. Since eIF4E-targeted agents will most likely find their greatest clinical utility in combination with standard breast cancer therapeutics, additional preclinical studies must now begin to determine what specific classes of chemotherapeutics or endocrine agents are best given in combination with eIF4E-targeted therapeutics. We foresee that analogs of ribavirin with improved eIF4E targeting and pharmacologic properties will be developed in the coming years and that this new class of targeted agents will not only become useful in the management of breast cancer patients, but they will also become part of an emerging group of breast cancer subtype-selective therapeutics.
Supplementary Material
eIF4E protein levels were assessed by western blot analysis.
Box plots depicting (A) reduced 4E-BP1 expression and (B) elevated ratio of eIF4E/4E-BP1 in luminal A cases in comparison to other subtypes.
Kaplan Meier analysis of tumor cohort dichotomized at median into High vs. Low expressing groups in (A) Luminal B, (B) Luminal A, (C) Basal, (D) Her2, (E) Normal. Black: high eIF4E/4EBP2, light grey: low eIF4E/4EBP2.
Kaplan Meier analysis of tumor cohort dichotomized at median into High vs. Low expressing groups in (A) Luminal B, (B) Luminal A, (C) Basal, (D) Her2, (E) Normal. Black: high eIF4E/PML, light grey: low eIF4E/PML.
Acknowledgments
Grant support: This research was supported in part by a BCRF-AACR Grant for Translational Breast Cancer Research (WM). Other financial support was received from the Canadian Institute for Health Research (CIHR MOP-12863, WM and FP; MOP-43979, WM), National Cancer Institute of Canada (NCIC #19202, WM), NIH-P50-CA58207, RL1-AG032113, and U24-CA14358 (CB and CY), NIH-98571(KB). Wilson Miller is a Chercheur National of Fonds de la Recherche en Santé du Québec (FRSQ) and Katherine Borden holds a Canada Research Chair.
The authors thank Dr. Louis Gaboury and the histology platform at IRIC for IHC staining of eIF4E in skin biopsies.
References
- 1.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Research. 2008;68:631–4. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 2.Holm N, Byrnes K, Johnson L, Abreo F, Sehon K, Alley J, et al. A Prospective Trial on Initiation Factor 4E (eIF4E) Overexpression and Cancer Recurrence in Node-Negative Breast Cancer. Annals of Surgical Oncology. 2008;15:3207–15. doi: 10.1245/s10434-008-0086-9. [DOI] [PubMed] [Google Scholar]
- 3.Byrnes K, White S, Chu QY, Meschonat C, Yu H, Johnson LW, et al. High eIF4E, VEGF, and microvessel density in stage I to III breast cancer. Annals of Surgery. 2006;243:684–92. doi: 10.1097/01.sla.0000216770.23642.d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman LJ, Peter MB, Teall TJ, Brannan RA, Hanby AM, Honarpisheh H, et al. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br J Cancer. 2009;100:1393–9. doi: 10.1038/sj.bjc.6605044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3′UTR. J Cell Biol. 2005;169:245–56. doi: 10.1083/jcb.200501019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem Cell Biol. 2008;86:178–83. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 7.Culjkovic B, Topisirovic I, Borden KL. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle. 2007;6:65–9. doi: 10.4161/cc.6.1.3688. [DOI] [PubMed] [Google Scholar]
- 8.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 9.Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 2001;20:4547–59. doi: 10.1093/emboj/20.16.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Topisirovic I, Borden KL. Homeodomain proteins and eukaryotic translation initiation factor 4E (eIF4E): an unexpected relationship. Histol Histopathol. 2005;20:1275–84. doi: 10.14670/HH-20.1275. [DOI] [PubMed] [Google Scholar]
- 11.Gingras AC, Kennedy SG, O’Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–13. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konings IR, Verweij J, Wiemer EA, Sleijfer S. The applicability of mTOR inhibition in solid tumors. Curr Cancer Drug Targets. 2009;9:439–50. doi: 10.2174/156800909788166556. [DOI] [PubMed] [Google Scholar]
- 13.Baselga J, Semiglazov V, van Dam P, Manikhas A, Bellet M, Mayordomo J, et al. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer. J Clin Oncol. 2009;27:2630–7. doi: 10.1200/JCO.2008.18.8391. [DOI] [PubMed] [Google Scholar]
- 14.Chan S, Scheulen ME, Johnston S, Mross K, Cardoso F, Dittrich C, et al. Phase II study of temsirolimus (CCI-779), a novel inhibitor of mTOR, in heavily pretreated patients with locally advanced or metastatic breast cancer. J Clin Oncol. 2005;23:5314–22. doi: 10.1200/JCO.2005.66.130. [DOI] [PubMed] [Google Scholar]
- 15.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 17:249–61. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun SY, Rosenberg LM, Wang XR, Zhou ZM, Yue P, Fu H, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Research. 2005;65:7052–8. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 18.Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–48. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soni A, Akcakanat A, Singh G, Luyimbazi D, Zheng YH, Kim DY, et al. eIF4E knockdown decreases breast cancer cell growth without activating Akt signaling. Molecular Cancer Therapeutics. 2008;7:1782–8. doi: 10.1158/1535-7163.MCT-07-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong K, Wang R, Wang X, Lin F, Shen JJ, Gao P, et al. Tumor-specific RNAi targeting eIF4E suppresses tumor growth, induces apoptosis and enhances cisplatin cytotoxicity in human breast carcinoma cells. Breast Cancer Research and Treatment. 2009;113:443–56. doi: 10.1007/s10549-008-9956-x. [DOI] [PubMed] [Google Scholar]
- 21.Tan K, Culjkovic B, Amri A, Borden KL. Ribavirin targets eIF4E dependent Akt survival signaling. Biochem Biophys Res Commun. 2008;375:341–5. doi: 10.1016/j.bbrc.2008.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101:18105–10. doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, et al. Molecular targeting of the oncogene eIF4E in AML: a proof-of-principle clinical trial with ribavirin. Blood. 2009;114:257–60. doi: 10.1182/blood-2009-02-205153. [DOI] [PubMed] [Google Scholar]
- 24.Borden KL, Culjkovic-Kraljacic B. Ribavirin as an anti-cancer therapy: acute myeloid leukemia and beyond? Leuk Lymphoma. 2010 doi: 10.3109/10428194.2010.496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–50. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersson F, Couture MC, Hanna N, Miller WH. Enhanced retinoid-induced apoptosis of MDA-MB-231 breast cancer cells by PKC inhibitors involves activation of ERK. Oncogene. 2004;23:7053–66. doi: 10.1038/sj.onc.1207956. [DOI] [PubMed] [Google Scholar]
- 30.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–26. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 32.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–86. [PubMed] [Google Scholar]
- 33.Culjkovic B, Tan K, Orolicki S, Amri A, Meloche S, Borden KL. The eIF4E RNA regulon promotes the Akt signaling pathway. J Cell Biol. 2008;181:51–63. doi: 10.1083/jcb.200707018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeFatta RJ, Nathan CO, De Benedetti A. Antisense RNA to eIF4E suppresses oncogenic properties of a head and neck squamous cell carcinoma cell line. Laryngoscope. 2000;110:928–33. doi: 10.1097/00005537-200006000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Strudwick S, Borden KL. The emerging roles of translation factor eIF4E in the nucleus. Differentiation. 2002;70:10–22. doi: 10.1046/j.1432-0436.2002.700102.x. [DOI] [PubMed] [Google Scholar]
- 37.Larsson O, Li S, Issaenko OA, Avdulov S, Peterson M, Smith K, et al. Eukaryotic translation initiation factor 4E-Induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Research. 2007;67:6814–24. doi: 10.1158/0008-5472.CAN-07-0752. [DOI] [PubMed] [Google Scholar]
- 38.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman D, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–63. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 39.Kerekatte V, Smiley K, Hu B, Smith A, Gelder F, De Benedetti A. The proto-oncogene/translation factor eIF4E: a survey of its expression in breast carcinomas. Int J Cancer. 1995;64:27–31. doi: 10.1002/ijc.2910640107. [DOI] [PubMed] [Google Scholar]
- 40.Makhlouf AA, Namboodiri AM, McDermott PJ. Transcriptional regulation of the rat eIF4E gene in cardiac muscle cells: the role of specific elements in the promoter region. Gene. 2001;267:1–12. doi: 10.1016/s0378-1119(01)00399-7. [DOI] [PubMed] [Google Scholar]
- 41.Topisirovic I, Siddiqui N, Orolicki S, Skrabanek LA, Tremblay M, Hoang T, et al. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol Cell Biol. 2009;29:1152–62. doi: 10.1128/MCB.01532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinonen M, Bono P, Narko K, Chang S-H, Lundin J, Joensuu H, et al. Cytoplasmic HuR Expression Is a Prognostic Factor in Invasive Ductal Breast Carcinoma. Cancer Research. 2005;65:2157–61. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 43.Sorrells DL, Black DR, Meschonat C, Rhoads R, De Benedetti A, Gao M, et al. Detection of eIF4E gene amplification in breast cancer by competitive PCR. Ann Surg Oncol. 1998;5:232–7. doi: 10.1007/BF02303778. [DOI] [PubMed] [Google Scholar]
- 44.Topisirovic I, Kentsis A, Perez JM, Guzman ML, Jordan CT, Borden KL. Eukaryotic translation initiation factor 4E activity is modulated by HOXA9 at multiple levels. Mol Cell Biol. 2005;25:1100–12. doi: 10.1128/MCB.25.3.1100-1112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 2000;19:3142–56. doi: 10.1093/emboj/19.12.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marty B, Maire V, Gravier E, Rigaill G, Vincent-Salomon A, Kappler M, et al. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res. 2008;10:R101. doi: 10.1186/bcr2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Knowles E, O’Toole SA, McNeil CM, Millar EK, Qiu MR, Crea P, et al. PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int J Cancer. 126:1121–31. doi: 10.1002/ijc.24831. [DOI] [PubMed] [Google Scholar]
- 48.Coleman LJ, Peter MB, Teall TJ, Brannan RA, Hanby AM, Honarpisheh H, et al. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. British Journal of Cancer. 2009;100:1393–9. doi: 10.1038/sj.bjc.6605044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Journal of the American Medical Association. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eIF4E protein levels were assessed by western blot analysis.
Box plots depicting (A) reduced 4E-BP1 expression and (B) elevated ratio of eIF4E/4E-BP1 in luminal A cases in comparison to other subtypes.
Kaplan Meier analysis of tumor cohort dichotomized at median into High vs. Low expressing groups in (A) Luminal B, (B) Luminal A, (C) Basal, (D) Her2, (E) Normal. Black: high eIF4E/4EBP2, light grey: low eIF4E/4EBP2.
Kaplan Meier analysis of tumor cohort dichotomized at median into High vs. Low expressing groups in (A) Luminal B, (B) Luminal A, (C) Basal, (D) Her2, (E) Normal. Black: high eIF4E/PML, light grey: low eIF4E/PML.