Abstract
Digital reconstruction of neuronal morphology is a powerful technique for investigating the nervous system. This process consists of tracing the axonal and dendritic arbors of neurons imaged by optical microscopy into a geometrical format suitable for quantitative analysis and computational modeling. Algorithmic automation of neuronal tracing promises to increase the speed, accuracy, and reproducibility of morphological reconstructions. Together with recent breakthroughs in cellular imaging and accelerating progress in optical microscopy, automated reconstruction of neuronal morphology will play a central role in the development of high throughput screening and the acquisition of connectomic data. Yet, despite continuous advances in image processing algorithms, to date manual tracing remains the overwhelming choice for digitizing neuronal morphology. We summarize the issues involved in automated reconstruction, overview the available techniques, and provide a realistic assessment of future perspectives.
Keywords: Automated Reconstruction, Neuronal Morphology, Neuron Tracing, Image Analysis, Vectorization
1. Digital Reconstruction of Neuronal Morphology
Neuronal morphology underlies nervous system connectivity (Buckmaster et al., 2004; Stepanyants et al., 2004) and single cell information processing (Kock and Segev, 2000; Häusser and Mel, 2003). By quantifying the shape and location of axons and dendrites, digital representations of neurons imaged by optical microscopy provide a powerful tool in investigating neural form and function. As a natural extension of hand drawn camera lucida representations of neuron shape, digital reconstructions allow for an almost unlimited variety of quantitative morphometrics (Vallotton et al., 2007; Scorcioni et al., 2008), as well as providing the basis for computational models of morphology (e.g. Hillman 1979; Burke et al., 1992; also reviewed in Donohue and Ascoli, 2005) and electrophysiology (Lazarewicz et al., 2002; Migliore et al., 2003; Davison et al., 2004).
Typical digital reconstructions represent neuronal morphology as a series of points along the neurites with their positions, radius, connectivity, and process type (e.g. soma, dendrite, and axon). While neuronal processes can be captured in more realistic detail by volume or surface encoding (Wearne et al., 2005), the above vector-style representation is very concise and sufficient for most purposes (Cannon et al., 1998; Ascoli et al., 2001).
The majority of publicly available digital neuronal reconstructions were traced by hand (Ascoli et al., 2007; Table 1) by virtually drawing the axonal and dendritic processes using a computer mouse. This operation can be carried out either by tracking the neuronal processes from the live feed of the microscope connected to the computer or by first capturing an image stack and then processing at a later time. In either way, the process is time consuming (Oberlander et al., 2007) and can be error prone (Ascoli et al., 2007). Automating neuromorphological reconstruction through the use of computer image analysis algorithms promises increased accuracy, reproducibility, and productivity. Currently, because of the time involved in manual reconstruction, many studies rely on multiple human operators to reconstruct enough neurons. The concerning source of noise and bias introduced by the substantial inter-operator variability (Scorcioni et al., 2004) would be eliminated by automated data collection.
Table 1.
Reconstruction methods represented in NeuroMorpho.org.
| Method | Cells | Type |
|---|---|---|
| Amira | 252 | Semi-Automated |
| Arbor | 99 | Manual |
| Automorph | 2 | Manual |
| Custom | 198 | Manual |
| Eutectic | 571 | Manual |
| Neurolucida | 4424 | Manual |
| NeuronMorpho | 33 | Manual |
| NeuroZoom | 80 | Manual |
| Tablet | 14 | Manual |
Of the 5673 neuronal reconstructions in verson 4.0 of the NeuroMorpho.org repository, less than 5% were reconstructed with semi-automated methods, and none were reconstructed with fully automated methods. While both Amira and Neurolucida now contain automated reconstruction tools, to the best of our knowledge these tools were not used to create any of the cells in NeuroMorpho.org.
Most importantly, large scale collection of morphological data will likely change the type of questions that can be addressed (Zhang et al., 2007a). For example, while most currently available neuronal reconstructions are from healthy mature animals, faster data collection could facilitate the comparison of different developmental ages or pathological conditions. High throughput analysis may also enable real time screening of pharmacological manipulations (Wu et al., 2010). The recent introduction of genetic approaches to image neurons in multiple distinct colors (Livet et al., 2007), in principle, allow for the collection of large data sets from individual animals. Similarly, the expanding possibility to extend image acquisition in live cells (Losavio et al., 2006) and over long temporal windows through time lapse microscopy (Li et al., 2010) will enable quantitative analysis of structural dynamics once digital tracing becomes automatic. Automated reconstructions will also constitute a powerful and necessary tool in light of the rapid progress in the optical detection of neuronal activity, which may soon enable the combined acquisition of both structural and functional data.
Digital encoding of neuronal morphology can be automated to varying degrees. At its simplest, automation can take the user guided tracing and re-compute the diameter and center lines (Meijering et al., 2008; Myatt and Nasuto, 2009). While these semi-automated algorithms only provide modest gains in speed, they can eliminate much of the user introduced variability. At the other end of the spectrum, fully automated algorithms are designed to take image stacks containing stained neurons and produce digital reconstructions with little to no user input (Zhang et al., 2008). Most current approaches fall somewhere in between. Algorithms also differ in whether they are designed to work on two or three dimensional data, and how comprehensively they detect every type of substructure, such as somata, spines, and even branch or end points.
In the following, we provide an overview of existing methodologies as a resource for both neuroscience researchers looking for tools to automate their data collection, and algorithm developers planning the implementation of new automated techniques. Table 2 is an unabridged list of automated reconstruction efforts. The included information on methodological details summarizes and distinguishes the different approaches and provides a central starting point to confront the issues involved in automating digital reconstruction of neuronal morphology.
Table 2.
Automated reconstruction algorithms.
| Pre-Processing | Volume/ Surface/ Vector a | Auto Path/ End pt./Diameter | Local/ Global Gradients | Binary Tree Enforced? | Automation d | Auto Soma Detection? | Spines? | License e | Validation | Imaging | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Algorithm name | Key Algorithm Details | |||||||||||
| Al-Kofahi et al., 2002, 2003, 2008; Tsai et al., 2004 | AutoNeuron by Microbrightfield Inc., RPI-Trace3D | N | V | P,E,D | L | Tracing with directional kernels | Y | F | Y | N | C | M | |
| Broser et al., 2008 | AxoQuant | Y | V | P,D | L,G | Length only | N | S | N | N | M | BF | |
| Cuntz et al., 2008 | 3D skeletonization | Y | V | P,E,D | G | Thresholding, skeleton | Y | S | N | N | M | 2P | |
| Evers et al., 2004 | Amira module | N | O,S,V | P,E,D | L | Active contour, snakes | Y | S,F | Y | N | F | M | LS |
| http://www.ams.sunysb.edu/~lindquis/3dma /3dma_neuron/3dma_neuron.html | 3DMA-Neuron | N | V | P,D | L,G | Local correlation | Y | S | N | Y | C | LS | |
| http://www.bitplane.com/go/products/filamenttracer | Filament Tracer, Imaris | Y | V | P,E,D | L | Local contrast, 3D isotropic dilation | Y | S,F | Y | C | M | ||
| Konstantinidis et al., 2005 | Frame-based denoising | N | O | Na | L | Robust edge detection | N | F | Y | S | F | ||
| Losavio et al., 2008 | ORION | Y | V | P,E,D | Probability volume | Y | S,F | N | N | F | A, M | LS,MF | |
| Lu et al., 2009 | Wildfire (Reconstruct extension) | Y | O | P | G | Region growing across sections | N | S | N | N | O | M | LS |
| Meijering et al., 2008 | NeuronJ | Y | V | P | L | Live wire, global optimization algorithm | N | S | N | N | F | M | F |
| Myatt and Nasuto, 2009 | Neuromantic | N | V | P | L | Gaussian filters, search graph | y | S | N | N | F | M | |
| Narro et al., 2007 | NeuroMetrics | Y | V | P,E | G | Skeleton thresholding, edge detection | N | S | N | N | F | M | F |
| Oberlaender et al., 2007 | Local threshold and thinning | Y | V | P,E | L | Thresholding and thinning | Y | S | N | N | C | M | BF |
| Pool et al., 2008 | NeuriteTracer | Y | V | P, E | G | Skeletonization, thresholding | N | F | Y | N | O | M | F |
| Schmitt et al., 2004 | Skeleton fit with active contours | N | V,S | P,E,D | L | Skeletonization, snakes | Y | S | N | Y | M | LS | |
| Selinummi et al., 2006 | Sobel edge detection | N | O | No | G | Sobel edge detection | N | F | Y | N | BF | ||
| Srinivasan et al., 2007 | Probabilistic region merging | N | V | P | L | Adaptive smoothing, watershed | N | S | N | N | F | M | LS |
| Streekstra and van Pelt, 2002 | Gaussian derivatives kernels | V | P,D | Gaussian derivative kernels | N | F | N | N | S | C | |||
| Takayuki et al., 2006 | SSDT | Y | V | P,D | G | Single seed distance transform | Y | F | N | ||||
| Vallotton et al., 2007 | HCA-Vision | Y | V | P,E | G | Watershed, dynamic programming | Y | F | Y | N | C | A | F |
| Vasilkoski and Stepanyants, 2009 | Voxel-coding | N | O,V | P,E,D | L | Modified active contours | Y | F | Y | N | S | Na | |
| Weaver et al., 2004; Wearne et al., 2005; Rodriguez et al., 2006, 2009 | Neuron Studio, Rayburst, voxel scooping | Y | O,S,V | P,E,D | L | Raybursts for diameters, ISODATA for segmentation | N | S | N | Y | F | S, M | LS,MF |
| Wouterlood, 2005 | Amira (tutorial) | Y | O,S,V | P,E,D | G | Wireframe ortholine | N | S | Y | Y | C | LS | |
| Xiong et al., 2006 | Multineurite | Y | V | P,E | G | Local Hessian matrix, Gaussian kernel | N | F | N | N | F | M | F |
| Zhang et al., 2007a | Edge-based tracing | Y | V | P,E | L | Approximation of Gaussian kernel | N | F | Y | N | M | F | |
| Zhang et al., 2007b; Zhang et al., 2008 | Dynamic programming, watershed | Y | V | P,E | L | Gaussian kernel, centerline by dynamic programming | N | F | N | N | M | LS |
Neurite representation: O, volume; S, surface; V, vector.
Automatically detected features: P, paths; E, end points; D, diameters.
Setting of gradient thresholds: L, local; G, global.
Automation extent: F, full; S, semi.
License type: C, commercial; F, freely available; O, open source.
Validation data: M, manual reconstructions; A, automated reconstructions; S, synthetic images.
Suitable imaging: M, multiple modalities; LS, laser scanning; F, fluorescence; BF, brightfield; MF, multi-photon; 2P, two-photon; C, confocal.
2. Automated Tracing Methodologies
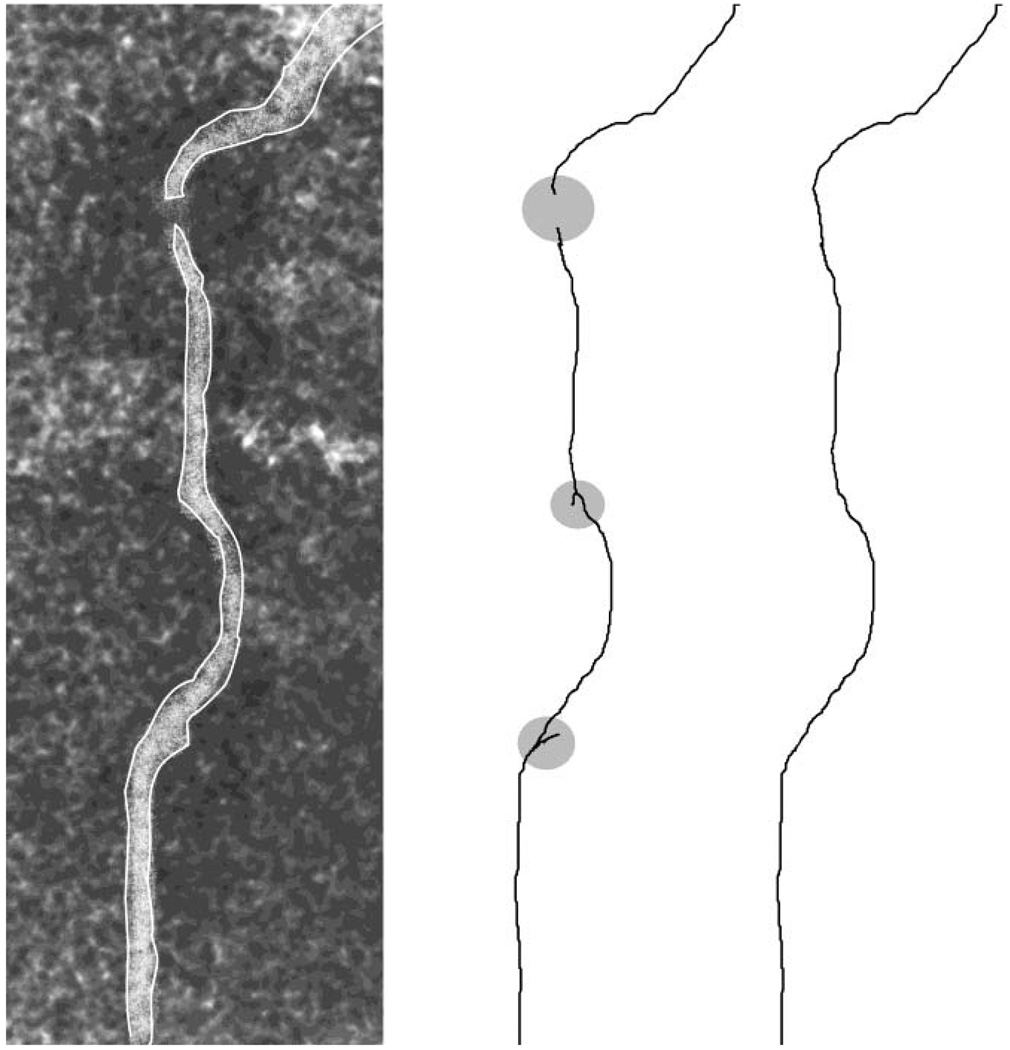
Skeletonization has been a popular solution to medial axis extraction (e.g. Pool et al., 2007), a fundamental step in neuronal reconstruction. Skeletonization consists of segregating the backbone of the arbor structure from the image stack. This step generally uses a global threshold (Cuntz et al., 2008) or edge detection algorithms (Selinummi et al., 2006) to identify potential neurites based on their intensity differences from the background. The extracted volumes are then iteratively thinned from the edges to find their centerlines (Fig. 1). Global (Pool et al., 2007) or local (Schmitt et al., 2004) thresholds or gradients can then be used to determine diameters. These methods are frequently used in preparations suitable for 2D imaging, such as neuronal cultures (Narro et al., 2007) or flat mounted thin tissues. Because this approach must process the entire image, it can be computationally intensive (i.e. slow) for large images (Al-Kofahi et al., 2002). While differences in staining intensity can produce gaps in the detected neurites, processing of the entire image allows these gaps to be detected and corrected by looking for unconnected neurites (Fig. 1; Narro et al., 2007; Xiong et al., 2006). Irregularities in the neurites may cause skeletonization algorithms to produce small spurious side branches, which can be removed by enforcing a minimum branch length (Fig. 1; Valotton et al., 2007).
Figure 1. Skeletonization process.
The neuronal process is identified against the background (left) and thinned by skeletonization algorithms to produce segments (center). Spurious side branches are then removed and disconnected segments connected (right).
A second class of automated reconstruction methods use a variety of snake, watershed, or other energy minimizing algorithms to connect seed points along the neurites (e.g. Xiong et al., 2006). End points, branch points, and sometimes intermediate points are either selected by the user (Narro et al., 2007) or detected algorithmically (Xiong et al., 2006; Zhang et al., 2007). These points are then connected in 2 or 3 dimensions by minimizing some energy function corresponding to a path along stained structures. Presuming that branch and end points are chosen correctly, these methods are very robust with regards to incomplete staining. Algorithmic seed point selection, however, can be tricky and is sensitive to staining irregularities. One solution is to allow the user to continuously select new seed points to guide the reconstruction in a semi-automated fashion (Meijering et al., 2008; Lu et al., 2009). User selection of terminal points is adopted in successful semi-automated solutions, such as Imaris’ Filament Tracer (http://www.bitplane.com/go/products/filamenttracer). Although following the neurite path from tip to root typically ensures greater robustness, this choice is not easily scalable to full automation.
One of the more recent developments in automated reconstruction are methodologies which trace neurites out from the soma (Al-Kofahi, 2002) or other points (Zhang et al., 2007) much as a human operator would. Kernels or templates corresponding to bits of segment are tested iteratively at different orientations at the end of the tracing until end points are detected. One prominent advantage of this approach is that only the immediate vicinity of extending neurite trace needs to be analyzed, greatly reducing the computational load (Al-Kofahi, 2002). Tracing algorithms can be sensitive to staining discontinuities and imaging non-uniformities, but methods have been developed to mitigate these issues (Al-Kofahi, 2003).
3. Validation Methods
The chief method for validating automated reconstruction algorithms is to compare the resulting digital representations with a gold standard reference reconstruction. Most often this gold standard is a manual reconstruction (Table 2). Some studies have used well established automated (Losavio et al., 2008) or semi-automated (Vallotton et al., 2007) methods to generate reference reconstructions. Some labs develop their image analysis methodologies using synthetic data, for example simplified tubular structures (Streekstra and van Pelt, 2002), convolved images of previously digitized neurons (Konstantinidis et al., 2005), or volumetric data created from existing digital reconstructions (Vasilkoski and Stepanyants, 2009). In these cases, the reference reconstruction used to generate the synthetic data typically constitutes the gold standard.
Once a reference gold standard is selected, there are several approaches for devising the comparison with the algorithmic reconstruction to be evaluated. As mentioned earlier, digital reconstructions allow for an almost unlimited number of morphometrics. Gross metrics, such as mean diameter (Schmitt et al., 2004), branch number (Narro et al., 2007), and total length (Pool et al., 2008) are commonly compared. Topological characterizations such as caulescence (Brown et al., 2008) or the tree edit distance (Heumann and Wittum, 2009) are also particularly useful in this regard. Occasionally more intricate measurements such as electrophysiological behavior (Losavio et al., 2008) or center line deviations (Zhang et al., 2007b) are used for validation. Some studies attempt to show that the measurements are within a certain percentage of the reference (Oberlander et al., 2007). Others employ multiple reference observations created by different users to assess whether the automated reconstruction falls within the between-user variability (Schmitt et al., 2004).
It is important as well to quantify not only the accuracy of a method, but also the time savings over alternative (manual) approaches (Oberlaender et al., 2007) or the amount of user interaction required to obtain complete reconstructions (Lu et al., 2009). A more general and unified procedure for validating new reconstruction methodologies in terms of both speed and accuracy could help advance the field.
4. Tissue Considerations
Different tissue preparations lend themselves to different image analysis approaches. Cultured neurons, for example, are generally two dimensional relative to their in vivo counterparts. An algorithm designed for three dimensional data could get very confused where neurites cross over each other (Lu et al., 2009). Cultured neurons also tend to be stained at high contrast and therefore may be less challenging to skeletonize. The types of neuronal processes one wishes to digitize also influence the selection of the most appropriate automated algorithms. Axons are often traced distal from the soma, and the exact branching topology is generally more difficult and less vital to determine than in dendrites. Axon branch diameters change less than in tapering dendrites, and are typically considered less critical to record precisely than lengths and spatial patterns (e.g. Broser et al., 2008; Srinivasan et al., 2007). The opposite holds true for dendrites, whereby characterizing spine shape is facilitated by algorithms, like Rayburst, which can capture irregular volumes (Wearne et al., 2005).
Histological and imaging methods also typically determine how many neurons are stained, as well as their contrast relative to the background. Intracellular injection offers the opportunity to fill single cells, or multiple cells with no overlap, and has the advantate of allowing the collection of electrophysiology data. Viral expression in principle provides information about synaptic pathways in addition to arbor morphology. However, intracellular injection and viral expression can be more time consuming and technically demanding to establish and execute than other staining methods, such as classic Golgi or tract tracing. The majority of methods listed in Table 2 were tailored for fluorescent imaging techniques, with laser scanning confocal microscopy being the most common. Also evident from Table 2, few automated reconstruction algorithms have been developed for and/or tested on multiple imaging modalities. While different image processing methods are better suited to various tissue preparations, development of more general algorithms would go a long way to increasing data reuse, standardization, and reproducibility.
5. Other Algorithm Properties
Many automated reconstruction methodologies require image pre-processing. Often, these are included as part of the reconstruction software (e.g. Vallotton et al., 2007). Pre-processing can include deconvolution (blind or imaging system specific), tiling of multiple high magnification fields of view to create single mosaic images (Cuntz et al., 2008, Weaver et al., 2004), and a variety of image intensity rescaling, signal normalization, and contrast enhancements. Some methods involve the removal of small artifacts (Losavio et al., 2008). Depending on the staining technique, imaging method, and arbor size, it is often necessary to recombine serial sections into a single volume before processing (e.g. Oberlander et al., 2007). Some automated reconstruction methodologies also incorporate post reconstruction analysis such as automated extraction of morphometircs (Al-Kofahi et al., 2002; Vallotton et al., 2007; Cuntz et al., 2008).
Other considerations when choosing an automated reconstruction approach for data generation or as a base for developing new methods are the technical and legal details. Automated algorithms range from proprietary commercial to open source (Table 2). Open source development has proven to be a powerful tool in addressing other scientific problems where the technical skills of those in the field are high but the commercial potential limited (e.g. the Bioconductor project for R, Gentleman et al., 2004). While open source algorithms can be adapted by subsequent users to address their specific scientific problems, they also hold the promise of collaborative expansion into general and feature rich programs. Open source programs also tend to use non-proprietary data structures, increasing interoperability between tracing, modeling, and analysis programs and encouraging standardized data types. However, sparse or idiosyncratic interfaces often make freely distributed programs more difficult to use than their commercial counterparts.
Not only does the format of the data vary from vector to volumes, but even within a format the details can be very different. Vector data, for example, may or may not include diameter information (Meijering et al., 2008). Actual file formats vary too, from ASCII text describing vector connectivity (e.g. Evers et al., 2004) to proprietary binary formats as used by the Neurolucida system (Microbrightfield, Inc.). Other practical considerations include the operating system the algorithm was developed for and the programming language it was written in.
6. Incorporating Statistical Models
It is telling that most reference reconstructions for automated algorithm validation and most publicly available neuronal morphologies are traced manually. Part of the reason for the current state of affairs is that automated reconstruction methodologies can not yet duplicate the intuition and expertise of an experienced human technician. One possible direction for improving automated algorithms is to use existing reconstructions to generate morphometric statistics for different cell classes, such as those used in morphological modeling (e.g. Samsonovich and Ascoli, 2005; Donohue and Ascoli, 2008). These statistics could be used to help disambiguate morphological alternatives and adjust local parameters as the arbors are being traced.
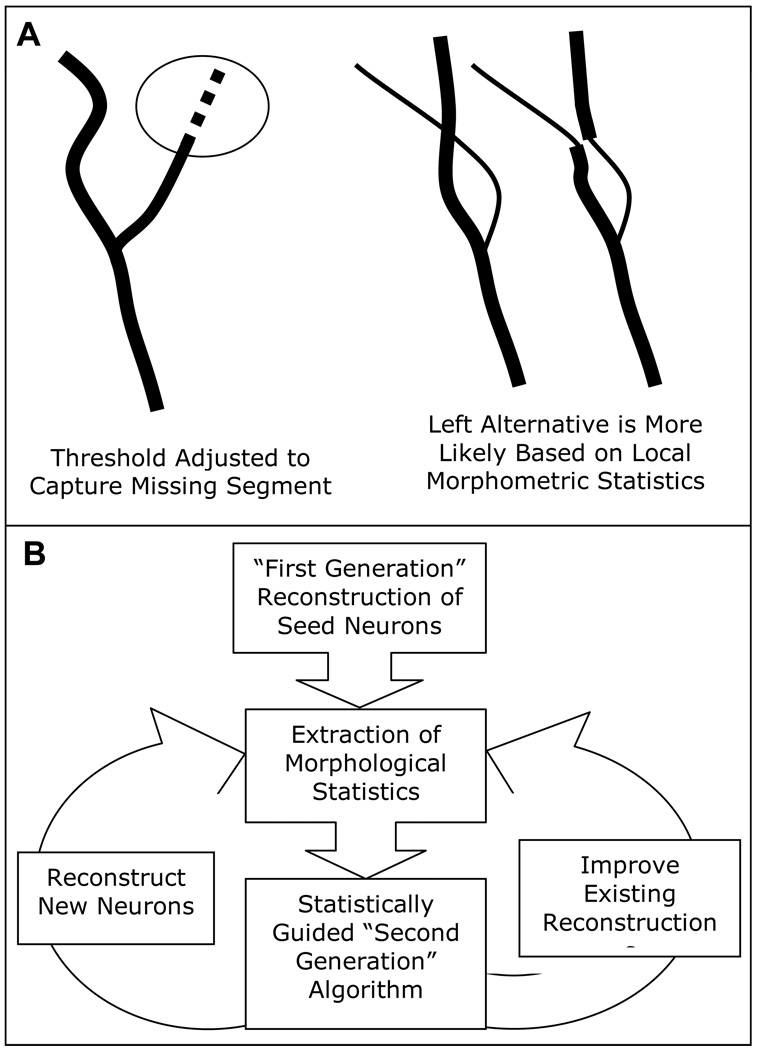
The details of incorporating statistical knowledge of branching morphology into automated reconstruction will vary according to the nature of the algorithm (Fig. 2A). Global image analysis approaches could have gradient or intensity thresholds adjusted until the number or total length of branches is within the observed range for the particular cell type. For algorithms which trace neurites out from a starting location, finer grain statistics could be utilized. For example, diameter changes, branch angles, and branch lengths of traced neurites could all be influenced at a local level by information from previously traced neurons. One can imagine a situation in which an apparent diameter change is found to be particularly unlikely for the given cell class and sub-cellular location (e.g. third order branch, or 145 microns from the soma), and is perhaps due to incomplete staining or optical occlusion. In locations with missing or unlikely staining information, probabilistic modeled data could be substituted.
Figure 2. Incorporation of statistical models into automated reconstructions of neuronal morphology.
A) Local or global thresholds can be adjusted if statistics of previously reconstructed neurons suggest that branches are being missed (left). Statistics of local parameters such as taper rates and branch lengths can be used to disambiguate reconstructions (right). B) Statistics from reconstructed neurons can be used to improve algorithms in an iterative fashion.
Manual error correcting is a major bottleneck in most automated reconstruction methodologies. Highlighting those areas of a reconstruction which are most unlikely based on previously reconstructed neurons of the same class could significantly speed the process. In cases where a small finite number of mutually exclusive alternative interpretations are possible, presenting those alternatives to a human reviewer in a systematic way (e.g. multiple choices ranked by likelihood or effect on tree size) could greatly speed the manual editing process. The selected choices by the human user, perhaps capturing an underlying idiosyncrasy of the tissue preparation or staining, could then be used to improve the rankings in subsequent presentation of alternatives.
Subjective judgments based on extensive experience are one of the crucial reasons that hand reconstructions remain the gold standard. Any success in capturing this experience algorithmically would greatly improve the accuracy of fully automated reconstructions. The incorporation of statistical models may thus be envisioned as an iterative process. The available data set is first digitally traced by the “first generation” algorithm. The resulting reconstructions are next analyzed to extract the statistical distributions of the appropriate morphometric parameters. The resulting model is then incorporated in a “second generation” algorithm, which is used to re-trace the original data set, ideally producing a more accurate pool of reconstructions to extract more reliable statistics, and so forth (Fig. 2B). In the same vein, as automated techniques become more powerful and ubiquitous, the increased numbers of reconstructed cells could also be continuously fed back into the algorithms to improve them.
7. Concluding Remarks
Increased speed of reconstruction promises to change the type of questions we can ask regarding neuronal morphology. Despite advances in automatic reconstruction techniques, however, they still have far to go before they surpass manual reconstructions as the method of choice. This is almost certain to change as algorithms become more robust and general. To help fill the gap between the current state of the art and the future of fully automated reconstructions, a scientific competition was recently organized, named DIADEM challenge (Ascoli, 2008), short for DIgital reconstructions of Axonal and DEndritic Morphology (http://diademchallenge.org). The original intent of the DIADEM challenge was to identify the most critical practical obstacles that still need to be overcome to advance this research area towards full automation. The format of the competition included a one year development period during which registered teams worked remotely to design and implement algorithms for tracing five data sets representative of the diversity of experimental conditions in cellular neuroscience. All contestants received feedback on their submissions, based on a custom-designed quantitative metric, the overall evaluation of expert judges, and an estimate of the time it would have taken the data owners to edit the submitted entries up to their standards. The best performing contestants qualified for the final tournament, where they competed on new data sets and interacted live with each others as well as with the judges, organizers, and data owners. In addition to achieving the goal of clearly demarking existing needs, this unprecedented initiative fostered novel and promising technical solutions. One of the key criteria for success in the DIADEM challenge was the generality of the algorithms, i.e. how well a given solution would perform on the diverse collection of data sets. As expected, some data sets proved to be more difficult than others to trace. Moreover, individual algorithms naturally tended to “specialize”, performing better on different data sets. However, no pattern emerged mapping algorithms to data sets one to one. Surprisingly, there was no clear divide between confocal fluorescence and brightfield data sets in terms of overall or individual algorithms performance.
Indeed, one possible reason for the multitide of different automated reconstruction techniques is that the best approach varies depending on the biological questions being asked and the properties of the tissue being analyzed. In many cases, researchers would rather develop their own algorithms than learn the idiosyncrasies of an existing method which may not precisely address their concerns. Custom methodologies have the added advantages of being potentially less expensive than commercial application, and giving the creator full control over file formats and process pipelines, allowing them to integrate more fully pre-processing (deconvolution, tiling, etc.) or post-processing (measurement, validation, etc.) steps. One consequence is that algorithms are rarely re-used by other labs. Adherence to open source development principles, adoption of universal data forms, and an organized effort to document and archive existing programs, could help alleviate this issue. Ultimately, increased standardization of definitions and methodologies could allow for a more interoperable ecosystem of tools to measure, model, and verify neuronal reconstructions.
Acknowledgments
NIH R01 NS39600, MURI ONR N00014-10-1-0198.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Duncan E. Donohue, Email: dedonohue@gmail.com.
Giorgio A. Ascoli, Email: ascoli@gmu.edu.
References
- Al-Kofahi KA, Can A, Lasek S, Szarowski DH, Dowell-Mesfin N, Shain W, Turner JN, Roysam B. Median-based robust algorithms for tracing neurons from noisy confocal microscope images. IEEE Trans. Inf. Technol. Biomed. 2003;7(4):302–317. doi: 10.1109/titb.2003.816564. [DOI] [PubMed] [Google Scholar]
- Al-Kofahi KA, Lasek S, Szarowski DH, Pace CJ, Nagy G, Turner JN, Roysam B. Rapid automated three-dimensional tracing of neurons from confocal image stacks. IEEE Trans. Inf. Technol. Biomed. 2002;6(2):171–187. doi: 10.1109/titb.2002.1006304. [DOI] [PubMed] [Google Scholar]
- Al-Kofahi Y, Dowell-Mesfin N, Pace C, Shain W, Turner JN, Roysam B. Improved detection of branching points in algorithms for automated neuron tracing from 3D confocal images. Cytometry A. 2008;73(1):36–43. doi: 10.1002/cyto.a.20499. [DOI] [PubMed] [Google Scholar]
- Ascoli GA. Neuroinformatics grand challenges. Neuroinformatics. 2008;6(1):1–3. doi: 10.1007/s12021-008-9010-5. [DOI] [PubMed] [Google Scholar]
- Ascoli GA, Donohue DE, Halavi M. NeuroMorpho.Org: a central resource for neuronal morphologies. J. Neurosci. 2007;27(35):9247–9251. doi: 10.1523/JNEUROSCI.2055-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli GA, Krichmar JL, Nasuto SJ, Senft SL. Generation, description and storage of dendritic morphology data. Philos. Trans. R. Soc. B. 2001;356(1412):1131–1145. doi: 10.1098/rstb.2001.0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broser PJ, Erdogan S, Grinevich V, Osten P, Sakmann B, Wallace DJ. Automated axon length quantification for populations of labeled neurons. J. Neurosci. Methods. 2008;169(1):43–54. doi: 10.1016/j.jneumeth.2007.11.027. [DOI] [PubMed] [Google Scholar]
- Brown KM, Gillette TA, Ascoli GA. Quantifying neuronal size: Summing up trees and splitting the branch difference. Sem. Cell Dev. Biol. 2008;19:485–493. doi: 10.1016/j.semcdb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckmaster PS, Alonso A, Canfield DR, Amaral DG. Dendritic morphology, local circuitry, and intrinsic electrophysiology of principal neurons in the entorhinal cortex of macaque monkeys. J. Comp. Neurol. 2004;470(3):317–329. doi: 10.1002/cne.20014. [DOI] [PubMed] [Google Scholar]
- Burke RE, Marks WB, Ulfhake B. A parsimonious description of motoneuron dendritic morphology using computer simulation. J. Neurosci. 1992;12(6):2403–2416. doi: 10.1523/JNEUROSCI.12-06-02403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon RC, Turner DA, Pyapali GK, Wheal HV. An on-line archive of reconstructed hippocampal neurons. J. Neurosci. Methods. 1998;84:49–54. doi: 10.1016/s0165-0270(98)00091-0. [DOI] [PubMed] [Google Scholar]
- Cuntz H, Forstner F, Haag J, Borst A. The morphological identity of insect dendrites. PLoS Comput. Biol. 2008;4(12) doi: 10.1371/journal.pcbi.1000251. e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AP, Morse TM, Migliore M, Shepherd GM, Hines ML. Semi-automated population of an online database of neuronal models (ModelDB) with citation information, using PubMed for validation. Neuroinformatics. 2004;2(3):327–332. doi: 10.1385/NI:2:3:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue DE, Ascoli GA. Models of neuronal outgrowth. In: Koslow Subramaniam., editor. Databasing the Brain: From Data to Knowledge. Hoboken, NJ: Wiley Press; 2005. pp. 303–323. [Google Scholar]
- Donohue DE, Ascoli GA. A comparative computer simulation of dendritic morphology. PLoS Comput. Biol. 2008;4(5) doi: 10.1371/journal.pcbi.1000089. e1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers JF, Schmitt S, Sibila M, Duch C. Progress in functional neuroanatomy: precise automatic geometric reconstruction of neuronal morphology from confocal image stacks. J. Neurophysiol. 2005;93(4):2331–2342. doi: 10.1152/jn.00761.2004. [DOI] [PubMed] [Google Scholar]
- Filament Tracer by Bitplane. 2010 http://www.bitplane.com/go/products/filamenttracer.
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leish F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10) doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusser M, Mel B. Dendrites: bug or feature? Curr. Opin. Neurobiol. 2003;13(3):372–383. doi: 10.1016/s0959-4388(03)00075-8. [DOI] [PubMed] [Google Scholar]
- Heumann H, Wittum G. The Tree-Edit-Distance, a Measure for Quantifying Neuronal Morphology. Neuroinformatics. 2009;7:179–190. doi: 10.1007/s12021-009-9051-4. [DOI] [PubMed] [Google Scholar]
- Hillman DE. Neuronal shape parameters and substructures as a basis of neuronal form. In: Schmitt F, editor. The Neurosciences, Fourth Study Program. Cambridge, MA: MIT Press; 1979. pp. 477–498. [Google Scholar]
- Koch C, Segev I. The role of single neurons in information processing. Nat. Neurosci. 2000;3 Suppl:1171–1177. doi: 10.1038/81444. [DOI] [PubMed] [Google Scholar]
- Konstantinidis I, Santamaria-Pang A, Kakadiaris I. Frames-Based Denoising in 3D Confocal Microscopy Imaging. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2005;1:290–293. doi: 10.1109/IEMBS.2005.1616401. [DOI] [PubMed] [Google Scholar]
- Lazarewicz MT, Boer-Iwema S, Ascoli GA. Practical aspects in anatomically accurate simulations of neuronal electrophysiology. In: Ascoli GA, editor. Computational Neuroanatomy: Principles and Methods. Totowa, NJ: Humana Press; 2002. pp. 127–148. [Google Scholar]
- Li J, Wang Y, Chiu SL, Cline HT. Membrane targeted horseradish peroxidase as a marker for correlative fluorescence and electron microscopy studies. Front. Neural Circuits. 2010;4:6. doi: 10.3389/neuro.04.006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist WB, Weaver CM. 3DMA-Neuorn online manual. 2010 http://www.ams.sunysb.edu/~lindquis/3dma/3dma_neuron/3dma_neuron.html.
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Losavio BE, Liang Y, Santamaría-Pang A, Kakadiaris IA, Colbert CM, Saggau P. Live neuron morphology automatically reconstructed from multiphoton and confocal imaging data. J. Neurophysiol. 2008;100(4):2422–2429. doi: 10.1152/jn.90627.2008. [DOI] [PubMed] [Google Scholar]
- Losavio BE, Reddy GD, Colbert CM, Kakadiaris IA, Saggau P. Combining optical imaging and computational modeling to analyze structure and function of living neurons. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006;1:668–670. doi: 10.1109/IEMBS.2006.259552. [DOI] [PubMed] [Google Scholar]
- Lu J, Fiala JC, Lichtman JW. Semi-automated reconstruction of neural processes from large numbers of fluorescence images. PLoS One. 2009;4(5):e5655. doi: 10.1371/journal.pone.0005655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58(2):167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Migliore M, Morse TM, Davison AP, Marenco L, Shepherd GM, Hines ML. ModelDB: making models publicly accessible to support computational neuroscience. Neuroinformatics. 2003;1(1):135–139. doi: 10.1385/NI:1:1:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myatt DR, Nasuto SJ. Three-Dimensional Reconstruction of Neurons with Neuromantic. AISB Quarterly. 2009;125:1–2. [Google Scholar]
- Narro ML, Yang F, Kraft R, Wenk C, Efrat A, Restifo LL. NeuronMetrics: software for semi-automated processing of cultured neuron images. Brain Res. 2007;1138:57–75. doi: 10.1016/j.brainres.2006.10.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlaender M, Bruno RM, Sakmann B, Broser PJ. Transmitted light brightfield mosaic microscopy for three-dimensional tracing of single neuron morphology. J. Biomed. Opt. 2007;12(6) doi: 10.1117/1.2815693. 064029. [DOI] [PubMed] [Google Scholar]
- Pool M, Thiemann J, Bar-Or A, Fournier AE. NeuriteTracer: a novel ImageJ plug-in for automated quantification of neurite outgrowth. J. Neurosci. Methods. 2008;168(1):134–139. doi: 10.1016/j.jneumeth.2007.08.029. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Hof PR, Wearne SL. Rayburst sampling, an algorithm for automated three-dimensional shape analysis from laser scanning microscopy images. Nat. Protoc. 2006;1(4):2152–2161. doi: 10.1038/nprot.2006.313. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Hof PR, Wearne SL. Three-dimensional neuron tracing by voxel scooping. J. Neurosci. Methods. 2009;184(1):169–175. doi: 10.1016/j.jneumeth.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonovich AV, Ascoli GA. Statistical determinants of dendritic morphology in hippocampal pyramidal neurons: A hidden Markov model. Hippocampus. 2005;15(2):166–183. doi: 10.1002/hipo.20041. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Evers JF, Duch C, Scholz M, Obermayer K. New methods for the computer-assisted 3-D reconstruction of neurons from confocal image stacks. Neuroimage. 2004;23(4):1283–1298. doi: 10.1016/j.neuroimage.2004.06.047. [DOI] [PubMed] [Google Scholar]
- Scorcioni R, Lazarewicz M, Ascoli GA. Quantitative morphometry of hippocampal pyramidal cells: differences between anatomical classes and reconstructing laboratories. J. Comp. Neurol. 2004;473:177–193. doi: 10.1002/cne.20067. [DOI] [PubMed] [Google Scholar]
- Scorcioni R, Polavaram S, Ascoli GA. L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nat. Protoc. 2008;3(5):866–876. doi: 10.1038/nprot.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinummi J, Ruusuvuori P, Lehmussola A, Huttunen H, Yli-Harja O, Miettinen R. Three-dimensional digital image analysis of immunostained neurons in thick tissue sections. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006;1:4783–4786. doi: 10.1109/IEMBS.2006.259419. [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Zhou X, Miller E, Lu J, Lichtman J, Wong ST. Automated axon tracking of 3D confocal laser scanning microscopy images using guided probabilistic region merging. Neuroinformatics. 2007;5(3):189–203. doi: 10.1007/s12021-007-0013-4. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Tamas G, Chklovskii DB. Class-specific features of neuronal wiring. Neuron. 2004;43(2):251–259. doi: 10.1016/j.neuron.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Streekstra GJ, van Pelt J. Analysis of tubular structures in three-dimensional confocal images. Network. 2002;13(3):381–395. [PubMed] [Google Scholar]
- Takayuki Y, Teijiro I, Nobuyuki M, Hidetoshi I, Ryohei K. Reconstruction and simulation for three-dimensional morphological structure of insect neurons. Neurocomputing. 2006;69(10–12):1043–1047. [Google Scholar]
- Tsai CL, Stewart CV, Tanenbaum HL, Roysam B. Model-based method for improving the accuracy and repeatability of estimating vascular bifurcations and crossovers from retinal fundus images. IEEE Trans. Inf. Technol. Biomed. 2004;8(2):122–130. doi: 10.1109/titb.2004.826733. [DOI] [PubMed] [Google Scholar]
- Vallotton P, Lagerstrom R, Sun C, Buckley M, Wang D, De Silva M, Tan SS, Gunnersen JM. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58(2):167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Vallotton P, Lagerstrom R, Sun C, Buckley M, Wang D, De Silva M, Tan SS, Gunnersen JM. Automated analysis of neurite branching in cultured cortical neurons using HCA-Vision. Cytometry A. 2007;71(10):889–895. doi: 10.1002/cyto.a.20462. [DOI] [PubMed] [Google Scholar]
- Vasilkoski Z, Stepanyants A. Detection of the optimal neuron traces in confocal microscopy images. J. Neurosci. Methods. 2009;178(1):197–204. doi: 10.1016/j.jneumeth.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience. 2005;136(3):661–680. doi: 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
- Weaver CM, Hof PR, Wearne SL, Lindquist WB. Automated algorithms for multiscale morphometry of neuronal dendrites. Neural Comput. 2004;16(7):1353–1383. doi: 10.1162/089976604323057425. [DOI] [PubMed] [Google Scholar]
- Wouterlood FG. 3-D reconstruction of neurons from multichannel confocal laser scanning image series. Curr. Protoc. Neurosci. 2005;Chapter 2(Unit 2.8) doi: 10.1002/0471142301.ns0208s32. [DOI] [PubMed] [Google Scholar]
- Wu C, Schulte J, Sepp KJ, Littleton JT, Hong P. Automatic robust neurite detection and morphological analysis of neuronal cell cultures in high-content screening. Neuroinformatics. 2010;8(2):83–100. doi: 10.1007/s12021-010-9067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G, Zhou X, Degterev A, Ji L, Wong ST. Automated neurite labeling and analysis in fluorescence microscopy images. Cytometry A. 2006;69(6):494–505. doi: 10.1002/cyto.a.20296. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhou X, Degterev A, Lipinski M, Adjeroh D, Yuan J, Wong ST. A novel tracing algorithm for high throughput imaging Screening of neuron-based assays. J. Neurosci. Methods. 2007a;160(1):149–162. doi: 10.1016/j.jneumeth.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhou X, Degterev A, Lipinski M, Adjeroh D, Yuan J, Wong ST. Automated neurite extraction using dynamic programming for high-throughput screening of neuron-based assays. Neuroimage. 2007b;35(4):1502–1515. doi: 10.1016/j.neuroimage.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhou X, Lu J, Lichtman J, Adjeroh D, Wong ST. 3D Axon structure extraction and analysis in confocal fluorescence microscopy images. Neural Comput. 2008;20(8):1899–1927. doi: 10.1162/neco.2008.05-07-519. [DOI] [PMC free article] [PubMed] [Google Scholar]