Abstract
Villin-type headpiece domains are ~70 residue motifs that reside at the C-terminus of a variety of actin-associated proteins. Villin headpiece (HP67) is a commonly used model system for both experimental and computational studies of protein folding. HP67 is made up of two subdomains that form a tightly packed interface. The isolated C-terminal subdomain of HP67 (HP35) is one of the smallest autonomously-folding proteins known. The N-terminal subdomain requires the presence of the C-terminal subdomain to fold. In the structure of HP67, a conserved salt bridge connects N- and C-terminal subdomains. This buried salt bridge between residues E39 and K70 is unusual in a small protein domain. We used mutational analysis, monitored by CD and NMR, and functional assays to determine the role of this buried salt bridge. First, the two residues in the salt bridge were replaced with strictly hydrophobic amino acids, E39M/K70M. Second, the two residues in the salt bridge were swapped, E39K/K70E. Any change from the wild-type salt bridge residues results in unfolding of the N-terminal subdomain, even when the mutations were made in stabilized variant of HP67. The C-terminal subdomain remains folded in all mutants and is stabilized by some of the mutations. Using actin sedimentation assays we find that a folded N-terminal domain is essential for specific actin binding. Therefore, the buried salt bridge is required for the specific folding of the N-terminal domain which confers actin-binding activity to villin-type headpiece domains, even though the residues required for this specific interaction destabilize the C-terminal subdomain.
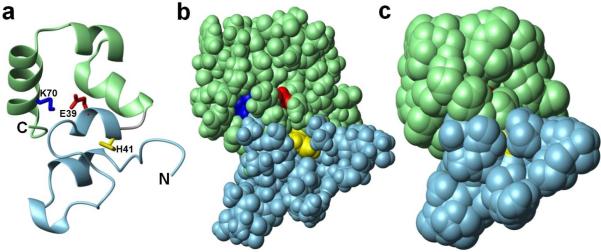
Villin is an actin bundling protein found in the brush border microvilli located at the apical surface of the cells that compose the gastrointestinal and renal absorptive epithelium. It contains two actin binding sites, one of which is contained in the C-terminal “headpiece” domain (1, 2). Headpiece domains are structurally and functionally independent modular domains (3). They are compact motifs containing ~70 residues. Villin headpiece (HP67) is 67 residues in length and contains two subdomains, an N-terminal subdomain (P10-H41), and a C-terminal subdomain (L42-F76) that folds independently from the N-terminal subdomain (3). Residues responsible for actin binding are found in both subdomains (4–7). Buried within the continuous hydrophobic core between the N- and C-terminal domains of HP67 is a salt bridge between E39 and K70 (Figure 1). This buried salt bridge is a highly conserved feature among essentially all known headpiece domain sequences. The presence of a buried polar interaction is unusual for a protein of this small size. It has been shown that as the size of a protein decreases, the probability of a charged group being buried in the hydrophobic core also decreases (8).
Figure 1.
The buried salt bridge in HP67. (a) Ribbon representation of the crystal structure of HP67 (1YU5). The C-terminal subdomain is colored in light green and the N-terminal subdomain is colored in light blue. Side chains of the buried salt bridge residues, glutamate 39 and lysine 70, are highlighted in red and blue, respectively. The buried histidine residue in the N-terminal subdomain is shown in yellow. (b) Space-filling representation using van der Waals radii. (c) Solvent accessible surface showing burial of E39, H41 and K70.
Despite its small size and continuous hydrophobic core, in vitro studies have shown this small protein exhibits multi-state unfolding. First, the less stable N-terminal subdomain unfolds, followed by the thermostable C-terminal subdomain (9). Indeed, the C-terminal subdomain is stable as an isolated 35-residue peptide (HP35) (10). Linking the two subdomains within the continuous hydrophobic core is the highly conserved buried salt bridge between E39 in the N-terminal subdomain and K70 in the C-terminal subdomain.
Previous studies have shown that Histidine 41, which is buried in the hydrophobic core of the N-terminal subdomain, acts as a pH-dependent unfolding switch (9, 11, 12). Protonation of H41 results in unfolding of the N-terminal subdomain while the C-terminal subdomain remains folded. Mutation of H41 to tyrosine (H41Y) eliminates the sequential pH-dependent unfolding of the N-terminal subdomain and stabilizes the C-terminal subdomain (9). Despite the increase in overall stability, the H41Y mutant still exhibits multistate unfolding, with unfolding of the N-terminal subdomain preceding the C-terminal subdomain (9).
Salt bridges in proteins can be energetically favorable or unfavorable (13–18). If the salt bridge is primarily solvent accessible, its contribution to stability may be small. If the salt bridge is largely inaccessible, its contribution can be large (19, 20). In a statistical analysis of salt bridges in proteins, Sarakatsannis and Duan have shown that the most frequent percentage solvent accessible surface area (PSASA) for salt bridges is greater than 20% with a maximum at 32 % (21). Using their criteria, the salt bridge in HP67 is much more buried than the average salt bridge with only 2.3% PSASA. Another trend noted by these authors and others (17) is that, by far, the most common separation for partners involved in salt bridges was four residues (e.g. one turn of α-helix), where as the residue separation in HP67 is 31. Thus, while most salt bridges in proteins are very local in the sequence, whether buried or exposed, the salt bridge in HP67 involves residues widely separated in the primary sequence.
In addition to their role in protein stabilization, salt bridges can provide specificity for a unique fold (8, 9, 13). There are a limited number of orientations for favorable charged residue packing. In terms of achieving energetically equivalent structures, packing of hydrophobic residues is generally less stringent. Structural specificity provided by a salt bridge can result in the proper formation of a discrete protein fold resulting in the proper ligand binding surface (13, 15, 22, 23).
To investigate the role of the highly conserved buried salt bridge in HP67, we created a series of mutants that targeted the E39-K70 interaction. These mutants include replacement of both of residues of the salt bridge with flexible, hydrophobic methionine residues, single residue replacements and a double mutant which swapped the partners of the salt bridge. We used NMR to assay the structure, circular dichroism spectroscopy (CD) to measure the stability, and actin sedimentation assays to determine the effect on the biological function of these mutants. We find that the wild-type salt bridge is absolutely essential for folding of the N-terminal subdomain and no mutations were tolerated. The salt bridge is not essential for the C-terminal subdomain fold. Sedimentation assays, reveal that none of the mutants exhibited significant actin binding activity. Thus, proper folding of the N-terminal subdomain is essential for activity. Furthermore, the headpiece-actin binding energy is insufficient to fold the N-terminal subdomain.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Proteins
The HP67- and H41Y- containing pET24a vectors used for cloning and expression of the salt bridge mutants have been previously described (3, 11). They encodes the last 67 residues of villin headpiece (HP67) corresponding to residues 760–826 of chicken villin. All salt bridge mutants were made with the QuikChange mutagenesis kit (Stratagene). Pairs of primers were designed to mutate: E39M, E39K, K70M, and K70E. All mutations were confirmed by DNA sequencing. All constructs were expressed in Escherichia coli BL21(DE3) cells (Novagen) as described (3). Labeling with 15N isotope was accomplished as described by Marley et al. (24). Protein concentrations determined by absorbance at 280 nm using extinction coefficient of 5690 M−1cm−1 (6990 M−1cm−1 for the H41Y constructs) (25). Supervillin headpiece was a gift from Jeffrey W. Brown (26).
F-actin was purified from chicken pectoral muscle using standard procedures and stored dialyzing against F-buffer (10 mM TRIS, 1 mM MgCl2, 100 mM NaCl, 0.1 mM ATP, 0.2 mM DTT, 3 mM NaN3, 0.1 mM CaCl2, pH 8.0) at 4°C (7, 27).
Nuclear Magnetic Resonance (NMR) spectroscopy
All NMR experiments were conducted on a Bruker DMX 500 MHz spectrometer. All samples were prepared in 10% D2O containing 10 mM sodium phosphate at pH 7, uncorrected for the effects of D2O. Chemical shifts were referenced to trimethylsilyl-propionic acid. One-dimensional NMR spectra were acquired using Watergate pulse sequences to suppress water (28). For 15N-heteronuclear single quantum coherence (HSQC) spectra, protein concentrations were approximately 1 mM and all spectra were taken at 25°C with 128 t1 increments of 2048 data points. The data were processed using NMRPipe (29) and analyzed in NMRView (30).
Circular Dichroism (CD) Spectroscopy and Unfolding Experiments
CD spectra were acquired on an Aviv 62DS spectrometer equipped with a Peltier temperature controller and automated titration apparatus. A 10 mm path length cell was used for all CD experiments. Wavelength spectra are the average of three scans of 20 μM protein in 10 mM phosphate buffer, pH 7, from 200–250 nm with an averaging time of 20 seconds at each wavelength in 1 nm steps at 25 °C. For thermal, GuHCl, and urea denaturation experiments the CD signal was monitored at 222 nm. Thermal denaturation was carried out from 20–98 °C with 2 degree increments, and protein concentrations of 5 μM in 10 mM phosphate buffer, pH 7.0. The first derivative of the smoothed thermal unfolding data was used to determine the Tm value.
GuHCl and urea unfolding experiments were performed with a titrator unit interfaced with the spectrometer at 25 °C and protein concentrations of 10 μM in 10 mM phosphate buffer at pH 7.0. Urea unfolding was done in the same buffer at pH 6.5. Concentrations of urea and GuHCl were determined from the refractive index of the solution (31). Concentrations of urea and GuHCl increased from 0–9.8 M and 0–8.5 M in 0.2 M increments, respectively. To determine thermodynamic parameters ΔGo, Cm, and m-values, chemical denaturation curves were fit as described by Santoro and Bolen (32) with Origin 6.0 (Microcal Software, Inc. Northampton, MA).
Actin Sedimentation Assays
Actin sedimentation assays were performed at 4°C as previously described with 15 μM F-actin and 0–200 μM headpiece domain (7, 12). After incubation for 1 hour in F-buffer (10 mM TRIS, 1 mM MgCl2, 100 mM NaCl, 0.1 mM ATP, 0.2 mM DTT, 3 mM NaN3, 0.1 mM CaCl2, pH 8.0), the binding reactions (50 μL) were spun at 100,000 g for 1 hour and the supernatants were decanted. Pellets were washed with 48 μL F-actin buffer and then resuspended in 48 μL of 7% acetic acid. Samples were stored at −20°C. The bound protein concentrations in the actin pellets were determined by UV quantitation during reverse phase high performance liquid chromatography (HPLC) (33). A C18 column (Vydac) 4.6 × 200 mm was run at 1 ml/min in water-acetonitrile gradients with 0.1 % trifluoroacetic acid and monitored at 220 nm. A standard curve was constructed by running samples of known concentrations of headpiece. Binding data was fit with Origin 6.0 (Microcal Software, Inc. Northampton, MA) as previously described (7). Three parameters were obtained by fitting: (1) equilibrium dissociation constant (KD), (2) maximal binding (Bmax), (3) non-specific (NS) binding. The data points used in fitting were the average of three separate sedimentation experiments.
RESULTS
To investigate the role of the highly conserved buried salt bridge in villin headpiece (Figure 1) we made a series of point mutations at positions E39 and K70. To determine if the buried salt bridge could be replaced by a hydrophobic interaction without introducing much steric hindrance, we created E39M/K70M, or “MetMet,” a double mutant. To determine whether the salt bridge could still be formed in the opposite orientation we created E39K/K70E, termed “SWAP.” In the process of making the double mutants, all possible single mutants were also constructed (i.e. E39K, E39M, K70E, K70M).
NMR Indicates that the Salt Bridge is Essential for the N-terminal Subdomain Fold
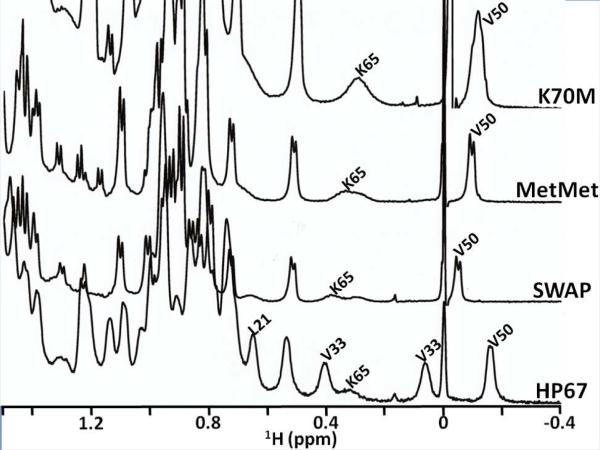
1D proton NMR spectra were obtained to determine the effect of double mutations on the headpiece fold. The upfield region of the 1H 500 MHz NMR spectrum of HP67 contains well-resolved resonances from both the N- and C- terminal subdomains (Figure 2). The methyl groups of L21 and V33 from the N-terminal subdomain and V50 as well as the Hβ of K65 from the C-terminal subdomain act as probes for folded structure within the two individual domains. In all of the salt bridge mutant spectra there is a loss of the L21 and V33 methyl peaks, indicating that the N-terminal subdomain is unfolded. The retention of the V50 and K65 peaks indicates the C-terminal subdomain remains folded. The small changes in chemical shift of the V50 and K65 peaks are due to localized changes in conformation within the C-terminal subdomain and are consistent with the chemical shifts observed in the isolated HP35 domain.
Figure 2.
Upfield region of the 1H 500 MHz 1D NMR spectra of HP67, SWAP, MetMet, and K70M. The methyl resonance of V50 and the Hβ of K65 in the C-terminal subdomain and the methyl groups of V33 and L21 in the N-terminal subdomain are labeled. Spectra were recorded at 25 °C in 10% D2O, 10 mM sodium phosphate buffer at pH 7.0.
NMR Indicates that the C-terminal Subdomain Retains the Native Fold
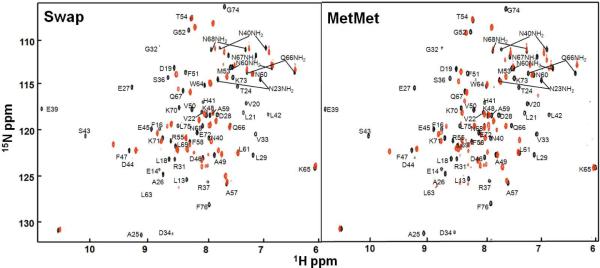
To further define the folded and unfolded regions within the salt bridge mutants we mapped the chemical shift changes of backbone amide protons relative to HP67 in the 1H-15N HSQC spectra. Figure 3 shows the 15N-1H HSQC spectra of SWAP and MetMet (red) overlaid with HP67 (black). The peak labels indicate the assignments for HP67. Unlike the spectrum of HP67, the spectra of the mutants contain more resonances than expected from their sequences, indicating some regions are slowly exchanging between two states. In addition, there are more peaks in the region corresponding to random coil chemical shifts (8–9 ppm) in the mutants than in HP67. What is clear from these spectra is that the well dispersed peaks from residues in the N-terminal of HP67 are absent in the spectra of salt bridge mutants (e.g. L13, E14, L19, V20, L21, A25, E27, L29, G32, D34, R37, L42, S43). In contrast, most of the disperse peaks arising from residues within the C-terminal subdomain have a corresponding peak directly under, or very close to, those from HP67 in the spectra of the mutants (e.g. A49, A57, T54, L61, K65, Q66). The resonances from residues corresponding to the end of the C-terminal helix of HP67 are also shifted (e.g. K70, G74, F76). However, these shifts show a close correspondence to those seen in the isolated HP35 subdomain (10). These observations are consistent with the unfolding of the N-terminal domain while the C-terminal domain adopts the structure the HP35 subdomain.
Figure 3.
Comparison of the 15N-HSQC spectra of Swap and MetMet with HP67. The spectrum of HP67 is shown in black and the assignments are labeled (3). The spectrum of SWAP and MetMet are shown in red on the left and right spectrum, respectively. The spectra were acquired at pH 7.0, 20 °C in 10 mM phosphate buffer. The protein concentrations for both SWAP and MetMet were 1 mM.
Although Unfolded, the N-Terminal Subdomain Influences the Stability of the C-Terminal Subdomain
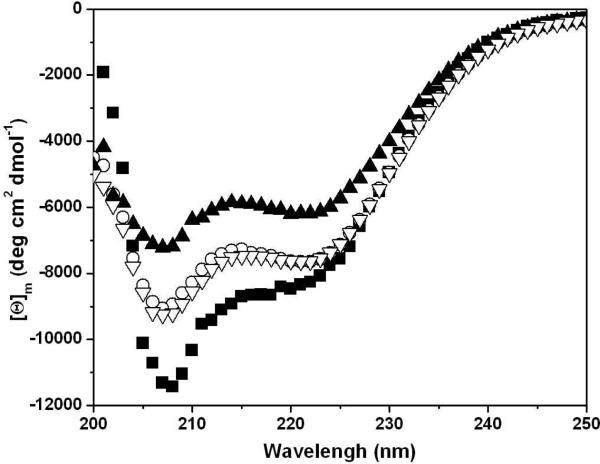
The C-terminal subdomain of HP67 contains most of the helical structure in HP67 and therefore the signal from the C-terminal subdomain dominates the CD spectrum. The far-UV CD spectra for all the salt bridge mutants exhibit significant helical content, similar to that observed for HP67, which is consistent with an independently folded C-terminal subdomain (Figure 4).
Figure 4.
CD spectra of HP67 and salt bridge mutants. Far-UV scans of 10 μM protein at 25 °C in 10 mM sodium phosphate buffer at pH 7.0 in a 10 mm path-length cell. HP67 (■), MetMet (▲), SWAP (◯), and K70M (▽).
The thermal stability of the C-terminal subdomain was determined by monitoring the helical CD signal at 222 nm as a function of temperature. HP67 exhibits a thermal unfolding temperature (Tm) of 79.8 °C. Despite having unfolded N-terminal subdomains, both mutants containing the K70M mutation (MetMet, and K70M) exhibit increases in Tm compared to that of wild-type (93.6 °C and 83.5 °C, respectively). In contrast, SWAP exhibits a Tm of 67.5 °C, a decrease of 12.3 °C in Tm. The hierarchy of Tm values is MetMet > K70M > HP67 > SWAP.
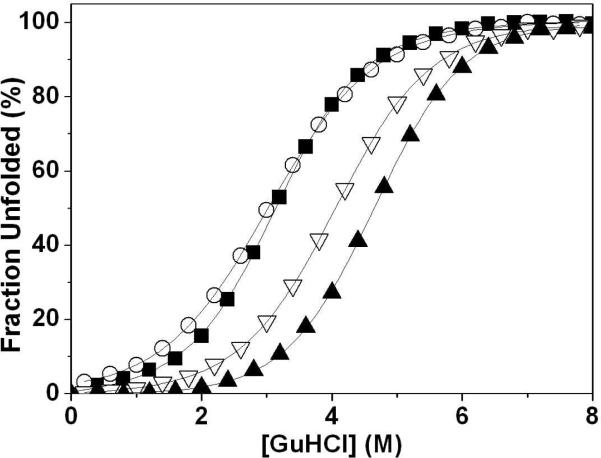
To determine the thermodynamic stability of the folded C-terminal subdomain of each construct, we performed chemical denaturation as a function of increasing guanidine hydrochloride (GuHCl, Figure 5) and urea (Supplemental Figure 1). Helical content was monitored by the CD signal at 222 nm. As expected from their elevated Tm values, MetMet and K70M mutants both show an increase in stability relative to HP67. All constructs, except SWAP, show cooperative unfolding with similar m-values. In the SWAP mutant, unfolding cooperativity was slightly, but significantly, reduced.
Figure 5.
Guanidine hydrochloride denaturation of headpiece constructs. GuHCl-induced denaturation of HP67 (■), MetMet (▲), SWAP (◯), and K70M (▽). Experiments were performed with 10 μM protein samples at 25 °C in 10 mM sodium phosphate buffer, pH 7.0. CD signal was monitored at 222 nm in a 10 mm cell. Data was best fit to the Santoro and Bolen equation (34) and then plotted as percent unfolded.
The GuHCl unfolding curves were fit to obtain ΔGo, Cm, and m-values (Table 1) (31, 34). SWAP was the only mutant which exhibits a decrease in both ΔGo and Cm. The relative trends in ΔGo and Cm values for these mutants and HP67 are the same as for the thermal unfolding experiment: MetMet > K70M > HP67 > SWAP. Thus, as has been observed for the isolated HP35 subdomain, replacement of lysine 70 with methionine increases its stability (35).
Table 1.
Thermodynamic Data for the HP67 and H41Y Constructs (GuHCl)
| ΔG°GuHCl (kcal/mol) | mvalue, GuHCl (kcal/mol M−1) | Cm, GuHCl (mol) | Tm (°C) | |
|---|---|---|---|---|
| HP67 | 2.6 | 0.8 | 3.4 | 79.8 |
| SWAP | 2.2 | 0.7 | 3.0 | 67.5 |
| MetMet | 4.0 | 0.9 | 4.6 | 93.6 |
| K70M | 3.1 | 0.8 | 4.0 | 83.5 |
| H41Y | 3.9 | 1.1 | 3.4 | 83.4 |
| H41Y/SWAP | 2.4 | 0.8 | 3.4 | 68.9 |
| H41Y/MetMet | 2.8 | 0.7 | 3.4 | 92.2 |
| H41Y/K70M | 3.5 | 0.8 | 4.4 | 84.4 |
A concern with using GuHCl unfolding data to calculate the stability of these salt bridge mutants is the potential ionic screening arising from the high concentrations of GuHCl. To address this concern we performed the same experiments in urea (Supplemental Figure 1) and the results from the urea experiments are in qualitative agreement with those in GuHCl, and the stability of the constructs following the same hierarchy: MetMet > K70M > HP67 > SWAP. However, a complete thermodynamic analysis in urea is complicated due to the lack of unfolded baselines for most of these constructs.
The Conserved Buried Salt Bridge is Essential for Specific Actin Binding
To test whether these salt bridge mutations affected the actin-binding activity of villin headpiece, we measured the apparent actin-headpiece affinity of Swap, MetMet, and K70M using a sedimentation assay. Non-actin binding Supervillin headpiece (SVHP) was used as a negative control (12, 36). The binding constant (KD), maximum of amount of protein specifically bound (Bmax), and non-specific binding (NS) were determined by fitting the binding curves as described previously (12) (Figure 6).
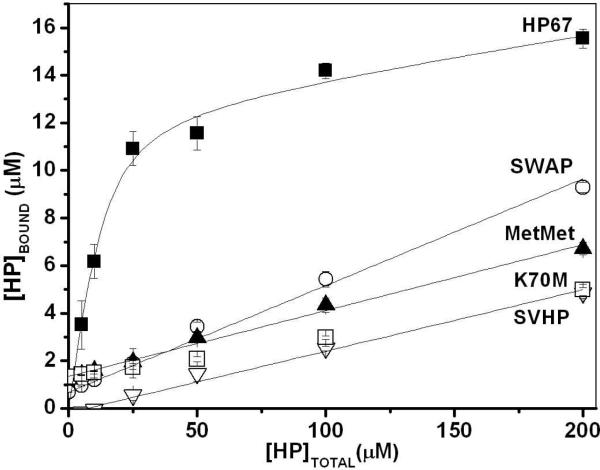
Figure 6.
F-actin Sedimentation Assay. The concentration of the headpiece constructs cosedimenting with F-actin (HPbound) is plotted against total headpiece construct concentration (HPtotal) in each reaction. The data points are the average of the three separate experiments. Error bars indicate the standard deviations of the three experiments. The curves and lines are best fit to the average data points. For HP67 (■), a saturating binding equation, which includes nonspecific binding, was used. For MetMet (▲), SWAP (◯), K70M (▽), and SVHP (□) the data is best fit to a linear equation corresponding to non-specific binding of 3%, 4%, 2%, and 2%, respectively.
Under the conditions of this assay HP67 has a KD of 5 μM, consistent with previous results (7, 12). The Bmax value (15 μM) is the same as the actin monomer concentration used in the reaction and is consistent with a 1:1, protein: actin stoichiometry. Unlike the wild-type sequence, mutant headpiece constructs bound only weakly to actin. Mutants accumulated in the pellet fractions in an essentially linear fashion as a function of concentration, without displaying signs of saturation. MetMet, SWAP and K70M binding data gave poor fits for specific binding. When fit to a straight line, non-specific binding was determined to be 3%, 4%, and 2%, for MetMet, SWAP and K70M, respectively. Note that the wild-type HP67 domain has similar non-specific value of 1%, yet exhibited saturation of binding to F-actin (7). SVHP had a 2% non-specific binding in this experiment. Therefore, the salt bridge mutants exhibited similar non-specific binding values compared to a protein previously known to not bind F-actin. Thus, the correctly folded N-terminal subdomain is essential for specific F-actin binding. Furthermore, the lack of specific binding by any salt bridge mutant indicates that the binding energy of the headpiece-actin interface is insufficient to induce proper folding of the N-terminal subdomain
Salt Bridge Mutants Remain Unfolded in the More Stable HP67/H41Y Background
In an attempt to induce folding of the N-terminal domain of the salt bridge mutants, the mutations were also made in the background of a stabilized mutant of HP67, H41Y (HP67/H41Y). H41 is highly conserved in headpiece domains, no other substitutions have been found at this site except tyrosine. Based on hydrogen exchange experiments, H41Y has increased stability of ~0.7 kcal/mol (9, 11, 37). The thermal unfolding (Tm) values for the salt bridge mutants in the H41Y background follow the same hierarchy as found for the wild-type HP67 background (Table 1). The ΔG°, m-value and Cm determined by GuHCl unfolding follow a slightly different pattern (Supplemental Figure 2). H41Y exhibits the highest ΔG° value followed by K70M/H41Y, MetMet/H41Y, and then SWAP/H41Y. Thus, the increased stability of H41Y background cannot compensate for the destabilizing effect of the salt bridge mutations to allow proper folding of the N-terminal subdomain.
DISCUSSION
Buried salt bridges are unusual in small proteins and the observation of a highly conserved and buried salt bridge in HP67 is unexpected (17, 21). To test the constraints of the salt bridge, we replaced the two residues with hydrophobic interactions or swapped their polarity.
1D and 2D NMR data show that any mutation of this salt bridge results in unfolding of most of the N-terminal subdomain. Residues in the region linking the N- and C-terminal subdomains can still affect the thermostability of the C-terminal subdomain, as seen in the MetMet double mutant, which is more stable than the K70M single mutant. Thus, it appears that when the buried E39 residue is replaced with methionine in the MetMet double mutant, the hydrophobic side chain can form stabilizing contacts with the C-terminal subdomain.
In this study, we found that any mutations to the residues participating in the buried salt bridge are not tolerated and result in partial unfolding of most of the N-terminal subdomain. Further, unfolding of the N-terminal subdomain disrupts the actin binding face. Thus, the buried salt bridge serves a vital role by specifying the actin-binding conformation of the N-terminal residues.
There appears to be a competition between the requirement for a highly specific salt bridge needed to ensure proper folding of the N-terminal subdomain on the one hand, and the destabilization of the C-terminal subdomain by K70 on the other. The equilibrium unfolding of HP67 is known to be multistate. The first state is the canonical HP67 fold with both subdomains folded. In the second state, the N-terminal subdomain is unfolded while the C-terminal subdomain adopts the structure of isolated HP35, which contains slight alterations in the structure and dynamics, especially of the C-terminal residues Another state, in which both domains are unfolded, is populated under more strongly denaturing conditions. Any mutation to the salt bridge favors the competition towards the second state. Thus, the presence of the native E39-K70 salt bridge stabilizes the N-terminal subdomain, but destabilizes the C-terminal subdomain. This has some interesting consequences for the study of small domains derived from larger proteins. A commonly employed strategy to stabilize a domain is to use a sequence alignment to derive consensus residues at core positions and to then replace residues in the sequence of interest that deviate from the consensus. Such an approach would erroneously fail to predict that mutation of K70 stabilizes the isolated C-terminal subdomain, HP35. In the present case, K70 is conserved because its stabilizing intersubdomain interactions are more important for function than its destabilizing intrasubdomain interactions.
Supplementary Material
ACKNOWLEDGMENT
We thank Jeffrey W. Brown for the gift of Supervillin headpiece and help with the HPLC-based actin-binding assay. Thanks to Robert Sauer and Andreas Martin for generous use of CD time.
This work was supported by funding from NIH grant GM62886 to CJM, and NSF grant MCB0919860 to DPR
ABBREVIATIONS
- ATP
adenosine triphosphate
- Cm
the midpoint of the denaturant induced unfolding transition
- CD
circular dichroism
- E39K
glutamate 39 to lysine mutation in HP67
- E39M
glutamate 39 to methionine mutation of HP67
- ΔG°
apparent free energy of unfolding
- D2O
deuterium dioxide
- DTT
dithiothreitol
- F-actin
filamentous actin
- GuHCl
guanidine hydrochloride
- H41Y
mutant of HP67 replacing residue histidine 41 with tyrosine
- HP35
residues 42–76 of the isolated C-terminal domain of HP67
- HP67
67 amino acid polypeptide encoding residues 10–76 of villin headpiece (residues 760–826 of intact chicken villin)
- HPLC
high-pressure liquid chromatography
- HSQC
heteronuclear single quantum coherence
- K70E
lysine 70 to glutamate mutation in HP67
- K70M
lysine 70 to methionine mutation in HP67
- MetMet
HP67 which contains two mutations, E39M and K70M
- NS
non-specific binding
- NMR
nuclear magnetic resonance
- PSASA
percent solvent accessible surface area
- SVHP
Supervillin headpiece
- SWAP
HP67 which contains two mutations, E39K and K70E
- Tm
the midpoint of thermal unfolding
- TRIS
tris(hydroxymethylamino)methane
- UV
ultraviolet
- WT
wild-type.
Footnotes
SUPPORTING INFORMATION AVAILABLE Denaturant unfolding of SWAP, MetMet, and K70M mutations in the more stable H41Y background shown in Supplemental Figures S1 (urea) and S2 (GuHCl). This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- (1).Hesterberg LK, Weber K. Isolation of a domain of villin retaining calcium-dependent interaction with G-actin, but devoid of F-actin fragmenting activity. Eur J Biochem. 1986;154:135–140. doi: 10.1111/j.1432-1033.1986.tb09368.x. [DOI] [PubMed] [Google Scholar]
- (2).Hampton CM, Liu J, Taylor DW, DeRosier DJ, Taylor KA. The 3D structure of villin as an unusual F-Actin crosslinker. Structure. 2008;16:1882–1891. doi: 10.1016/j.str.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Vardar D, Buckley D, Frank B, McKnight C. NMR structure of an F-actin binding “headpiece” motif from villin. Journal of Molecular Biology. 1999;249:1299–1310. doi: 10.1006/jmbi.1999.3321. [DOI] [PubMed] [Google Scholar]
- (4).Friederich E, Vancompernolle K, Huet C, Goethals M, Finidori J, Vandekerckhove J, Louvard D. An actin-binding site containing a conserved motif of charged amino acid residues is essential for the morphogenic effect of villin. Cell. 1992;70:81–92. doi: 10.1016/0092-8674(92)90535-k. [DOI] [PubMed] [Google Scholar]
- (5).Doering DS, Matsudaira PT. Cysteine scanning mutagenesis at 40 of 76 positions in villin headpiece maps the F-actin binding site and structural features of the domain. Biochemistry. 1996;35:12677–12685. doi: 10.1021/bi9615699. [DOI] [PubMed] [Google Scholar]
- (6).Vermeulen W, Vanhaesebrouck P, Van Troys M, Verschueren M, Fant F, Goethals M, Ampe C, Martins JC, Borremans FAM. Solution structures of the C-terminal headpiece subdomains of human villin and advillin, evaluation of headpiece F-actin-binding requirements. Protein Science. 2004;13:1276–1287. doi: 10.1110/ps.03518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Meng J, Vardar D, Wang Y, Guo HC, Head JF, McKnight CJ. High-resolution crystal structures of villin headpiece and mutants with reduced F-actin binding activity. Biochemistry. 2005;44:11963–11973. doi: 10.1021/bi050850x. [DOI] [PubMed] [Google Scholar]
- (8).Bolon DN, Mayo SL. Polar residues in the protein core of Escherichia coli thioredoxin are important for fold specificity. Biochemistry. 2001;40:10047–10053. doi: 10.1021/bi010427y. [DOI] [PubMed] [Google Scholar]
- (9).Tang Y, Grey MJ, McKnight J, Palmer AG, 3rd, Raleigh DP. Multistate folding of the villin headpiece domain. J Mol Biol. 2006;355:1066–1077. doi: 10.1016/j.jmb.2005.10.066. [DOI] [PubMed] [Google Scholar]
- (10).McKnight CJ, Doering DS, Matsudaira PT, Kim PS. A thermostable 35-residue subdomain within villin headpiece. Journal of Molecular Biology. 1996;260:126–134. doi: 10.1006/jmbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- (11).Meng J, McKnight CJ. Crystal structure of a pH-stabilized mutant of villin headpiece. Biochemistry. 2008;47:4644–4650. doi: 10.1021/bi7022738. [DOI] [PubMed] [Google Scholar]
- (12).Vardar D, Chishti AH, Frank BS, Luna EJ, Noegel AA, Oh SW, Schleicher M, McKnight CJ. Villin-type headpiece domains show a wide range of F-actin-binding affinities. Cell Motil Cytoskeleton. 2002;52:9–21. doi: 10.1002/cm.10027. [DOI] [PubMed] [Google Scholar]
- (13).Hendsch ZS, Tidor B. Do Salt Bridges Stabilize Proteins - a Continuum Electrostatic Analysis. Protein Science. 1994;3:211–226. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lumb KJ, Kim PS. Measurement of Interhelical Electrostatic Interactions in the Gcn4 Leucine-Zipper. Science. 1995;268:436–439. doi: 10.1126/science.7716550. [DOI] [PubMed] [Google Scholar]
- (15).Waldburger CD, Schildbach JF, Sauer RT. Are Buried Salt Bridges Important for Protein Stability and Conformational Specificity. Nature Structural Biology. 1995;2:122–128. doi: 10.1038/nsb0295-122. [DOI] [PubMed] [Google Scholar]
- (16).Wimley WC, Gawrisch K, Creamer TP, White SH. Direct measurement of salt-bridge solvation energies using a peptide model system: Implications for protein stability. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:2985–2990. doi: 10.1073/pnas.93.7.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kumar S, Nussinov R. Salt bridge stability in monomeric proteins. Journal of Molecular Biology. 1999;293:1241–1255. doi: 10.1006/jmbi.1999.3218. [DOI] [PubMed] [Google Scholar]
- (18).Luisi DL, Snow CD, Lin JJ, Hendsch ZS, Tidor B, Raleigh DP. Surface salt bridges, double-mutant cycles, and protein stability: an experimental and computational analysis of the interaction of the Asp 23 side chain with the N-terminus of the N-terminal domain of the ribosomal protein L9. Biochemistry. 2003;42:7050–7060. doi: 10.1021/bi027202n. [DOI] [PubMed] [Google Scholar]
- (19).Takano K, Tsuchimori K, Yamagata Y, Yutani K. Contribution of salt bridges near the surface of a protein to the conformational stability. Biochemistry. 2000;39:12375–12381. doi: 10.1021/bi000849s. [DOI] [PubMed] [Google Scholar]
- (20).Schneider JP, Lear JD, DeGrado WF. A Designed Buried Salt Bridge in a Heterodimeric Coiled Coil. Journal of the American Chemical Society. 1997;119:5742–5743. [Google Scholar]
- (21).Sarakatsannis JN, Duan Y. Statistical characterization of salt bridges in proteins. Proteins-Structure Function and Bioinformatics. 2005;60:732–739. doi: 10.1002/prot.20549. [DOI] [PubMed] [Google Scholar]
- (22).Kretsinger JK, Schneider JP. Design and Application of Basic Amino Acids Displaying Enhanced Hydrophobicity. Journal of the American Chemical Society. 2003;125:7907–7913. doi: 10.1021/ja029892o. [DOI] [PubMed] [Google Scholar]
- (23).Schueler O, Margalit H. Conservation of salt bridges in protein families. J Mol Biol. 1995;248:125–135. doi: 10.1006/jmbi.1995.0206. [DOI] [PubMed] [Google Scholar]
- (24).Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- (25).Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Brown JW, Vardar-Ulu D, McKnight CJ. How to arm a supervillin: designing F-actin binding activity into supervillin headpiece. J Mol Biol. 2009;393:608–618. doi: 10.1016/j.jmb.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pardee JD, Spudich JA. Purification of muscle actin. Methods in Enzymology. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- (28).Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- (29).Delaglio F, Grzesiek G, Vuister G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6:227–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- (30).Johnson B, Blevins R. NMRView: A computer program for the visualization and analysis of NMR data. Journal of Biomolecular NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- (31).Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods in Enzymology. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- (32).Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- (33).Brown JW, McKnight CJ. Identifying competitive protein antagonists for F-actin with reverse-phase high-performance liquid chromatography. Anal Biochem. 398:117–119. doi: 10.1016/j.ab.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bolen DW, Santoro MM. Unfolding free energy changes determined by the linear extrapolation method. 2. Incorporation of delta G degrees N-U values in a thermodynamic cycle. Biochemistry. 1988;27:8069–8074. doi: 10.1021/bi00421a015. [DOI] [PubMed] [Google Scholar]
- (35).Bi Y, Cho JH, Kim EY, Shan B, Schindelin H, Raleigh DP. Rational design, structural and thermodynamic characterization of a hyperstable variant of the villin headpiece helical subdomain. Biochemistry. 2007;46:7497–7505. doi: 10.1021/bi6026314. [DOI] [PubMed] [Google Scholar]
- (36).Wulfkuhle JD, Donina IE, Stark NH, Pope RK, Pestonjamasp KN, Niswonger ML, Luna EJ. Domain analysis of supervillin, an F-actin bundling plasma membrane protein with functional nuclear localization signals. J Cell Sci. 1999;112(Pt 13):2125–2136. doi: 10.1242/jcs.112.13.2125. [DOI] [PubMed] [Google Scholar]
- (37).Grey MJ, Tang Y, Alexov E, McKnight CJ, Raleigh DP, Palmer AG., 3rd Characterizing a partially folded intermediate of the villin headpiece domain under non-denaturing conditions: contribution of His41 to the pH-dependent stability of the N-terminal subdomain. J Mol Biol. 2006;355:1078–1094. doi: 10.1016/j.jmb.2005.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.