Binding of histone H2A.Z to the SUN family member Mps3 is chromatin independent.
Abstract
Understanding the relationship between chromatin and proteins at the nuclear periphery, such as the conserved SUN family of inner nuclear membrane (INM) proteins, is necessary to elucidate how three-dimensional nuclear architecture is established and maintained. We found that the budding yeast SUN protein Mps3 directly binds to the histone variant H2A.Z but not other histones. Biochemical and genetic data indicate that the interaction between Mps3 and H2A.Z requires the Mps3 N-terminal acidic domain and unique sequences in the H2A.Z N terminus and histone-fold domain. Analysis of binding-defective mutants showed that the Mps3–H2A.Z interaction is not essential for any previously described role for either protein in nuclear organization, and multiple lines of evidence suggest that Mps3–H2A.Z binding occurs independently of H2A.Z incorporation into chromatin. We demonstrate that H2A.Z is required to target a soluble Mps3 fragment to the nucleus and to localize full-length Mps3 in the INM, indicating that H2A.Z has a novel chromatin-independent function in INM targeting of SUN proteins.
Introduction
The influence of nuclear architecture on gene expression and genomic stability has been explored in a variety of systems and experimental contexts, and it is clear that the nonrandom positioning of chromosomes plays a pivotal role in the regulation of multiple nuclear functions, including transcription, replication, DNA damage repair, and mRNA processing (Mekhail and Moazed, 2010; Taddei et al., 2010). What is less well understood is how the spatial arrangement of chromosomes is established and maintained. Recruitment of particular regions of the genome to the nuclear periphery requires interactions between chromatin and nuclear envelope (NE) factors, such as nuclear pore complexes (NPCs) and inner nuclear membrane (INM) proteins (Strambio-De-Castillia et al., 2010; Taddei et al., 2010). The SUN (Sad1-UNC-84) domain family of INM proteins plays an essential role in tethering of specific chromatin domains to the nuclear periphery during both meiotic and mitotic growth (Hiraoka and Dernburg, 2009; Starr, 2009).
SUN proteins contain an N-terminal head domain that extends into the nucleus, allowing for interactions with chromatin, whereas the C-terminal tail, including the SUN domain, is located in the lumenal space between the INM and the outer nuclear membrane (ONM; Starr, 2009). In budding yeast, the sole SUN protein, Mps3, is a structural component of the spindle pole body (SPB), the Saccharomyces cerevisiae centrosome-equivalent organelle (Jaspersen et al., 2002, 2006; Nishikawa et al., 2003). Mps3 is also present throughout the INM where it functions in the localization of telomeres and nonrepairable DNA double-stranded breaks (DSBs) to the NE (Antoniacci and Skibbens, 2006; Bupp et al., 2007; Oza et al., 2009; Schober et al., 2009). The mps3Δ75–150 allele, which lacks the N-terminal acidic domain and fails to localize to the nuclear periphery, displays defects in nuclear organization (Bupp et al., 2007; Oza et al., 2009; Schober et al., 2009), suggesting that proper targeting of SUN proteins to the INM is a critical aspect of nuclear architecture.
Like other INM proteins, SUN proteins are synthesized on the rough ER membrane, which is continuous with the ONM. The classical model of INM localization involves lateral diffusion along the ONM and movement through the pore membrane (Smith and Blobel, 1993; Soullam and Worman, 1995; Holmer and Worman, 2001; Ohba et al., 2004). Because of the small size (<30 kD) of their extralumenal domains, SUN proteins were initially proposed to reach the INM using this passive diffusion pathway (Lusk et al., 2007). However, this mechanism of localization raises a key question: how are SUN proteins retained in the INM? One possibility is that the extralumenal domain contains a nuclear localization sequence (NLS) or other targeting domain, which would result in continuous greater net flux toward the INM, which is similar to the karyopherin-mediated transport of Heh1/2 to the INM in budding yeast (King et al., 2006) or the INM sorting motifs described in baculovirus (Braunagel et al., 2004). Consistent with this possibility, Turgay et al. (2010) identified an NLS and a Golgi retrieval signal in hSUN2 that are involved in its NE localization. A second possibility, which is not mutually exclusive, is that SUN proteins bind to nuclear factors to prevent diffusion back out of the nucleus. Candidate anchors include lamins, although these proteins are not required for SUN protein localization in many cell types and are not present in plants and fungi (Padmakumar et al., 2005; Crisp et al., 2006; Haque et al., 2006, 2010; Hasan et al., 2006; Turgay et al., 2010) or chromatin and chromatin-binding proteins. Lastly, SUN proteins may use ONM proteins for anchorage in the INM through the formation of a linker complex that spans the lumenal space between INM and ONM. Binding to ONM KASH (Klarsicht, ANC-1 and Syne homology) proteins has recently been shown to be important for Sun1/2 localization in mammals (Haque et al., 2010), but it is unclear whether this mechanism of INM localization is applicable to budding yeast, which appear to lack KASH proteins (Starr, 2009).
Here, we examine the role of Mps3 binding to the conserved H2A variant H2A.Z in chromosome positioning, gene regulation, genomic stability, and INM localization using genetic, biochemical, and cell biological methods. We discovered that overexpression of HTZ1, which encodes the H2A.Z variant in budding yeast, but not other combinations of yeast histones, was able to rescue the growth defect of certain mps3 mutants, suggesting that the proteins may interact in vivo. H2A.Z is present nonrandomly throughout eukaryotic chromatin, is deposited into chromatin throughout the cell cycle by the multisubunit SWR1 ATPase complex, and functions in transcriptional control and chromosome stability (Zlatanova and Thakar, 2008). H2A.Z is involved in NE recruitment of activated promoters such as GAL1–10 and nonrepairable DSBs (Brickner et al., 2007; Kalocsay et al., 2009; Light et al., 2010) and plays a role in telomeric and mating-type silencing (Dhillon and Kamakaka, 2000; Meneghini et al., 2003; Zhang et al., 2005). However, we show that binding to Mps3 does not appear to be critical for any of these processes. Rather, our results suggest that Mps3–H2A.Z binding is direct and specific, requires the Mps3 N terminus and unique sequences within both the H2A.Z N-terminal tail domain and histone-fold region, and functions to target Mps3 to the INM during an unperturbed cell cycle.
Results
Overexpression of HTZ1 suppresses mps3-F592S mutants
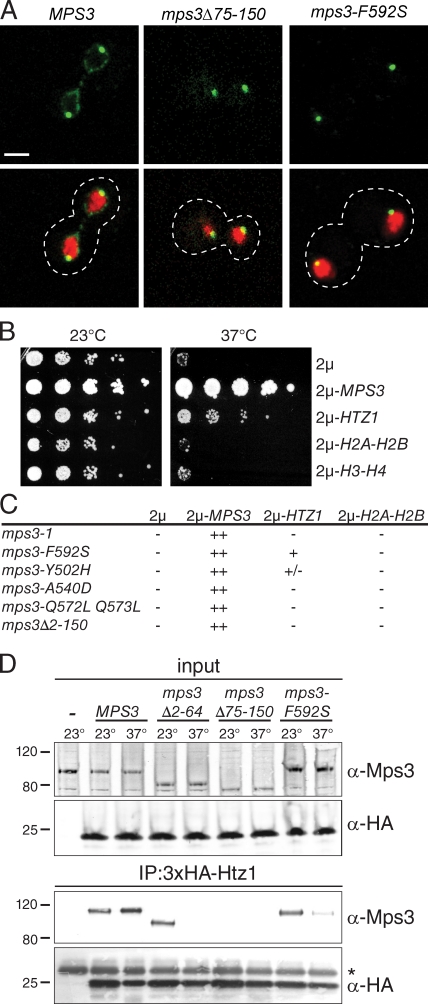
Mps3 may have an additional function in mitotic progression that is separate from its role in SPB duplication as indicated by the phenotype of a temperature-sensitive mutant in the Mps3 SUN domain, mps3-F592S (Jaspersen et al., 2006). Localization of mps3-F592S–GFP revealed that the mutant protein is present at the SPB but fails to localize to the INM even at the permissive temperature of 23°C (Fig. 1 A). This pattern of localization is similar to mps3Δ75–150-GFP (Fig. 1 A), a mutant that lacks residues in the N-terminal acidic domain and has defects in nuclear organization (Bupp et al., 2007; Oza et al., 2009; Schober et al., 2009). mps3-F592S mutants also exhibit a defect in telomere tethering and partial defect in silencing (Fig. S1). Therefore, we hypothesized that the mps3-F592S mutant protein may be unable to bind to proteins involved in the three-dimensional organization of the nucleus.
Figure 1.
HTZ1 is a dosage suppressor of mps3-F592S. (A) Representative single-plane confocal images showing the localization of Mps3-GFP, mps3-F592S-GFP, or mps3Δ75–150-GFP (green) together with H2B-mCherry (red). The cell is outlined in white based on the differential interference contrast (DIC) image. Bar, 2 µm. (B) mps3-F592S mutants (SLJ1751) were transformed with a 2µ plasmid containing no insert, MPS3, HTZ1, HTA1-HTB1 (H2A-H2B), or HHT1-HHF1 (H3-H4) and tested for their ability to grow at 23 and 37°C in a serial dilution assay. (C) Similarly, growth of each mps3 mutant containing the indicated gene on a 2µ plasmid was analyzed at 37°C for 2 d in a serial dilution assay. Suppression was scored as follows: ++, growth at the fourth dilution and beyond; +, growth at the second or third dilution; +/−, growth only at the first dilution; −, no growth above the background. (D) Liquid nitrogen ground lysates were prepared from wild-type (SLJ001), 3×HA-HTZ1 (SLJ2082), 3×HA-HTZ1 mps3Δ2–64 (SLJ2326), 3×HA-HTZ1 mps3Δ75–150 (SLJ2322), and 3×HA-HTZ1 mps3-F592S (SLJ2324) strains grown at 23°C or shifted to 37°C for 3 h. The protein composition of lysates and anti-HA immunoprecipitates (IP) was analyzed by Western blotting with anti-Mps3 and anti-HA antibodies. Positions of molecular mass markers (kilodaltons) are indicated next to each blot, and the asterisk marks the position of the cross-reacting band present in the control sample. The minus sign represents the negative control strain used.
We isolated dosage suppressors of mps3-F592S to identify putative proteins that interact with Mps3 at the INM. One of these suppressors included the ORF encoding the histone variant H2A.Z (the HTZ1 gene; hereafter referred to as Htz1), which had previously been identified as an Mps3-interacting protein in the global yeast two-hybrid analysis and is a component of yeast chromatin (Dhillon and Kamakaka, 2000; Uetz et al., 2000; Wu et al., 2005). Overproduction of HTZ1 using a 2µ plasmid partially rescued the growth defect of mps3-F592S mutants at 37°C (Fig. 1 B) but not other mps3 alleles, including the SUN domain mutants mps3-Q572LQ573L and mps3-A540D and the N-terminal mutant mps3Δ2–150 (Fig. 1 C; Jaspersen et al., 2006; Bupp et al., 2007). Interestingly, suppression was specific to the variant histone Htz1, as we were unable to restore growth by either independent or coordinate overexpression of genes encoding the canonical histones H2A, H2B, H3, or H4 (Fig. 1 B).
Htz1 binds to the Mps3 acidic domain
To determine whether Mps3 and Htz1 form a complex that might be required for nuclear organization at the INM, we created a strain containing 3×HA-HTZ1 expressed from the endogenous promoter as the sole copy of HTZ1 and prepared lysates by cryolysis, a method of protein extraction and solubilization that has been particularly effective at preserving delicate interactions with membrane-associated complexes (Jaspersen et al., 2006; Bupp et al., 2007). We analyzed anti-HA pull-downs by immunoblotting using polyclonal anti-Mps3 antibodies and found that Mps3 and Htz1 coimmunoprecipitate (Fig. 1 D). Consistent with our genetic prediction, we found that the mutant mps3-F592S protein shows reduced binding to Htz1, especially at the nonpermissive temperature (Fig. 1 D).
Htz1 is predominately a nuclear protein because of its incorporation into chromatin (Dhillon and Kamakaka, 2000; Wu et al., 2005), so we expected that Htz1 would bind to the N-terminal nucleoplasmic region of Mps3. We tested binding between Htz1 and two deletion mutants in the Mps3 N terminus that do not affect viability or SPB duplication (Bupp et al., 2007; Conrad et al., 2007): mps3Δ2–64 lacks residues 2–64, whereas mps3Δ75–150 contains a deletion in the N-terminal acidic domain located between amino acids 75 and 150. At 23°C, the first half of the N terminus is not required for Htz1 binding, whereas the acidic region plays an essential role in the interaction (Fig. 1 D). However, at 37°C, the entire N terminus of Mps3 is essential for Htz1 binding. These results indicate that Htz1 primarily binds to the acidic region of Mps3, although other sites may be important for stabilizing the interaction at higher temperatures.
The role of chromatin in Mps3–Htz1 binding
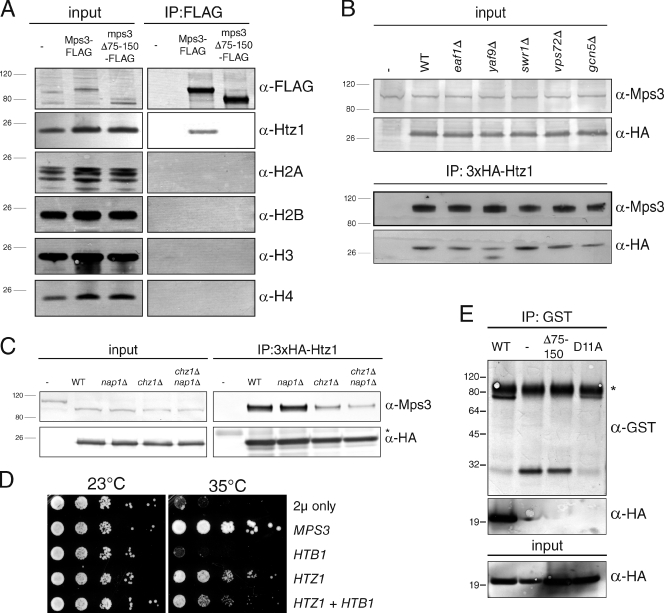
Chromatin is thought to play an important role in tethering INM proteins and in transcriptional regulation at the periphery (Akhtar and Gasser, 2007; Lusk et al., 2007), so we tested whether Mps3 bound to Htz1-containing nucleosomes. Mps3-3×FLAG as well as mps3Δ75–150-3×FLAG was immunoprecipitated from total nuclear extracts, and blots were probed using antibodies against the canonical histones H2A, H2B, H3, and H4 (Fig. 2 A). We were unable to detect H2A, H2B, H3, or H4 in our Mps3 immunoprecipitates, although Htz1 was readily observed. This suggests that Mps3 primarily interacts with Htz1 or that Htz1 is the only binding reaction robust enough to withstand a coimmunoprecipitation. Importantly, both FLAG-H2A and FLAG-H2B were able to immunoprecipitate Htz1 as well as other histone subunits under identical conditions (Fig. S2 A). Therefore, it is highly unlikely that lack of Mps3 binding to core histones is caused by disruption of histone–histone binding during our cryolysis and immunoprecipitation procedure.
Figure 2.
Mps3 binds directly to soluble Htz1. (A) Liquid nitrogen ground lysates were prepared from wild-type (WT; SLJ001), Mps3-3×FLAG (SLJ2002), and mps3Δ75–150-3×FLAG (SLJ2840) strains. Proteins present in each lysate and bound to anti-FLAG beads were analyzed by immunoblotting with the indicated antibodies. (B and C) We also analyzed coimmunoprecipitation of Mps3 and Htz1 in wild-type (SLJ2082), swr1Δ (SLJ3310), vps72Δ (SLJ4250), yaf9Δ (SLJ3742), eaf1Δ (SLJ3313), gcn5Δ (SLJ3307), nap1Δ (SLJ3225), chz1Δ (SLJ3227), and nap1Δ chz1Δ (SLJ5244) strains using anti-HA beads, and binding to Mps3 was assayed by Western blotting. (D) mps3-F592S mutants (SLJ1751) were transformed with 2µ-URA3, 2µ-URA3-MPS3 or 2µ-URA3-HTZ1 as well as CEN-HIS3 or CEN-HIS3-GAL-HTB1 and tested for their ability to grow on synthetic complete-URA-HIS plus 2% galactose/raffinose at 23 and 35°C in a serial dilution assay. (E) 100 ng of purified GST (−) or the indicated GST fusion protein to the Mps3 N terminus was immobilized on anti-GST Sepharose beads, and then 200 ng of purified HA-Htz1 was added to each. After a 1-h incubation at 4°C, bound proteins were analyzed by Western blotting with anti-GST and anti-HA antibodies. Positions of molecular mass markers (kilodaltons) are indicated next to each blot, and the asterisk marks the position of the cross-reacting band present in the control sample. Minus signs represent the negative control strains used. IP, immunoprecipitation.
To further test whether Mps3 binds to soluble Htz1 or a chromatin-associated form of the protein, we took advantage of the fact that deletion of various components of the SWR1 or NuA4 chromatin-remodeling complexes results in cells containing different levels of chromatin-associated Htz1 (Kobor et al., 2004; Krogan et al., 2004; Mizuguchi et al., 2004; Zhang et al., 2004; Wu et al., 2005). We immunoprecipitated Htz1 from swr1Δ, vps72Δ, yaf9Δ, eaf1Δ, and gcn5Δ mutants, and in all cases, we observed robust binding between Mps3 and Htz1 (Fig. 2 B). We also examined Mps3 binding to the htz1-M6 mutant protein, which contains a replacement of certain residues in the C terminus with those from H2A that renders it unable to incorporate into chromatin (Wu et al., 2005). However, Mps3 was able to bind to the htz1-M6 mutant protein as well as wild-type Htz1 (see Fig. 4 B). We also found that Mps3 binding to Htz1 was not altered by treatment of lysates with DNaseI (Fig. S2 B). CHZ1 and NAP1 encode two partially redundant histone chaperones that stimulate nucleosome replacement by the SWR1 complex and are involved in the nuclear localization of Htz1 (Mizuguchi et al., 2004; Luk et al., 2007; Zhou et al., 2008; Straube et al., 2010). We showed that these were not essential for the Mps3–Htz1 interaction, although reduced binding between Mps3 and Htz1 in cells lacking Chz1 may suggest a role for the chaperone in stabilizing the Mps3–Htz1 complex (Fig. 2 C). Co-overexpression of HTB1, which encodes one copy of H2B, along with HTZ1 did not enhance the ability to rescue mps3-F592S growth at 35°C (Fig. 2 D). Thus, both biochemical and genetic data strongly suggest that incorporation of Htz1 into chromatin is not required for Mps3 binding.
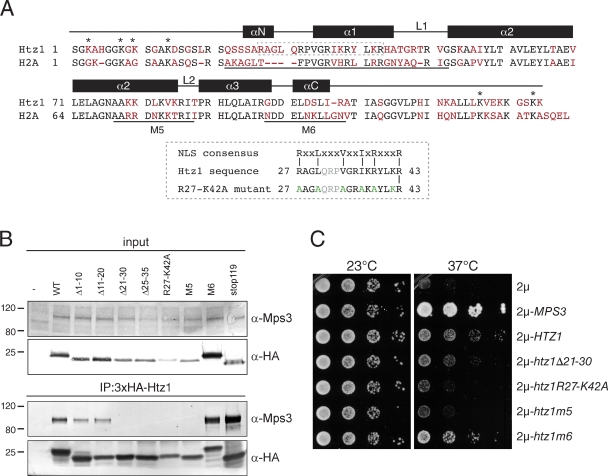
Figure 4.
Mps3 binds to two distinct and unique regions in Htz1. (A) Alignment of budding yeast Htz1 and H2A showing identical residues in black and unique residues in red. Regions of H2A involved in α helix and loop formation within the histone octamer are shown schematically above the alignment (Chakravarthy et al., 2004). Lysines in Htz1 that are important for acetylation or sumoylation are marked with an asterisk. The predicted H2A NLS, M5, and M6 regions are underlined. The NLS present in Htz1 is boxed in gray, and residues mutated in R27-K42 are shown in green. Light gray letters are in the repeat region of variable length in the NLS sequence. (B) Liquid nitrogen ground lysates were prepared from wild-type (SLJ001), 3×HA-HTZ1 (SLJ2082), 3×HA-htz1Δ1–10 (SLJ2805), 3×HA-htz1Δ11–20 (SLJ2806), 3×HA-htz1Δ21–30 (SLJ2807), 3×HA-htz1Δ25–35 (SLJ3210), 3×HA-htz1-R27-K42A (SLJ3211), 3×HA-htz1-m5 (SLJ3757), 3×HA-htz1-m6 (SLJ3758), and 3×HA-htz1stop119 (SLJ3762) strains, and the protein composition of anti-HA immunoprecipitates (IP) was analyzed by Western blotting with anti-HA and anti-Mps3 antibodies. (C) mps3-F592S mutants (SLJ1751) were transformed with a 2µ plasmid containing no insert, MPS3, HTZ1, or htz1 alleles and tested for their ability to grow at 23 and 37°C in a serial dilution assay. Positions of molecular mass markers (kilodaltons) are indicated next to each blot. The minus sign represents the negative control strain used. WT, wild type.
To show that Mps3 and Htz1 directly interact in vitro in the absence of chromatin or other factors, we purified amino acids 1–150 of the Mps3 N terminus (wild type), a version of Mps3 lacking the acidic domain (Δ75–150; amino acids 1–75), and a version of the Mps3 N terminus that had 11 conserved acidic amino acids mutated to alanine (D11A: D88A, D89A, D90A, D91A, D92A, D93A, D104A, D110A, D113A, E114A, and D116A) as GST fusion proteins and incubated equal amounts of each, or GST alone, with purified HA-Htz1. The ability of each GST protein to bind HA-Htz1 was assayed by pull-down using GST resin followed by Western blotting with antibodies to recognize HA. As shown in Fig. 2 E, the full-length Mps3 N terminus directly binds HA-Htz1, whereas GST, GST-mps3Δ75–150, and GST–mps3-D11A show little to no binding. Therefore, we conclude that Mps3 directly interacts with Htz1 and that the association does not require Htz1 assembly into chromatin.
Role of Mps3–Htz1 binding in transcriptional regulation and DNA damage repair
Our observation that Mps3 interacts with Htz1 outside of the context of chromatin leads to the prediction that loss of Mps3–Htz1 binding should not have any effect on transcriptional regulation or the maintenance of genomic integrity. That is to say, mps3Δ75–150 mutants that are unable to bind to Htz1 would not be expected to affect Htz1-dependent transcriptional regulation, recruitment of active genes to the nuclear membrane, or chromatin structure. Similarly, Mps3 binding to Htz1 should not be regulated by posttranslational modifications of Htz1 that occur during transcription and the DNA damage response. In the experiments that follow, we tested this hypothesis using multiple lines of investigation; in all cases, our results are consistent with a chromatin-independent function for Mps3–Htz1 binding.
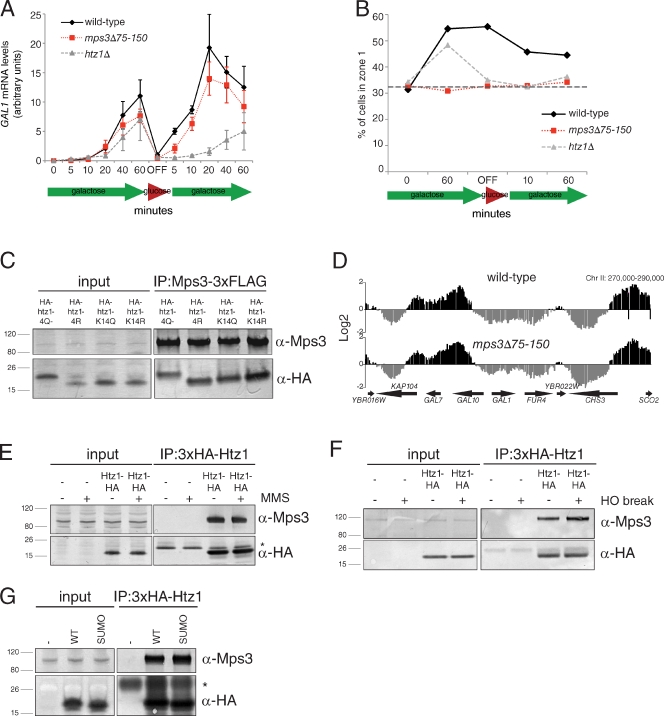
Certain genes, such as GAL1, are recruited to the NE where they undergo transcriptional activation and can be rapidly reactivated after a brief period of repression (Casolari et al., 2004; Brickner et al., 2007; Kundu et al., 2007; Zacharioudakis et al., 2007; Halley et al., 2010; Kundu and Peterson, 2010). We examined GAL1 expression using quantitative real-time PCR (qRT-PCR) and found that wild-type and mps3Δ75–150 mutants have virtually identical GAL1 expression profiles, being induced rapidly and to high levels in galactose (Fig. 3 A). In contrast, expression of GAL1 was not as robust in htz1Δ mutants and was severely reduced during reinduction after a period of glucose repression (Fig. 3 A). Examination of the subnuclear position of the GAL1 locus (marked by tandem integrated arrays of tetracycline operator repeats [TetOR] in cells expressing a GFP fusion to the tetracycline repressor [TetR] DNA binding domain [GFP-Tetr] and the nucleoporin Nup49 fused to GFP) showed that it was not localized to the NE in mps3Δ75–150 cells (Fig. 3 B). This result indicates that a specific subnuclear localization of GAL1 is not absolutely required for proper transcriptional control of this gene. We further confirmed that Mps3 is not involved in Htz1-mediated transcriptional regulation by showing that mutations of N-terminal lysine residues (K4, K8, K11, and K14) in Htz1, which undergo acetylation at promoters of active genes (Babiarz et al., 2006; Keogh et al., 2006; Millar et al., 2006; Halley et al., 2010), do not affect Mps3 binding (Fig. 3 C).
Figure 3.
Analysis of Mps3–Htz1 binding in transcriptional regulation and after DNA damage. (A) Wild-type (SLJ001), htz1Δ (SLJ2080), and mps3Δ75–150 (SLJ2836) cells were grown overnight in YEP (yeast extract peptone) + 2% raffinose to early log phase, then switched into YEP + 2% galactose/raffinose media for the indicated time points (induction). Cells were pelleted, washed, and resuspended into YPD and allowed to grow for 1 h at 30°C (OFF) before transfer back to YEP + 2% galactose/raffinose (reinduction). At the indicated times, samples were taken to analyze GAL1 transcript levels compared with ACT1 using qRT-PCR. Shown are mean values and standard deviation from four independent experiments. (B) Wild-type (SLJ2546), mps3Δ75–150 (SLJ2547), and htz1Δ (SLJ5191) cells in which ∼112 copies of the tetracycline operator (TetOr) were integrated in an intergenic region adjacent to GAL1–10 and expressing GFP-Tetr (Cabal et al., 2006) and NUP49-GFP were grown as described in A. Subnuclear position of the GAL1–10 locus was scored with respect to the distance from the NE in a single-plane image and assigned a position into one of three zones of equal volume as previously described (Bupp et al., 2007). The percentage of foci found in the outermost zone of the nucleus at various time points is plotted. The gray horizontal bar at 33% corresponds to a random distribution. (C) Point mutations in the four acetylation sites in the N terminus of Htz1 (K3, K8, K10, and K14) to mimic or block acetylation (Q or R, respectively) were introduced either individually (K14Q, SLJ2848; K14R, SLJ2849) or as a group (K4Q, SLJ2808; K4R, SLJ2809) into yeast as the sole copy of HTZ1 in the cell, and binding to Mps3-3×FLAG was tested by immunoprecipitation (IP) with anti-HA beads from cells prepared by liquid nitrogen lysis. Protein binding was assayed by Western blotting with the indicated antibodies. (D) The high-resolution map of Htz1 localization throughout the genome in wild-type (SLJ2082) and mps3Δ75–150 (SLJ2322) cells containing 3×HA-HTZ1 was determined by ChIP followed by microarray analysis. Shown is a 20-kb chromosomal region on chromosome II (Chr II; x axis) that includes the GAL1–10 locus. Enrichment of a particular sequence within each region in the ChIP compared with total DNA is plotted on the y axis. Gaps are caused by elements not present on the array. (E) Wild-type (SLJ001) and 3×HA-HTZ1 (SLJ2082) cells were arrested for 2 h in S phase with 10 mM hydroxyurea and then 0.1% methyl methanesulfonate (MMS) was added to half of each culture to induce DNA damage. Total levels of 3×HA-Htz1 and Mps3 as well as binding in anti-HA immunoprecipitates were analyzed by Western blotting with anti-HA and anti-Mps3 antibodies. (F) Similarly, DNA damage was induced in these same strains by induction of the HO endonuclease, which results in a single nonrepairable DSB, and Mps3–Htz1 was analyzed as in E. (G) Two C-terminal sumoylation (SUMO) sites of Htz1 were mutated to arginine (K126R and K133R; SLJ3241) and binding to Mps3 was compared with an isogenic strain with a wild-type (WT) 3×HA-HTZ1 (SLJ3234) in anti-HA immunoprecipitates. An untagged control was also used (SLJ3232). Positions of molecular mass markers (kilodaltons) are indicated next to each blot, and asterisks mark the position of the cross-reacting bands present in the control sample. Minus signs represent the negative control strains used.
Chromatin immunoprecipitation (ChIP) followed by microarray analysis showed that the position of Htz1-containing nucleosomes is not significantly affected in mps3Δ75–150 mutants compared with wild-type: the Pearson correlation coefficient between the two datasets was 0.755. This value is similar to that observed between biological replicates. In both our wild-type and mps3Δ75–150 strains, 3×HA-Htz1 was observed at promoters, similar to its previously described pattern of deposition (Figs. 3 D and S3; Krogan et al., 2004; Mizuguchi et al., 2004; Guillemette et al., 2005; Li et al., 2005; Zhang et al., 2005). These experiments provide compelling evidence that Mps3 binding to Htz1 is not important for the recruitment of chromatin domains to the NE during transcription and support our coimmunoprecipitation data, which point to a novel chromatin-independent function for Htz1.
We also considered the possibility that Mps3–Htz1 binding is important for the localization of nonrepairable DSBs to the NE. If the interaction is regulated by this mechanism, Mps3 should not bind to htz1-K126R K133R or htz1stop119 proteins, which lack critical C-terminal lysines involved in sumoylation, tethering of break sites at the NE, and recruitment of Mps3 to the region of DNA damage (Kalocsay et al., 2009). However, both mutants showed no change in Mps3 binding compared with wild-type Htz1 (Figs. 3 E and 4 B). Furthermore, the amount of Mps3 bound to Htz1 did not increase under conditions of DNA damage (Fig. 3, F and G), which we would expect if the Mps3–Htz1 association were controlled by being part of the cellular response to DNA damage. Again, these results support our hypothesis that Mps3–Htz1 binding has a novel chromatin-independent function.
Mps3 binds to the Htz1 N terminus
A comparison of Htz1 with H2A shows sequence divergence between the two histones in the C-terminal tail and also in their N termini (Fig. 4 A), making these the most likely regions for Mps3 to specifically interact with Htz1. Using a series of deletion and point mutants, we mapped the region of Htz1 required for binding to Mps3 so that we might better understand the role that the Mps3–Htz1 interaction plays in vivo. Deletion of amino acids 1–10 and 11–20 of Htz1 did not affect Mps3 binding, whereas removal of residues 21–30 or 25–35 resulted in complete loss of the Mps3 association (Fig. 4 B), indicating that an Mps3 binding site is located between amino acids 21 and 35. Examination of the Htz1 sequence between amino acids 21 and 35 revealed that it is similar to the highly basic NLS recognized by multiple karyopherins, (R/KxxL(x)nV/YxxV/IxK/RxxxK/R, in single letter amino acid code, where x represents any amino acid and n signifies a repeat of amino acids of variable length; Fig. 4 A; Lange et al., 2008). This region was recently shown to be involved in Htz1 localization to the nucleus, although an NLS was not reported (Straube et al., 2010). To confirm that these residues are important for Mps3 binding, we generated the htz1R27-K42A mutant that contains arginine 27, leucine 30, valine 34, isoleucine 37, arginine 39, and lysine 42 all changed to alanine and showed it also does not interact with Mps3 by coimmunoprecipitation (Fig. 4 B).
We also found that replacement of a region in the histone-fold domain of Htz1 to the corresponding sequence of H2A, creating htz1-m5, abolished Mps3 binding (Fig. 4 B). A similar replacement in the adjacent region (htz1-m6) or other mutations in the C-terminal tail had no effect on binding (Fig. 4 B). The lack of binding is not caused by mislocalization of the Htz1 mutant proteins because all localize to the nucleus (Wu et al., 2005; Straube et al., 2010). Collectively, these data indicate that Mps3 interacts with at least two distinct regions of Htz1: a region in the N terminus that has similarity to a basic NLS and a region in the histone-fold domain that is divergent between Htz1 and H2A.
Mps3 localization to the INM requires Htz1
Another explanation for HTZ1 suppression of mps3-F592S mutants is that Htz1 overproduction increases mps3-F592S protein levels at the NE and/or SPB to an amount sufficient for cell growth because of a role in targeting Mps3 to the INM. If this is the case, overexpression of HTZ1 mutants unable to bind to Mps3 should fail to rescue the growth defect of mps3-F592S at 37°C. In addition, these mutants should have a defect in Mps3-GFP localization to the INM that is similar to cells lacking HTZ1 and mps3-F592S–GFP or mps3Δ75–150-GFP mutants. Finally, we would predict that targeting Mps3 to the INM is a novel function of Htz1 that does not depend on its association with chromatin; mutants that affect assembly of Htz1 should not result in mislocalization of Mps3-GFP from the INM.
We first tested the ability of different htz1 alleles to rescue growth of mps3-F592S mutants at 37°C. Unlike wild-type HTZ1 or htz1-m6, which partially restored growth, htz1R27-K42A and htz1-m5 were unable to suppress the growth defect of mps3-F592S above the background (Fig. 4 C). htz1Δ21–30 was also unable to rescue growth, but its defect was less pronounced than htz1R27-K42A and htz1-m5 (Fig. 4 C). The inability of specific htz1 mutants to restore growth to mps3-F592S is consistent with our data that these domains mediate the Mps3–Htz1 interaction and support the possibility that Mps3–Htz1 binding is important for INM localization of Mps3.
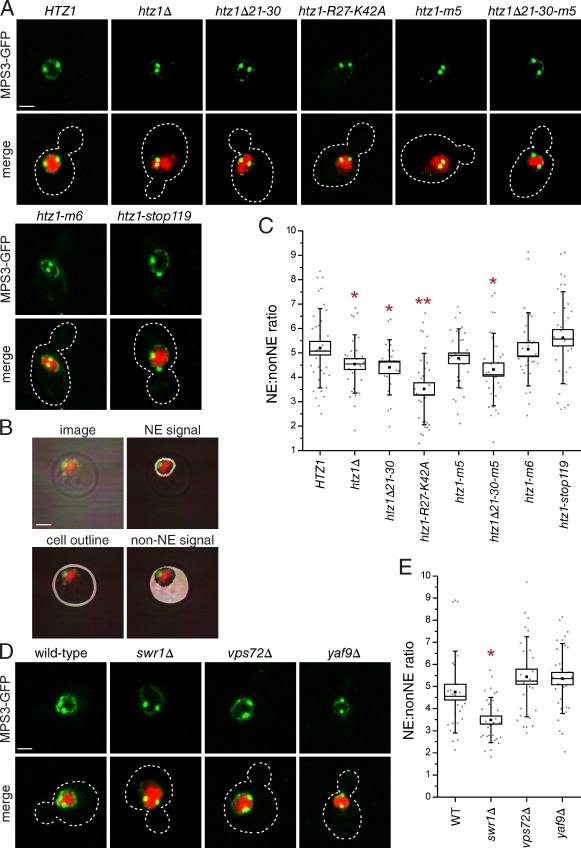
Next, we examined Mps3-GFP localization using confocal imaging to test our hypothesis that Htz1 is required for Mps3 localization. In wild-type cells, we detected Mps3-GFP at the SPB and the nuclear membrane as previously described (Fig. 5 A; Bupp et al., 2007). The signal at the INM was greatly reduced in cells lacking HTZ1 (Fig. 5 A). A similar phenotype was also observed in htz1Δ21–30, htz1R27-K42A, and htz1-m5 mutant backgrounds but not in htz1-m6 or htz1stop119 strains, suggesting that loss of Htz1 binding results in mislocalization of Mps3 from the INM.
Figure 5.
Htz1 is required for Mps3 localization to the INM. (A) Localization of Mps3-GFP (green) and H2B-mCherry (red) in wild-type HTZ1 (SLJ4818), htz1Δ (SLJ3408), htz1Δ21–30 (SLJ3536), htz1-R27-K42A (SLJ4826), htz1-m5 (SLJ4822), htz1Δ21–30-m5 (SLJ4824), htz1-m6 (SLJ4820), or htz1stop119 (SLJ4827). The cell is outlined in white based on the DIC image. (B) Schematic showing how fluorescence intensity of GFP at the INM (NE) and mislocalized protein on membranes throughout the cell (non-NE) were determined using the nuclear signal and transmitted light image. (C) The quantitation of the NE/non-NE ratio in Mps3-GFP in images from A was performed as described in the Materials and methods. (D) Similarly, Mps3-GFP was localized in wild-type (SLJ3529), swr1Δ (SLJ4759), vps72Δ (SLJ4915), and yaf9Δ (SLJ4916) cells. (E) Quantitation of the signal at the INM compared with other membranes is shown. In C and E, a single or double asterisk indicates mutants that displayed a statistically significant (P < 0.05) or highly statistically significant (P < 10−4) change, respectively, in the mean NE/non-NE ratio compared with the wild-type (WT) control. Values for each data point (gray circles), the mean (black square) and median values (line), standard error (box), and standard deviation (lines) for each sample are depicted. Bars, 2 µm.
To quantify the extent of Mps3-GFP mislocalization from the INM in our mutants, we used a fusion of one copy of histone 2B with mCherry (H2B-mCherry) to fluorescently label the nucleus and used transmitted light to visualize the cell boundary, which allowed us to determine the mean fluorescence intensity of Mps3-GFP on nonnuclear membranes (the signal inside the boundary defined by transmitted light but outside that of the nucleus) and at the nuclear membrane (the signal defined by the outer boundary of the nuclear marker; Fig. 5 B). A threshold cutoff enabled us to exclude the SPB signal, which is unaffected in most mutants (Fig. S4 A). Because of the tight coupling of nuclear and cell size in budding yeast (Jorgensen et al., 2007), changes in the NE/non-NE ratio between strains will reflect an increase or decrease in localization of Mps3-GFP at the INM. Using this method, we were able to show that htz1Δ, htz1Δ21–30, and htz1Δ21–30-m5 showed a statistically significant reduction (P < 0.05) in the amount of Mps3-GFP at the INM. In addition, the extent of Mps3-GFP mislocalization in htz1R27-K42A is as severe as that of mps3Δ75–150-GFP and mps3-F592S–GFP cells (a ∼1.5-fold reduction in the NE/non-NE ratio compared with wild type; Figs. 5 C and 6 B and not depicted). htz1-R27-42A is a semidominant mutant that has a more severe effect on Mps3 localization than htz1Δ (Fig. 5 C). We suspect that the htz1-R27-42A protein titrates out other factors important for targeting or tethering Mps3 in the INM.
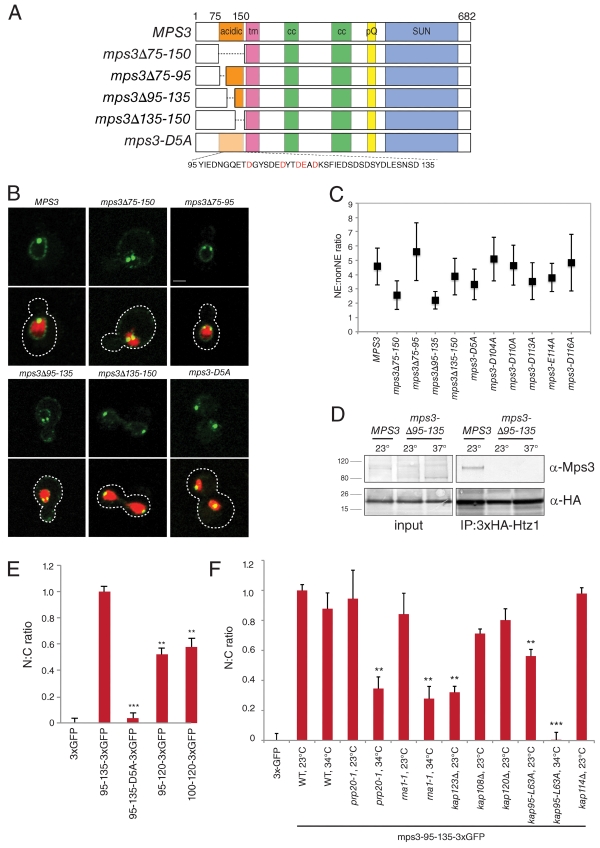
Figure 6.
Amino acids 95–135 of Mps3 are necessary and sufficient for nuclear localization. (A) Schematic of wild-type MPS3 showing the acidic domain from amino acids 75–150, the transmembrane domain (tm), regions of coiled-coil (cc), poly-glutamine repeat (pQ), and SUN domain as well as deletion mutants in the N terminus. mps3-D5A contains alanines in place of conserved acidic amino acids (red). (B) Representative single-plane image of wild-type and mutant versions of MPS3 fused to GFP (green) together with H2B-mCherry (red). The cell is outlined in white based on the DIC image. Bar, 2 µm. (C) The mean NE/non-NE value and standard deviation for each sample are depicted. (D) The protein composition of anti-HA immunoprecipitates (IP) from 3×HA-HTZ1 (SLJ2082) and 3×HA-HTZ1 mps3Δ95–135 (SLJ4855) strains grown at 23°C or shifted to 37°C for 3 h was analyzed by Western blotting with anti-Mps3 (top) and anti-HA (bottom) antibodies. Positions of molecular mass markers (kilodaltons) are indicated next to the blot. (E) The indicated amino acids were fused to three tandem copies of GFP and expressed from the GAL1 promoter in wild-type cells by addition of 2% galactose for 1 h at 23°C. The distribution of 3×GFP fragments in the nucleus and cytoplasm was quantitated in ≥100 cells of each genotype to generate a ratio of nuclear to cytoplasmic signal and then normalized so that the value for cells expressing 3×GFP only and mps3-95-135-3×GFP were 0 and 1, respectively. (F) The distribution of 3×GFP or mps3-95-135-3×GFP in the nucleus and cytoplasm was quantitated and normalized as in E in cells induced for 1 h at 23 or 34°C. In E and F, error bars represent the standard deviation from the mean. Statistical significance compared with mps3-95-135-3×GFP in each experiment of P < 10−6 (double asterisks) and P < 10−9 (triple asterisks) is shown. N:C, nucleus/cytoplasmic ratio.
When we examined Mps3-GFP localization in vps72Δ, yaf9Δ, and swr1Δ mutants, we found little change in its distribution compared with wild-type cells (Figs. 5, D and E; and S4 B). The defect in Mps3-GFP localization that we observe in swr1Δ mutants is most likely caused by an overall reduction in levels of Htz1 in the nucleus rather than a defect in chromatin incorporation because we do not see INM localization defects in other components of the SWR1 complex or in htz1-m6 mutants (Fig. 5, A and D). Therefore, it appears that Htz1 has a novel chromatin-independent function in INM localization of Mps3.
Htz1 is required for nuclear targeting of a soluble N-terminal fragment of Mps3
Analysis of a series of deletions allowed us to narrow the region of Mps3 required for nuclear organization, INM localization and Htz1 binding to amino acids 95–135 within the N-terminal acidic domain (Figs. 2 D; 6, A–D; S1; and S4 C; and not depicted). Not only are these residues necessary for localization of full-length Mps3-GFP to the peripheral NE but also they are sufficient to target three tandem copies of GFP to the nucleus (Fig. 6 E). Removal of additional residues decreases nuclear targeting (Fig. 6 E), suggesting that residues 95–135 are the minimal nuclear targeting domain (NTD). The most notable feature of the NTD is the presence of conserved acidic amino acids, which are important for Htz1 binding, telomere tethering, and localization of Mps3 to the INM as demonstrated by our mutational analysis (Figs. 2 E, S1, and 6, B and C). However, this region does not contain any apparent NLS as judged by several different prediction algorithms.
Using the NTD, we developed an Mps3 nuclear targeting assay to show that Htz1 binding is important for translocation of Mps3 into the nucleus as opposed to tethering Mps3 in the INM, which has previously been predicted for chromatin components (Lusk et al., 2007). At ∼85 kD, the mps3-95-135-3×GFP fragment exceeds the diffusion limit of the NPC (Görlich and Kutay, 1999), and it presumably reaches the nucleus by active transport despite the lack of an NLS. Consistent with this possibility, we found that blocking active transport by inactivation of either Prp20 or Rna1, which affect the GTPase cycle of Ran (Belhumeur et al., 1993; Corbett et al., 1995), or loss of function mutations in the karyopherins Kap123 or Kap95 resulted in a significant decrease in nuclear accumulation of mps3-95-135-3×GFP (Fig. 6 F).
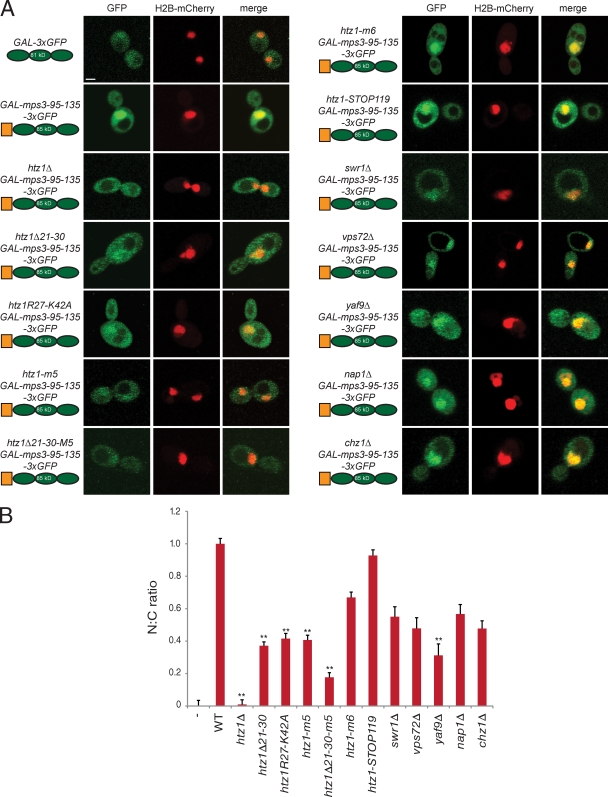
In cells lacking HTZ1 or containing a mutant version of htz1 unable to bind to Mps3, such as htz1Δ21–30, htz1R27-K42A, or htz1-m5, accumulation of the soluble mps3-95-135-3×GFP fragment in the nucleus was greatly diminished compared with wild-type cells (Fig. 7 A). To compare the distribution of the protein between samples, we quantified the mean GFP intensity in the nucleus and cytoplasm to generate a nucleus/cytoplasm (N/C) ratio and then normalized this value such that cells expressing 3×GFP and mps3-95-135-3×GFP were assigned values of 0 and 1, respectively. The N/C ratio of 0.01 ± 0.03 in htz1Δ cells indicates that the mps3-95-135-3×GFP protein showed a nuclear and cytoplasmic distribution similar to 3×GFP (Fig. 7 B). Although deletion or mutation of the NLS region of Htz1 or the M5 domain did not have as dramatic of an effect, we observed a statistically significant (P < 10−6) drop in N/C ratio in all three strains to ∼0.4 (Fig. 7). Moreover, these two regions appear to have an additive effect on localization, as htz1Δ21–30-m5 results in a decrease in the N/C ratio to 0.17 ± 0.03. In contrast, mutations in the C-terminal region of Htz1 that block its incorporation into chromatin have only a minor defect in the localization of the soluble Mps3 fragment, which is similar to what we observed in cells lacking SWR1, VPS72, NAP1, or CHZ1 (Fig. 7).
Figure 7.
Nuclear transport of a soluble N-terminal Mps3 fragment requires Htz1. (A) A schematic of each 3×GFP fragment along with its predicted size and strain background is shown next to representative images of its localization (green) following addition of 2% galactose for 1 h at 23°C. H2B-mCherry (red) was used to highlight the nucleus and show the degree of nuclear colocalization in the merged image. Bar, 2 µm. (B) The distribution of 3×GFP fragments in the nucleus and cytoplasm was quantitated as described in Fig 6 E. n ≥100 cells of each genotype. Error bars represent the standard deviation from the mean. Statistical significance compared with mps3-95-135-3×GFP in each experiment of P < 10−6 (double asterisks) is shown. The minus sign represents the negative control strain used. N:C, nucleus/cytoplasmic ratio. WT, wild type.
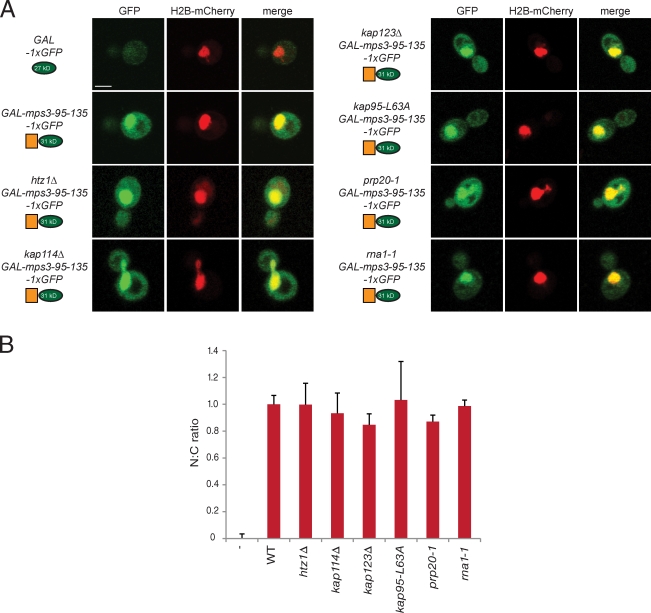
Fusion of amino acids 95–135 to a single copy of GFP results in an ∼31-kD protein that is smaller than the diffusion limit of the NPC (Görlich and Kutay, 1999). Nuclear localization of mps3-95-135-1×GFP occurs by passive diffusion as indicated by the fact that mutations in multiple transport factors do not exhibit defects in its nuclear accumulation (Fig. 8, A and B). In addition, we found that this fragment is still enriched in the nucleus in cells lacking HTZ1, resulting in an N/C ratio of 0.99 ± 0.16 (Fig. 8, A and B). The requirement for Htz1 in localization of mps3-95-135-3×GFP, but not mps3-95-135-1×GFP, can be best explained by a role of Htz1 in the nuclear transport of Mps3.
Figure 8.
Htz1 is not required for tethering Mps3 in the nucleus. (A) Amino acids 95–135 were also fused to a single copy of GFP (green) and localized in the strains shown after addition of 2% galactose for 1 h at 30°C. H2B-mCherry (red) was used to highlight the nucleus and show the degree of nuclear colocalization in the merged image. Bar, 2 µm. (B) The distribution of 1×GFP fragments in the nucleus and cytoplasm was quantitated as described in Fig 6 E. None of the mutants are statistically significant from wild-type. n ≥100 cells of each genotype. Error bars represent the standard deviation from the mean. The minus sign represents the negative control strain used. N:C, nucleus/cytoplasmic ratio. WT, wild type.
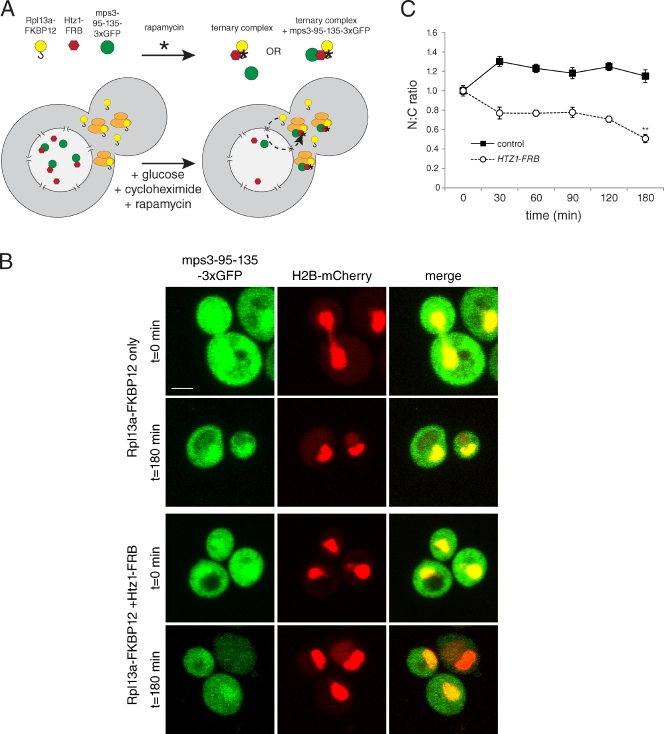
To further demonstrate that Htz1 is involved in targeting Mps3, we took advantage of the anchor-away technique, which is designed to tether specific nuclear proteins in the cytoplasm (Haruki et al., 2008). In this assay, the human FKBP12 protein is anchored in the cytoplasm by fusion to the ribosomal protein Rpl13A, which transits through the nucleus and back to the cytoplasm in the process of ribosome assembly (Fig. 9 A). A nuclear target protein, such as Htz1, is fused to the FKBP12–rapamycin binding (FRB) domain of human mammalian target of rapamycin that interacts with FKBP12 in the presence of rapamycin, resulting in Htz1-FRB translocation to the cytoplasm. Addition of rapamycin to Htz1-FRB cells before addition of galactose to induce expression of mps3-95-135-3×GFP should mimic an htz1Δ mutant if Htz1 binding is critical for Mps3 localization. Moreover, Htz1-FRB should also be able to target the soluble fragment to the cytoplasm if rapamycin is added to cells after mps3-95-135-3×GFP production. Consistent with these hypotheses, mps3-95-135-3×GFP did not accumulate in the nucleus in Htz1-FRB cells that had been treated with rapamycin for 30 min before addition of galactose (unpublished data). In addition, we observed an ∼50% decrease in the N/C ratio in Htz1-FRB cells treated with rapamycin for 180 min to 0.51 ± 0.04 but no decrease in the N/C ratio in control cells (Fig. 9, B and C), indicating that Htz1 is able to shuttle the soluble Mps3 fragment out of the nucleus and tether it in the cytoplasm. When taken together with our biochemical experiments and our localization analysis of full-length Mps3-GFP, we conclude that Htz1 has an active chromatin-independent function in targeting Mps3 to the nucleus.
Figure 9.
Targeting Mps3 to the cytoplasm through Htz1. (A) Schematic of the anchor-away technique (adapted from Haruki et al., 2008). The ribosomal protein Rpl13A fused to FKBP12 and Htz1 fused to FRB form a ternary complex in the presence of rapamycin (asterisks). Htz1-FRB and the soluble mps3-95-135-3×GFP fragment are predominantly nuclear, but because ribosomal proteins transiently transit through the nucleus (dashed line) during the process of 40S and 60S subunit assembly, Rpl13A-FKPB12 is able to deplete Htz1-FRB from the nucleus and target it to the cytoplasm. Proteins bound to Htz1-FRB should also be depleted from the nucleus and localize to the cytoplasm. (B) mps3-95-135-3×GFP expression was induced using 2% galactose in cells containing HTZ1-FRB (SLJ5029) or an isogenic control lacking the FRB fusion (SLJ5028). After 30 min at 23°C, cells were transferred to prewarmed media containing 10 µg/ml rapamycin plus 2% dextrose and 100 µg/ml cycloheximide to repress transcription and translation of mps3-95-135-3×GFP and transferred to 34°C. Representative images of mps3-95-135-3×GFP localization (green) before (t = 0) and after (t = 180) addition of rapamycin for 3 h at 34°C in control and Htz1-FRB cells are shown. H2B-mCherry (red) was used to highlight the nucleus and show the degree of nuclear colocalization in the merged image. Bar, 2 µm. (C) The nucleus/cytoplasmic (N:C) ratio was determined as described in Fig 6 E. Values were normalized such that cells expressing 3×GFP only and cells at t = 0 were assigned 0 and 1, respectively. At least 50 images were quantitated for each time point in two independent experiments. Error bars represent the standard deviation from the mean. Statistical significance compared with the cells at t = 0 of P < 10−6 (double asterisk) is shown.
Discussion
In this paper, we examine the interaction between the SUN protein Mps3 and the histone variant H2A.Z. We initially anticipated that Mps3–Htz1 binding would play a direct role in the recruitment of particular regions of the genome to the NE. Much to our surprise, our data do not support this hypothesis. Instead, multiple lines of evidence indicate that Mps3 binding to Htz1 does not require Htz1 incorporation into chromatin, leading us to propose that Htz1 has a novel chromatin-independent function in targeting Mps3 to the INM.
The interaction between Mps3 and Htz1 is highly specific and was identified by several different methods: two-hybrid, genetic, and biochemical analysis. In all three approaches, Htz1 was the only histone that specifically interacted with Mps3. The fact that we have been unable to detect enrichment for other histones in our Mps3-3×FLAG preparations by Western blotting or mass spectroscopic analysis further confirms that Mps3 and Htz1 form a unique chromatin-independent complex. Additional evidence that Mps3 binding to Htz1 occurs outside of the context of chromatin comes from our observations that mutations in the SWR1 complex or Htz1 itself, which abolish its deposition into chromatin, do not affect the physical interaction with Mps3 or genetic suppression of mps3 mutants. Based on the regions of Htz1 that are essential for Mps3 binding, it is perhaps not surprising that Mps3 does not bind to chromatin-associated Htz1 because both the NLS and M5 domains are buried within the histone octamer and would presumably not be accessible in the context of a nucleosome (Chakravarthy et al., 2004; Zlatanova and Thakar, 2008).
Htz1-binding partners have been analyzed using a variety of genetic and biochemical methods (for an example see Krogan et al., 2004; Wu et al., 2005, 2009; Luk et al., 2007; Lambert et al., 2009; Straube et al., 2010), raising the question of why Mps3 was not previously detected. Because of the essential function of Mps3 in SPB duplication, it is not present in the yeast deletion collection that has been used in systematic genetic analyses of Htz1-containing complexes. Using traditional methods for chromatin preparation, many membrane-associated nuclear components, including SPBs, NPCs, and other nuclear matrix factors, are not efficiently solubilized. The use of cryolysis allowed us to isolate intact protein complexes associated with the yeast nuclear membrane and to identify proteins that only associate in minor quantities with Mps3. Our choice of methods is particularly critical because Mps3 is not abundant, and only a small fraction of Htz1 is likely bound to the SUN protein (unpublished data).
Consistent with our finding that Mps3–Htz1 binding is chromatin independent, we found that transcriptional regulation is unaffected in mps3Δ75–150 mutants that abolish its association with Htz1. We show that GAL1 expression is fully induced and reinduced after a brief period of repression despite the fact that it is not localized to the NE. This suggests that peripheral localization is not absolutely essential for transcriptional control of this locus despite the requirement for Htz1. Mps3 may be directly involved in tethering the GAL1 locus at the NE, or it could be required for proper localization of other NE proteins, such as nucleoporins, import/export factors, or Mlp1/2. Our observation that the position of Htz1-containing nucleosomes is unaffected in mps3Δ75–150 mutants also supports the hypothesis that Mps3 binding to Htz1 is not essential for transcriptional control, including establishment and maintenance of boundary regions near telomere ends and the mating-type loci. When combined with our data showing that the presence of DNA damage or mutations in Htz1 that abolish recruitment of certain types of DSBs to the nuclear membrane does not affect Mps3–Htz1 binding, these data strongly suggest that the Mps3–Htz1 interaction is involved in a novel aspect of nuclear organization.
Our hypothesis that Mps3–Htz1 binding functions to target Mps3 to the INM stems from the following set of observations. First, deletion of the Htz1-binding region in Mps3 results in a version of the protein that is defective in peripheral localization. Second, deletion of HTZ1 or mutation of the Mps3-binding regions in Htz1 causes a similar mislocalization of Mps3-GFP. Third, we found that these same mutants are also unable to target a large soluble reporter construct containing the Mps3 N-terminal region to the nucleus. However, a smaller version of this reporter does accumulate in the nucleus. Lastly, artificially targeting Htz1 to the cytoplasm results in the redistribution of the soluble Mps3 fragment from the nucleus to the cytoplasm. Collectively, these results indicate that Htz1 binding actively localizes Mps3 to the INM.
We propose that Mps3 is able to interact with Htz1 in the cytoplasm, and using the NLS, it is cotransported into the nucleus. Whether binding to both the NLS and/or M5 region of Htz1 is required for nuclear translocation is unknown. However, the fact that we can remove the C-terminal tail of Htz1, which is involved in binding to several nuclear proteins (Wu et al., 2005, 2009; Luk et al., 2007), suggests that this region is dispensable. The Nap1 chaperone interacts with the Htz1 NLS region and might bridge its interaction with karyopherins (Straube et al., 2010), yet neither Mps3 binding to Htz1 nor its localization to the INM requires Nap1 or the other Htz1 chaperone Chz1. However, localization of the soluble Mps3 fragment as well as full-length Mps3 is partially dependent on Kap123, one of the major karyopherins involved in Htz1 transport (Straube et al., 2010). Our data showing that mps3-95-135-1×GFP is enriched in the nucleus in cells lacking HTZ1 suggest that Htz1 binding is not essential for Mps3 tethering Mps3 in the INM. Thus, we hypothesize that, once Mps3 is targeted to the nucleus by Htz1, additional nuclear factors anchor it in the INM. Candidate tethers include components of the silent information regulator complex, yKu70/80, or other telomere-binding proteins that have previously been shown to bind to Mps3 (Uetz et al., 2000; Antoniacci et al., 2004, 2007; Bupp et al., 2007; Schober et al., 2009).
Amino acids between 95 and 135 are both necessary and sufficient for Mps3 targeting to the nucleus. Mutation of multiple acidic residues to alanine completely blocked INM localization of full-length Mps3 protein and nuclear accumulation of a soluble reporter. When we mutated fewer residues, the effect on localization was less pronounced, and in fact, no single amino acid was absolutely essential for Mps3 localization. Although it is possible that a change in the charge of the Mps3 N terminus is what is affected in our mutants, our observation that INM localization is most significantly reduced in E114A and D116A suggests that this region does in fact contain a specific sequence that serves as a NTD. However, we do not predict that the Mps3-NTD is an NLS or an INM-sorting motif, which are typically short lysine-rich sequences that mediate karyopherin binding (Braunagel et al., 2004; Lange et al., 2007). The Mps3-NTD has the opposite molecular characteristics, and furthermore, we were unable to observe a direct interaction between Mps3 and any of the major yeast karyopherins (unpublished data). Sequence analysis of several yeast INM proteins reveals the presence of highly acidic patches, one of which is adjacent to the reported NLS of Heh1 and in the region of Heh1 that is required for its INM localization (King et al., 2006). It will be interesting to test whether import of these proteins might also involve Htz1.
In general, loss of Htz1 binding abolished Mps3 localization to the INM but did not significantly affect its levels at the SPB. Changes in Mps3-GFP SPB levels in some mutants are most likely caused by cell cycle– and size-dependent effects on SPB area (Byers and Goetsch, 1974; Winey et al., 1995; O’Toole et al., 1997). Localization to the SPB, but not the nuclear periphery, is similar to mps3Δ75–150-GFP and mps3-F592S–GFP and suggests that there are two pools of Mps3 at the INM: one population at the peripheral NE is affected by loss of Htz1 binding, and a second at the SPB is very stable and is not Htz1 dependent. This could be because SPB-associated Mps3 might be targeted and anchored by additional factors, such as Mps2 (Jaspersen et al., 2006). Curiously, a previous study suggested that Mps2 and another SPB component, Bbp1, interact with Yaf9, a shared component between the SWR1 complex required for Htz1 deposition and NuA4, an acetyltransferase essential for histone H4 modification (Le Masson et al., 2003). The molecular details behind Mps2–Bbp1–Yaf9 binding are not understood, but disruption of this interaction may explain why nuclear localization of the mps3-95-135-3×GFP fragment was reduced in yaf9Δ cells. Localization of full-length Mps3-GFP to the SPB and the INM is not affected by loss of Yaf9. Rather, deletion of YAF9 or VPS72, which likely increases soluble pools of Htz1 by blocking its deposition into chromatin (Krogan et al., 2004; Zhang et al., 2004; Wu et al., 2005), appears to result in a slight increase in INM levels of Mps3-GFP.
In conclusion, we have demonstrated a novel chromatin-independent role for the yeast H2A.Z variant Htz1 in the localization of Mps3. Although chromatin has been proposed to be especially important in yeast for tethering INM proteins, which lack nuclear matrix proteins, such as the nuclear lamins, our results favor a simpler model to explain the binding of Htz1 to Mps3—the histone variant is required to target the SUN protein to the INM. Although INM targeting of SUN proteins is likely more complex in metazoans, several studies have shown INM localization is lamin independent and requires aspects of active transport and the NPC as well as other sorting sequences in the C terminus and the SUN domain (Hodzic et al., 2004; Padmakumar et al., 2005; Crisp et al., 2006; Haque et al., 2006, 2010; Hasan et al., 2006; Turgay et al., 2010). The role of H2A.Z and chromatin in INM localization of other SUN proteins has never been rigorously tested, and based on our examination of the Mps3–Htz1 interaction, binding to chromatin factors does not necessarily translate into a requirement for chromatin, per se, in INM tethering. It is unclear why cells use a H2A.Z histone variant for INM localization of Mps3—an important future question will be to determine whether this role is specific to Mps3 or whether H2A.Z is also involved in targeting other proteins, including the mammalian SUN proteins, at the nuclear periphery.
Materials and methods
Yeast strains and plasmids
All strains are derivatives of W303 (ade2-1 trp1-1 leu2-3,112 ura3-1 his3-11,15 can1-100 RAD5+) unless indicated and are listed in Table S1. Strains containing htz1Δ::KANMX, GAL-HO, and TetOR adjacent to GAL1–10 were gifts from S. Biggins (Fred Hutchinson Cancer Research Center, Seattle, WA), S. Jentsch (Max Planck, Martinsried, Germany), and E. Hurt (Heidelberg University, Heidelberg, Germany), respectively. In addition, plasmids containing the histone genes were provided by F. Winston (Harvard Medical School, Boston, MA), and reagents for the anchor-away system were shared by U. Laemmli (University of Geneva, Geneva, Switzerland). Standard techniques were used for DNA and yeast manipulations, including C-terminal tagging of MPS3 and alleles with 3×FLAG or GFP and HTB2 with mCherry or deletion of genes with KANMX, NATMX, HIS3MX, or HYGMX. Site-directed mutagenesis was performed using a mutagenesis kit (QuikChange; Agilent Technologies), and all mutations were confirmed by sequence analysis. For dilution assays, 5 OD600 cells were serially diluted 10-fold in sterile growth media and stamped onto agar plates.
To construct pRS304-3×HA-HTZ1, an ∼1,500-bp region of the HTZ1 promoter was ligated into the KpnI and XhoI sites of pRS304 followed by three copies of the HA epitope ligated at the XhoI and EcoRI sites and the HTZ1 ORF and ∼250 bp of the 3′ untranslated region ligated at EcoRI and SacI sites. The final product was sequenced on both strands using T7, T3, and several internal primers and was integrated into TRP1 with BstZ17I. Point and deletion mutants were made by mutagenesis of this plasmid.
The MPS3 ORF and ∼500 bp of the promoter were amplified by PCR and cloned into the XhoI and BamHI sites of a pRS306 vector containing GFP to construct pSJ650 (pRS306-MPS3-GFP; Bupp et al., 2007). Mutants and deletions were made in this plasmid by site-directed mutagenesis. Plasmids were digested with EcoRV for integration at the URA3 locus.
The 1×GFP or 3×GFP sequence was subcloned into pDK20 (Jaspersen et al., 2002) to create pRS306-GAL-1×GFP (pSJ114) or pRS306-GAL-3×GFP (pSJ994), and then double-stranded DNA oligos corresponding to the Mps3 N-terminal region encoding amino acids 95–135, 95–120, and 100–120 were cloned into the XhoI and BamHI sites. These plasmids were digested with PstI to direct integration into the URA3 locus.
Cytological techniques
Localization of Mps3-GFP and H2B-mCherry was visualized in cultures grown to mid–log phase in minimal media supplemented with 3× adenine. 1 ml culture was centrifuged, washed with 1 ml of double-distilled H2O, and resuspended in ∼100 µl of double-distilled H2O, and 10 µl was placed on a 25% gelatin pad for image analysis (Bupp et al., 2007). At least two independent transformants of each genotype were analyzed by fluorescence microscopy in at least three independent experiments. Analysis of the subnuclear position of TetOR and lactose operator repeat (LacOR) was also performed as previously described (Bupp et al., 2007). In brief, a microscope (Axio Imager; Carl Zeiss) with a 100× α-Plan Fluar objective, NA of 1.45, (Carl Zeiss) and a digital camera (Orca-ER; Hamamatsu Photonics) were used to capture 15 image stacks of 200-nm step size through nuclei of log phase cells at room temperature. The spot-to-periphery distance and the nuclear diameter were determined in a single z-stack image where the spot was most concentrated using Axiovision 4.6.3 software (Carl Zeiss). By dividing the spot-to-periphery distance by the diameter, each spot fell into one of three zones of equal surface (Fig. S1 A). Zone 1 has a width of 0.184× the nuclear radius (r), zone 2 has a width of 0.184–0.422× r, and zone 3 has a width of 0.422× r.
Fluorescence was performed using a confocal microscope (LSM 510 Meta; Carl Zeiss) equipped with a module with avalanche photodiode detectors (ConforCor 3; Carl Zeiss), which allow single-photon counting, with a 100×, 1.46 NA α-Plan Fluar objective. GFP was excited using a 488-nm Ar laser line, whereas mCherry was excited with a 561-nm HeNe laser line with the appropriate filter sets. Data were acquired using AIM v.4.2 software (Carl Zeiss). Images were collected with 8–10 image stacks with a 0.3-µm step size through the cells at room temperature. Images were processed using ImageJ software (National Institutes of Health).
To quantify the degree of mislocalization of Mps3-GFP at the NE in mutant strains, we created a mask based on the transmitted light image of cells using Axiovision software. Next, we selected all pixels above a predefined threshold of the H2B-mCherry signal to create an additional mask that describes the nucleus. The outer pixels of this mask are considered the NE. These masks are used to quantify the amount of Mps3-GFP localized in the NE (INM signal) compared with the amount of protein that is mislocalized (non-NE/ER signal = non-NE) and to determine a ratio of mean intensity per unit area for each. The very bright SPB signal from Mps3-GFP is excluded from this calculation using an upper intensity cutoff. At least 30 images of each genotype were quantified per experiment, and the NE/non-NE ratio for each was plotted using OriginPro software (OriginLab). Outliers of greater than two standard deviations were excluded from analysis. In addition, a representative single section that contains the SPBs in plane from the z stack is shown. The level of Mps3-GFP at the SPB was quantitated in ≥100 cells using a 0.35 × 0.35–µm box using ImageJ and is shown separately in Fig. S4.
For quantification of the nuclear import of 1×GFP and 3×GFP, a projection image was created using ImageJ, in which the mean intensity of a 0.72 × 0.72–µm box was measured in both the nucleus that overlapped with the H2B-mCherry signal and a region of the cytoplasm that did not include the vacuole. At least 100 cells for wild-type and mutant strains were analyzed, and the N/C of the GFP signal was normalized such that 1×GFP or 3×GFP only was equal to zero and mps3-95-135-1×GFP or mps3-95-135-3×GFP in wild type was equal to one. The statistical significance of these values under different conditions was calculated using the Student’s t test.
Immunoprecipitation and Western blotting
Liquid nitrogen ground lysates were prepared from 500 OD600 mid–log phase cells as previously described (Jaspersen et al., 2006). In brief, cells were frozen in liquid nitrogen and ground with a ball mill (Retsch). Ground cell powder was allowed to thaw on ice and then resuspended in 9 ml extraction buffer (20 mM Hepes-NaOH, pH 7.5, 300 mM NaCl, 1 mM EDTA, 5 mM EGTA, 50 mM NaF, 50 mM β-glycerophosphate, 0.5% Triton X-100, 1 mM DTT, 1 mM PMSF, and 1 mg/ml each of pepstatin A, aprotinin, and leupeptin). After homogenization with a homogenizer (Polytron 10/35; Brinkman) for 30 s, lysates were centrifuged at 3,000 g for 10 min at 4°C, and the resulting supernatant was used for immunoprecipitations. 100 µl anti-HA resin (Roche) or 100 µl anti-FLAG resin (Sigma-Aldrich) was added to lysates to immunoprecipitate tagged proteins. After a 2-h incubation at 4°C, beads were washed five times, and 1/10 of the bound protein and 1/50–1/100 of the input lysate were analyzed by SDS-PAGE followed by Western blotting. In all figures, positions of molecular mass markers (kilodaltons) are indicated next to each blot, and asterisks mark the position of cross-reacting bands present in the control sample.
The following primary antibody dilutions were used in this study: 1:1,000 anti-HA 3F10 (Roche), 1:2,000 anti-FLAG M2 (Sigma-Aldrich), 1:1,000 anti-yH2A (Abcam), 1:1,000 anti-yH2B (Abcam), 1:1,000 anti-yH3 (Abcam), 1:1,000 anti-yH4 (Abcam), 1:1,000 anti-Htz1 (Abcam), 1:1,000 anti-GST (GE Healthcare), 1:2,500 anti-Mps3 (Jaspersen et al., 2006), 1:2,000 anti–glucose-6-phosphate dehydrogenase (Sigma-Aldrich), and 1:2,000 anti-GFP (Takara Bio Inc.). Alkaline phosphatase–conjugated secondary antibodies were used at 1:10,000 (Promega).
In vitro binding assay
The N-terminal region of MPS3 was amplified by PCR and cloned into pGEX-3X for expression in bacteria as a GST fusion protein. GST-Mps3–N terminus (amino acids 1–150) was purified from BL21(DE3) pLysE bacteria transformed with pGEX-3X-Mps3–N terminus (pSJ859) using GST resin (GE Healthcare). Similarly, mutant versions of the Mps3 N terminus were expressed and purified. Up to 10 µg of each protein was exchanged into binding buffer (PBS containing 0.1% Triton X-100 and an additional 250 mM NaCl) using spin columns (Microcon; Millipore).
20 µl anti-GST magnetic beads (Invitrogen) was blocked in 20% BSA and 5% milk in binding buffer for 1 h at 23°C and then incubated with 100 µg of purified GST protein for 1 h at 4°C with end-over-end rocking. The beads were washed, and 200 µg Htz1-HA (a gift from B. Li, University of Texas Southwestern Medical Center, Dallas, TX) in binding buffer was added to each, rotated for 1 h at 4°C, and then washed extensively in binding buffer. 2× SDS sample buffer was added, samples were heated to 100°C for 5 min, and binding reactions were analyzed by SDS-PAGE followed by Western blotting with anti-HA and anti-GST antibodies. 1/50 of the Htz1-HA aliquot added to each reaction was run as a control.
Analysis of RNA levels
Total RNA for qRT-PCR analysis was isolated from yeast cells with a scaled-down and simplified version of the hot phenol extraction procedure as previously described (Brickner et al., 2007). A total of 2–4 µg of DNase-treated total RNA was reverse transcribed using 5 µM oligo dT and 20 units of reverse transcriptase (Superscript III; Invitrogen) at 42°C for 1 h. The reaction was diluted fivefold, and 1/20 was used for qRT-PCR on a real-time PCR machine (iCycler) with iQ SYBR Green Supermix (Bio-Rad Laboratories). The sequences of the primers used for real time PCR have been previously described (Brickner et al., 2007). The relative concentration of cDNA templates for both the target gene (GAL1) and the control gene (ACT1) was calculated for each sample using standard curves for each primer set that were defined by linear regression analysis of cycle threshold values using a series of fivefold dilutions of yeast genomic DNA covering a 3,125-fold range. At least four biological and two technical replicates were performed on each sample.
ChIP and microarray analysis
ChIP followed by microarray analysis was performed essentially as previously described (Glynn et al., 2004). In brief, 225 ml of cells was grown at 30°C in YPD (yeast peptone dextrose) to an OD600 of 0.8, cross-linked for 15 min with 1% formaldehyde, and washed twice before chromatin extraction. Cells were lysed in a mini beadbeater (Mini-Beadbeater-16; Biospec), and chromatin was sonicated (eight cycles, 0.5 s pulse on/0.3 s off for 15 s, and 35% power; Digital Sonifier; Branson) to yield DNA fragments 200–1,000 bp in length. Chromatin was incubated overnight at 4°C with 4 µl α-HA antibody (Roche) and then with 40 µl blocked Protein G beads (GE Healthcare) at 23°C for 2 h. Samples were washed extensively, and proteins were eluted twice in a buffer containing 1% SDS. After reversal of the cross-linking and purification, DNA was randomly amplified to 3–5 µg (Bohlander et al., 1992). The input and immunoprecipitated DNA were labeled with Cy3 and Cy5 dyes, respectively, and hybridized to whole-genome microarrays (244K; Agilent Technologies) by MOgene. Microarray data were analyzed in R using the Limma package (Smyth, 2005). Within-array median normalization and between-array loess normalization were performed. Track files were viewed, and figures were generated using the University of California, Santa Cruz Genome Browser (Kent et al., 2002).
Online supplemental material
Fig. S1 shows telomere tethering and silencing defects in mps3 mutants. Fig. S2 shows the use of cryolysis to analyze Htz1-binding proteins. Fig. S3 shows that loss of Mps3 binding does not affect Htz1 localization to chromatin. Fig. S4 shows Mps3 protein levels and SPB localization in htz1 mutants. Table S1 shows yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201011017/DC1.
Acknowledgments
We thank F. Winston, E. Hurt, S. Jentsch, U. Laemmli, and S. Biggins for strains and plasmids and are grateful to B. Li, J. Workman, R. Camahort, J. Gerton, S. Hawley, K. Delventhal, K. Weaver, C. Seidel, B. Slaughter, and members of the Jaspersen laboratory for helpful discussions, reagents, protocols, and comments on the manuscript.
S.L. Jaspersen is supported by a March of Dimes Basil O’Connor Award, the M.R. and Evelyn Hudson Foundation, and funding from the Stowers Institute for Medical Research.
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- DIC
- differential interference contrast
- DSB
- double-stranded break
- FRB
- FKBP12–rapamycin binding
- INM
- inner nuclear membrane
- LacOR
- lactose operator repeat
- NE
- nuclear envelope
- NLS
- nuclear localization sequence
- NPC
- nuclear pore complex
- NTD
- nuclear targeting domain
- ONM
- outer nuclear membrane
- qRT-PCR
- quantitative real-time PCR
- SPB
- spindle pole body
- TetOR
- tetracycline operator repeats
- TetR
- tetracycline repressor
References
- Akhtar A., Gasser S.M. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8:507–517 10.1038/nrg2122 [DOI] [PubMed] [Google Scholar]
- Antoniacci L.M., Skibbens R.V. 2006. Sister-chromatid telomere cohesion is nonredundant and resists both spindle forces and telomere motility. Curr. Biol. 16:902–906 10.1016/j.cub.2006.03.060 [DOI] [PubMed] [Google Scholar]
- Antoniacci L.M., Kenna M.A., Uetz P., Fields S., Skibbens R.V. 2004. The spindle pole body assembly component Mps3p/Nep98p functions in sister chromatid cohesion. J. Biol. Chem. 279:49542–49550 10.1074/jbc.M404324200 [DOI] [PubMed] [Google Scholar]
- Antoniacci L.M., Kenna M.A., Skibbens R.V. 2007. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 6:75–79 10.4161/cc.6.1.3647 [DOI] [PubMed] [Google Scholar]
- Babiarz J.E., Halley J.E., Rine J. 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20:700–710 10.1101/gad.1386306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhumeur P., Lee A., Tam R., DiPaolo T., Fortin N., Clark M.W. 1993. GSP1 and GSP2, genetic suppressors of the prp20-1 mutant in Saccharomyces cerevisiae: GTP-binding proteins involved in the maintenance of nuclear organization. Mol. Cell. Biol. 13:2152–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlander S.K., Espinosa R., III, Le Beau M.M., Rowley J.D., Díaz M.O. 1992. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics. 13:1322–1324 10.1016/0888-7543(92)90057-Y [DOI] [PubMed] [Google Scholar]
- Braunagel S.C., Williamson S.T., Saksena S., Zhong Z., Russell W.K., Russell D.H., Summers M.D. 2004. Trafficking of ODV-E66 is mediated via a sorting motif and other viral proteins: facilitated trafficking to the inner nuclear membrane. Proc. Natl. Acad. Sci. USA. 101:8372–8377 10.1073/pnas.0402727101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D.G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P.C., Widom J., Brickner J.H. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp J.M., Martin A.E., Stensrud E.S., Jaspersen S.L. 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179:845–854 10.1083/jcb.200706040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. 1974. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 38:123–131 [DOI] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.-C., Hurt E.C., Nehrbass U. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S., Bao Y., Roberts V.A., Tremethick D., Luger K. 2004. Structural characterization of histone H2A variants. Cold Spring Harb. Symp. Quant. Biol. 69:227–234 10.1101/sqb.2004.69.227 [DOI] [PubMed] [Google Scholar]
- Conrad M.N., Lee C.Y., Wilkerson J.L., Dresser M.E. 2007. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 104:8863–8868 10.1073/pnas.0606165104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett A.H., Koepp D.M., Schlenstedt G., Lee M.S., Hopper A.K., Silver P.A. 1995. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J. Cell Biol. 130:1017–1026 10.1083/jcb.130.5.1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N., Kamakaka R.T. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell. 6:769–780 10.1016/S1097-2765(00)00076-9 [DOI] [PubMed] [Google Scholar]
- Glynn E.F., Megee P.C., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L. 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2:E259 10.1371/journal.pbio.0020259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Kutay U. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607–660 10.1146/annurev.cellbio.15.1.607 [DOI] [PubMed] [Google Scholar]
- Guillemette B., Bataille A.R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3:e384 10.1371/journal.pbio.0030384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley J.E., Kaplan T., Wang A.Y., Kobor M.S., Rine J. 2010. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 8:e1000401 10.1371/journal.pbio.1000401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F., Lloyd D.J., Smallwood D.T., Dent C.L., Shanahan C.M., Fry A.M., Trembath R.C., Shackleton S. 2006. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 26:3738–3751 10.1128/MCB.26.10.3738-3751.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F., Mazzeo D., Patel J.T., Smallwood D.T., Ellis J.A., Shanahan C.M., Shackleton S. 2010. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 285:3487–3498 10.1074/jbc.M109.071910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., Laemmli U.K. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 31:925–932 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hasan S., Güttinger S., Mühlhäusser P., Anderegg F., Bürgler S., Kutay U. 2006. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 580:1263–1268 10.1016/j.febslet.2006.01.039 [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Dernburg A.F. 2009. The SUN rises on meiotic chromosome dynamics. Dev. Cell. 17:598–605 10.1016/j.devcel.2009.10.014 [DOI] [PubMed] [Google Scholar]
- Hodzic D.M., Yeater D.B., Bengtsson L., Otto H., Stahl P.D. 2004. Sun2 is a novel mammalian inner nuclear membrane protein. J. Biol. Chem. 279:25805–25812 10.1074/jbc.M313157200 [DOI] [PubMed] [Google Scholar]
- Holmer L., Worman H.J. 2001. Inner nuclear membrane proteins: functions and targeting. Cell. Mol. Life Sci. 58:1741–1747 10.1007/PL00000813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Giddings T.H., Jr, Winey M. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159:945–956 10.1083/jcb.200208169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Martin A.E., Glazko G., Giddings T.H., Jr, Morgan G., Mushegian A., Winey M. 2006. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 174:665–675 10.1083/jcb.200601062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Edgington N.P., Schneider B.L., Rupes I., Tyers M., Futcher B. 2007. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 18:3523–3532 10.1091/mbc.E06-10-0973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M., Hiller N.J., Jentsch S. 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 33:335–343 10.1016/j.molcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M.C., Mennella T.A., Sawa C., Berthelet S., Krogan N.J., Wolek A., Podolny V., Carpenter L.R., Greenblatt J.F., Baetz K., Buratowski S. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20:660–665 10.1101/gad.1388106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Lusk C.P., Blobel G. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 442:1003–1007 10.1038/nature05075 [DOI] [PubMed] [Google Scholar]
- Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131 10.1371/journal.pbio.0020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Baetz K., Keogh M.C., Datta N., Sawa C., Kwok T.C., Thompson N.J., Davey M.G., Pootoolal J., Hughes T.R., et al. 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA. 101:13513–13518 10.1073/pnas.0405753101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S., Peterson C.L. 2010. Dominant role for signal transduction in the transcriptional memory of yeast GAL genes. Mol. Cell. Biol. 30:2330–2340 10.1128/MCB.01675-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S., Horn P.J., Peterson C.L. 2007. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 21:997–1004 10.1101/gad.1506607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.P., Mitchell L., Rudner A., Baetz K., Figeys D. 2009. A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol. Cell. Proteomics. 8:870–882 10.1074/mcp.M800447-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282:5101–5105 10.1074/jbc.R600026200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A., Mills R.E., Devine S.E., Corbett A.H. 2008. A PY-NLS nuclear targeting signal is required for nuclear localization and function of the Saccharomyces cerevisiae mRNA-binding protein Hrp1. J. Biol. Chem. 283:12926–12934 10.1074/jbc.M800898200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Masson I., Yu D.Y., Jensen K., Chevalier A., Courbeyrette R., Boulard Y., Smith M.M., Mann C. 2003. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell. Biol. 23:6086–6102 10.1128/MCB.23.17.6086-6102.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Pattenden S.G., Lee D., Gutiérrez J., Chen J., Seidel C., Gerton J., Workman J.L. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA. 102:18385–18390 10.1073/pnas.0507975102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W.H., Brickner D.G., Brand V.R., Brickner J.H. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell. 40:112–125 10.1016/j.molcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk E., Vu N.D., Patteson K., Mizuguchi G., Wu W.H., Ranjan A., Backus J., Sen S., Lewis M., Bai Y., Wu C. 2007. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell. 25:357–368 10.1016/j.molcel.2006.12.015 [DOI] [PubMed] [Google Scholar]
- Lusk C.P., Blobel G., King M.C. 2007. Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 8:414–420 10.1038/nrm2165 [DOI] [PubMed] [Google Scholar]
- Mekhail K., Moazed D. 2010. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 11:317–328 10.1038/nrm2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini M.D., Wu M., Madhani H.D. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 112:725–736 10.1016/S0092-8674(03)00123-5 [DOI] [PubMed] [Google Scholar]
- Millar C.B., Xu F., Zhang K., Grunstein M. 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20:711–722 10.1101/gad.1395506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 303:343–348 10.1126/science.1090701 [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Terazawa Y., Nakayama T., Hirata A., Makio T., Endo T. 2003. Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J. Biol. Chem. 278:9938–9943 10.1074/jbc.M210934200 [DOI] [PubMed] [Google Scholar]
- Ohba T., Schirmer E.C., Nishimoto T., Gerace L. 2004. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 167:1051–1062 10.1083/jcb.200409149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E.T., Mastronarde D.N., Giddings T.H., Jr, Winey M., Burke D.J., McIntosh J.R. 1997. Three-dimensional analysis and ultrastructural design of mitotic spindles from the cdc20 mutant of Saccharomyces cerevisiae. Mol. Biol. Cell. 8:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P., Jaspersen S.L., Miele A., Dekker J., Peterson C.L. 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 23:912–927 10.1101/gad.1782209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar V.C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A.A., Gotzmann J., Foisner R., Karakesisoglou I. 2005. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118:3419–3430 10.1242/jcs.02471 [DOI] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L.R., Gasser S.M. 2009. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 23:928–938 10.1101/gad.1787509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Blobel G. 1993. The first membrane spanning region of the lamin B receptor is sufficient for sorting to the inner nuclear membrane. J. Cell Biol. 120:631–637 10.1083/jcb.120.3.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. 2005. Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W., Springer, New York: 397–420 [Google Scholar]
- Soullam B., Worman H.J. 1995. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J. Cell Biol. 130:15–27 10.1083/jcb.130.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A. 2009. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 122:577–586 10.1242/jcs.037622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M.P. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- Straube K., Blackwell J.S., Jr, Pemberton L.F. 2010. Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions. Traffic. 11:185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Schober H., Gasser S.M. 2010. The budding yeast nucleus. Cold Spring Harb. Perspect. Biol. 2:a000612 10.1101/cshperspect.a000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y., Ungricht R., Rothballer A., Kiss A., Csucs G., Horvath P., Kutay U. 2010. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 29:2262–2275 10.1038/emboj.2010.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627 10.1038/35001009 [DOI] [PubMed] [Google Scholar]
- Winey M., Mamay C.L., O’Toole E.T., Mastronarde D.N., Giddings T.H., Jr, McDonald K.L., McIntosh J.R. 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129:1601–1615 10.1083/jcb.129.6.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.H., Alami S., Luk E., Wu C.H., Sen S., Mizuguchi G., Wei D., Wu C. 2005. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 12:1064–1071 10.1038/nsmb1023 [DOI] [PubMed] [Google Scholar]
- Wu W.H., Wu C.H., Ladurner A., Mizuguchi G., Wei D., Xiao H., Luk E., Ranjan A., Wu C. 2009. N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J. Biol. Chem. 284:6200–6207 10.1074/jbc.M808830200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharioudakis I., Gligoris T., Tzamarias D. 2007. A yeast catabolic enzyme controls transcriptional memory. Curr. Biol. 17:2041–2046 10.1016/j.cub.2007.10.044 [DOI] [PubMed] [Google Scholar]
- Zhang H., Richardson D.O., Roberts D.N., Utley R., Erdjument-Bromage H., Tempst P., Côté J., Cairns B.R. 2004. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol. Cell. Biol. 24:9424–9436 10.1128/MCB.24.21.9424-9436.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Roberts D.N., Cairns B.R. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 123:219–231 10.1016/j.cell.2005.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Feng H., Hansen D.F., Kato H., Luk E., Freedberg D.I., Kay L.E., Wu C., Bai Y. 2008. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat. Struct. Mol. Biol. 15:868–869 10.1038/nsmb.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J., Thakar A. 2008. H2A.Z: view from the top. Structure. 16:166–179 10.1016/j.str.2007.12.008 [DOI] [PubMed] [Google Scholar]