Sorting signal-mediated oligomerization and localization of the viral protein TGBp3 to curved ER tubules is essential for viral movement between cells in plants.
Abstract
Many plant RNA viruses use their nonstructural proteins to target and move through the cortical endoplasmic reticulum (ER) tubules within the plant intercellular junction for cell-to-cell spreading. Most of these proteins, including the triple-gene-block 3 protein (TGBp3) of Potexvirus, are ER membrane proteins. We previously showed that TGBp3 of the Bamboo mosaic potexvirus partitions into tubular subdomains of the ER in both yeast and plants, but the mechanism and physiological significance of this localization is unclear. Here, we demonstrate that a sorting signal present in TGBp3 is necessary and sufficient for its oligomerization and for targeting integral membrane proteins into puncta within curved ER tubules. Mutations in the TGBp3 sorting signal impair viral spread, and plants infected with viruses harboring these mutants were either asymptomatic or had reduced symptoms. Thus, we propose that Potexvirus use the sorting signal in TGBp3 to target infectious viral derivatives to cortical ER tubules for transmission through the intercellular junctions in plants.
Introduction
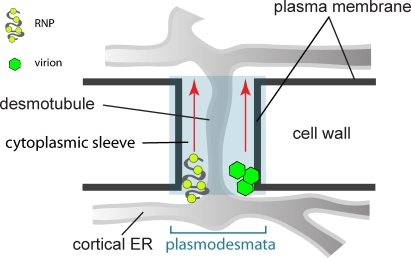
Viruses are intracellular pathogens that survive and multiply within host cells. For successful infection, viruses also adopt various strategies to spread from one cell to another. Animal viruses commonly use vesicular transport machineries in the secretory and endocytosis/exocytosis pathways for entry and exit (Pelkmans et al., 2001; Sieczkarski and Whittaker, 2002; Smith and Helenius, 2004; Greber and Way, 2006). However, plant viruses appear to spread via plasmodesmata, a communication channel between plant cells analogous to the gap junctions in animal cells. This specialized structure functions in selective transit of a wide range of macromolecules in plants, including RNAs and proteins (Overall and Blackman, 1996; Oparka, 2004). Primary plasmodesmata are formed during cytokinesis and subsequently undergo rearrangements to generate branches and central cavities to become secondary plasmodesmata (Ding et al., 1992b; Ehlers and Kollmann, 2001). The cortical ER tubules in the plasmodesmata, termed desmotubules, are separated from the plasma membrane by a cytoplasmic sleeve through which viral genome and other cellular signals are thought to pass and enter the adjacent cells (Fig. 1; Oparka, 2004; Lucas, 2006). Depending on the virus species, cell-to-cell movement may occur either in virion or nonvirion forms (Fig. 1). However, how plant viruses target to and move along the plasmodesmata remains poorly understood (Sanderfoot and Lazarowitz, 1996; Lucas, 2006; Hofmann et al., 2007).
Figure 1.
Schematic representation of plant plasmodesmata. Plasmodesmata are channels that connect two adjacent plant cells to each other, thus enabling viral infection to spread between cells. Plasmodesmata comprise the plasma membrane extension, cytoplasmic sleeve, and desmotubule that is continuous with the cortical ER networks. Depending on virus species, viral cell-to-cell movement through plasmodesmata can occur in virion or RNP complex forms.
Plant viruses encode nonstructural movement proteins to facilitate their own movement (Waigmann et al., 2004). The 30-kD protein (p30) of Tobacco mosaic virus (TMV) is the best known example, and studies of it have provided a paradigm for understanding viral spread in plants (Nishiguchi et al., 1978; Holt and Beachy, 1991; Hofmann et al., 2007). Many movement proteins, including p30, are able to modulate the size exclusion limits of the plasmodesmata by yet unknown mechanisms (Wolf et al., 1989; Howard et al., 2004). Although movement proteins of different plant viruses have distinct features, many share the ability to bind and transport together with the viral genome to the plasmodesmata (Waigmann et al., 2004). Intriguingly, most movement proteins are integral membrane proteins (Lucas, 2006), and many appear to localize in the ER. The highly dynamic plant ER is distributed throughout the cytoplasm (Staehelin, 1997) and may provide a track for efficiently moving macromolecules by lateral diffusion. Indeed, the studies of p30 suggest that this protein traffics to the plasmodesmata by lateral diffusion along the ER with the aid of actin and microtubule cytoskeletons (Boyko et al., 2000; Gillespie et al., 2002; Wright et al., 2007; Sambade et al., 2008; Christensen et al., 2009). Like most RNA viruses, TMV replicates in the ER (Más and Beachy, 1999). The ER association of p30 enables the sequestration of viral genome into a movement-competent ribonucleoprotein (RNP) complex, which is then transported to the desmotubules for intercellular viral transmission (Niehl and Heinlein, 2011). However, details of the mechanisms underlying these processes remain to be elucidated.
Movement proteins of viruses with the triple-gene-block (TGB) organization, such as Potexvirus, are also thought to target the plasmodesmata by association with the ER (Morozov and Solovyev, 2003; Verchot-Lubicz et al., 2010). The Potexvirus genome contains five open reading frames (ORFs), which encode the replicase, the three TGB movement proteins, and the capsid protein (CP), respectively (Fig. S5; Morozov and Solovyev, 2003). Spreading of Potexvirus requires all three TGB proteins and CP (Chapman et al., 1992; Forster et al., 1992), but it remains unclear whether viral cell-to-cell movement occurs in the form of virion or RNP complex (Cruz et al., 1998; Lough et al., 2000). The TGB 1 protein (TGBp1) is implicated in RNA binding, unwinding of the virion or RNP with its helicase activity (Morozov et al., 1999; Kiselyova et al., 2003), suppressing gene silencing of the host cells (Bayne et al., 2005; Senshu et al., 2009), and increasing plasmodesmata permeability during virus movement (Howard et al., 2004). The TGB 2 (TGBp2) and TGB 3 (TGBp3) proteins are small integral membrane proteins residing in the ER (Krishnamurthy et al., 2003; Mitra et al., 2003; Ju et al., 2005). Although these membrane proteins are difficult to study and many results are inconclusive (Verchot-Lubicz et al., 2010), most studies support the finding that TGBp3 targets TGBp2 and forms peripheral puncta in close proximity to the plasmodesmata (Solovyev et al., 2000; Schepetilnikov et al., 2005). Thus, TGBp2 and TGBp3 likely provide the membrane anchor for targeting Potexvirus to the plasmodesmata. The formation of peripheral puncta by TGBp3 is independent of the cytoskeletons in plants (Schepetilnikov et al., 2005, 2008), which implies a unique machinery for targeting Potexvirus to the plasmodesmata.
To better understand the nature and importance of these peripheral puncta formed during potexviral infection, we previously established a yeast system that recapitulates targeting of Bamboo mosaic potexvirus (BaMV) TGBp2 by TGBp3 to the peripheral puncta and demonstrated that TGBp3 binds to TGBp2 by forming a stoichiometric protein complex in the ER (Lee et al., 2010). In eukaryotic cells, including yeast, the ER is organized into flat membrane sheets and highly curved tubules (Voeltz et al., 2002; Shibata et al., 2006). Our results revealed that the peripheral puncta of TGBp3 in both yeast and plants are associated exclusively with tubular subdomains of the cortical ER marked by the ER-shaping protein reticulons and DP1/Yop1 (Lee et al., 2010). This feature is unique in light of the fact that most ER proteins characterized to date seem to move freely between the perinuclear ER sheets and the cortical ER that contains mostly tubules. It remains to be determined whether the partitioning mechanism for sorting TGBp3 into highly curved tubules of the cortical ER is crucial for potexviral movement. In this study, we have dissected the sorting requirements for targeting TGBp3 to the cortical ER tubules in both yeast and plants. Our study reveals a hydrophobic stretch in the cytoplasmic region of TGBp3 that serves as a signal for the oligomerization and sorting of TGBp3 and is essential for viral pathogenesis. Thus, our results support a model in which the plant Potexvirus uses the mechanism of sorting TGBp3 into the highly curved tubules of the cortical ER for viral cell-to-cell transmission.
Results
Identification of TGBp3 residues required for localization to cortical ER tubules
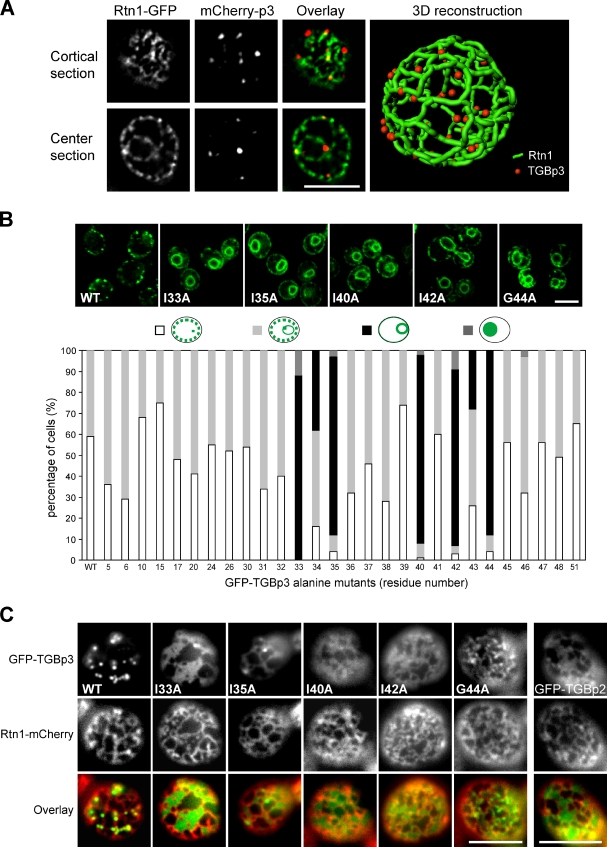
We recently demonstrated that TGBp3 of the BaMV recruits TGBp2 to the peripheral bodies near the cell cortex in yeast and plants (Lee et al., 2010). We also showed that these peripheral puncta are localized in cortical ER tubules labeled by the ER-shaping protein reticulons (Rtn1 and Rtn2) and DP1/Yop1 (Lee et al., 2010). 3D reconstruction results revealed that discrete TGBp3 puncta in yeast were embedded within cortical ER ribbons labeled by Rtn1 (Fig. 2 A and Video 1). About 40% of these puncta were localized at three-way junctions, and most of the remainder were associated with curved tubules (Fig. S1). Thus, the TGBp3 puncta appear to be stabilized at microdomains of the cortical ER tubules, preferably in curved areas.
Figure 2.
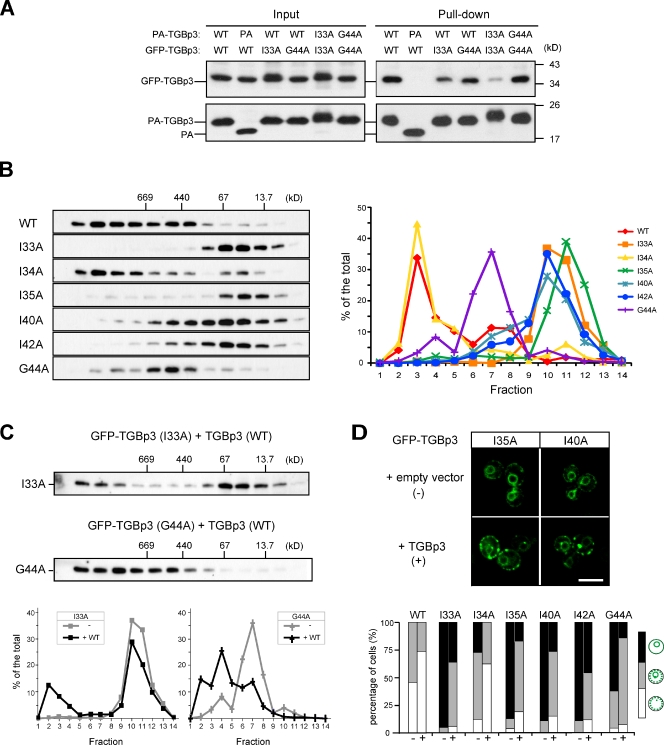
Several hydrophobic residues in the C-terminal region of TGBp3 are crucial for localization. (A) A yeast cell (CWY2765) expressing Rtn1-GFP and mCherry-TGBp3 was imaged for 3D reconstruction. (B) Localization of the indicated GFP-TGBp3 alanine mutants and quantification. (C) Peripheral view of yeast cells coexpressing Rtn1-mCherry and the indicated GFP-TGBp3 mutants or GFP-TGBp2. Bars, 5 µm.
To elucidate the mechanisms responsible for TGBp3 targeting, we performed alanine scanning mutagenesis of the entire TGBp3 sequence, followed by a visual screen for mutations that cause mislocalization of TGBp3 in yeast. Most of the alanine mutants were stably expressed at levels similar to that of the wild-type TGBp3 (Fig. S2 A). Confocal microscopy and quantification of subcellular localization revealed that mutations in the C-terminal region of TGBp3 affecting the hydrophobic residues I33A, I35A, I40A, I42A, and G44A no longer formed puncta in the cortical ER. Instead, they exhibited pronounced perinuclear ER localization, with some diffuse signals found near the cell cortex (Fig. 2 B). A peripheral view of the cells revealed that, except for G44A, most of these TGBp3 mutants partitioned into sheets of the cortical ER, as was also the case with GFP-TGBp2 (Fig. 2 C). Because TGBp3 first associates with the perinuclear ER before sorting into cortical ER tubules via the ER network (Lee et al., 2010), these results indicate that the C-terminal region of TGBp3 likely contains a sorting signal specifying cortical ER localization. These sorting-defective mutants interacted with TGBp2 at levels similar to the wild type (Fig. S2 B), which implies that the TGBp2 contact residues in TGBp3 may locate in other regions. In addition, mutants that lost the ability to form puncta also failed to sort to curved tubules, which suggests that the two processes may be coupled.
The cytoplasmic tail of TGBp3 is necessary and sufficient for targeting integral membrane proteins to cortical ER tubules
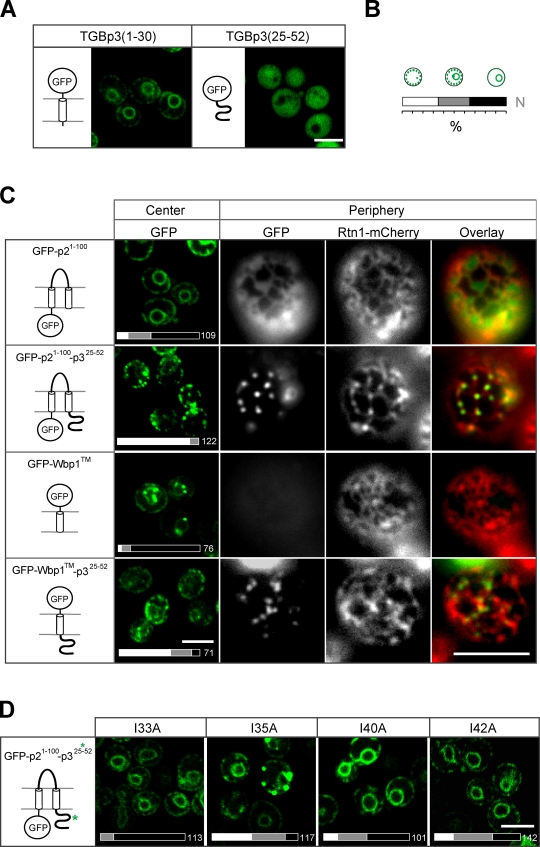
Because the critical mutations of TGBp3 reside in the C-terminal region, we next tested whether removal of this region alters TGBp3 localization. GFP-TGBp3 (amino acids 1–30) localized to the perinuclear ER (Fig. 3 A), which suggests that the N-terminal region of TGBp3 that contains a putative transmembrane domain is sufficient for localization to the perinuclear ER, but not cortical ER tubules. However, expressing the TGBp3 C-terminal region alone (amino acids 25–52) resulted in cytoplasmic localization (Fig. 3 A), which implies that the C-terminal region of TGBp3 may require a transmembrane domain to localize to cortical ER tubules. To test this notion directly, we examined whether the TGBp3 C-terminal region can confer localization to cortical ER tubules for the transmembrane domains of GFP-TGBp2 (amino acids 1–100), which alone is enriched at the perinuclear ER. GFP-TGBp2 (1–100) fused with the TGBp3 C-terminal region formed peripheral puncta residing within tubules labeled by Rtn1-mCherry (Fig. 3 C). Similarly, the TGBp3 C-terminal region relocalized the GFP-tagged transmembrane domain of yeast Wbp1 from the perinuclear ER to peripheral puncta (Fig. 3 C). Furthermore, TGBp2 (1–100) fused with the C-terminal region of TGBp3 sorting mutants failed to efficiently form peripheral puncta (Fig. 3 D). Together, these data support our hypothesis that the TGBp3 C-terminal region contains a putative sorting signal that is necessary and sufficient to localize transmembrane proteins to tubular subdomains of the ER.
Figure 3.
The C-terminal region of TGBp3 contains the sorting determinant for forming puncta within cortical ER tubules. (A) GFP fused to the N-terminal (amino acids 1–30) and C-terminal (25–52) regions of TGBp3 expressed in yeast (SEY6210) for fluorescence microscopy. (B) Quantification scheme for various subcellular localizations shown in this study. N indicates the total number of cells counted. (C) Hybrid constructs as indicated were expressed in yeast cells and imaged by confocal microscopy (center) or coexpressed with Rtn1-mCherry (CWY2754) and imaged by fluorescence microscopy (periphery). (D) The indicated residues in TGBp3 were mutated from the GFP-p21-100-p325-52 construct. Yeast cells expressing these proteins were imaged by confocal microscopy. The bars on the bottom indicate quantification data. Bars, 5 µm.
The sorting signal of TGBp3 is a conserved feature of Potexvirus
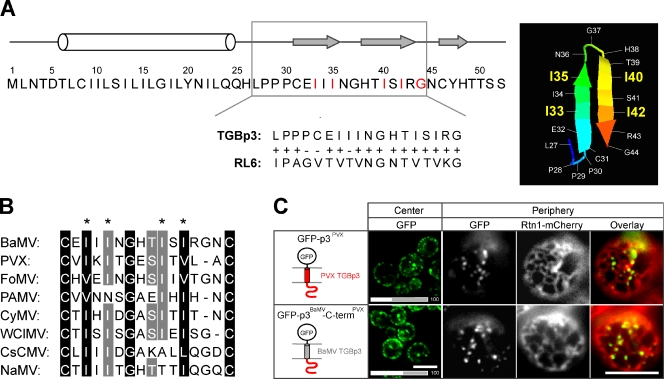
We next predicted the secondary and tertiary structures of the C-terminal regions of the potexviral TGBp3. According to the algorithm at the PSIPRED server (McGuffin et al., 2000), this region contains three consecutive β sheets (Fig. 4 A). The first two β sheets may fold into a hairpin structure that is most similar to a peptide within Geobacillus thermodenitrificans ribosomal protein L6 (RL6; Golden et al., 1993) according to the algorithm at the PHYRE server (Fig. 4 A; Kelley and Sternberg, 2009). When the hairpin sequence in RL6 was substituted for the putative hairpin sequence in BaMV TGBp3, the hybrid protein, like TGBp3, formed puncta in cortical ER tubules (unpublished data), which implies that the sorting signal may adopt a hairpin fold.
Figure 4.
The critical residues in the sorting signal region of TGBp3 are conserved among the Potexviruses. (A, left) Full-length amino acid sequence and secondary structure of TGBp3 predicted by PSIPRED algorithm (black line, coil; white bar, α helix; gray arrow, β strand; McGuffin et al., 2000). (A, right) The PHYRE algorithm (Kelley and Sternberg, 2009) predicts a hairpin structure in the TGBp3 sorting signal region based on RL6, a 50S ribosomal protein L6 of G. thermodenitrificans (available under GenBank/EMBL/DDBJ accession no. YP_001124252.1). The hairpin region of RL6 aligned with TGBp3 is shown below the TGBp3 sequence. (B) The predicted sorting signals flanked by two cysteine residues of several potexviral TGBp3 were aligned by GeneDoc. The asterisks indicate the four isoleucine residues crucial for BaMV TGBp3 localization. Viral sequences were acquired from the National Center for Biotechnology Information database: BaMV (accession no. NP_042586), PVX (accession no. AAF67821), FoMV (Foxtail mosaic virus, accession no. ABW25057), PAMV (Potato aucuba mosaic virus, accession no. NP_619749), CyMV (Cymbidium mosaic virus, accession no. ABO41880.1), WClMV (White clover mosaic virus, accession no. NP_620718), CsCMV (Cassava common mosaic virus, accession no. NP_042698), and NaMV (Nandina mosaic virus, accession no. AAX19934). (C) Yeast cells expressing GFP-tagged PVX TGBp3 and the hybrid BaMV TGBp3 whose C-terminal region was replaced with that of PVX were imaged by confocal microscopy (center) or coexpressed with Rtn1-mCherry (CWY2754) and imaged by fluorescence microscopy (periphery). The bars on the bottom indicate quantification data. Bars, 5 µm.
Sequence alignment results showed that the critical isoleucine residues at positions 33, 35, 40, and 42 of BaMV TGBp3 are highly conserved in Potexvirus (Fig. 4 B). To determine whether TGBp3 in Potexvirus other than BaMV has similar targeting mechanisms, we examined the localization of GFP-tagged TGBp3 of Potato virus X (PVX). GFP-TGBp3 of PVX, like BaMV, was targeted to the cortical ER tubules as labeled by Rtn1-mCherry in yeast (Fig. 4 C). Furthermore, the putative sorting signal of PVX TGBp3 can functionally replace the one in BaMV in targeting the hybrid protein to the cortical ER tubules (Fig. 4 C). Thus, the sorting signal is likely a common strategy for the localization of potexviral TGBp3 proteins to the curved ER microdomains.
We next expressed the hybrid constructs shown in Figs. 3 and 4 together with an additional plasmid expressing mCherry-TGBp3 of BaMV or PVX and examined whether the peripheral puncta colocalize. Indeed, all hybrid constructs with the sorting signals originally from BaMV TGBp3 colocalized with BaMV mCherry-TGBp3, with Pearson’s colocalization coefficients of ∼0.45, which is similar to the control with BaMV GFP-TGBp3 (Fig. S3 A). In contrast, all hybrid constructs with the sorting signals originally from PVX colocalized with PVX mCherry-TGBp3 (Fig. S3 B) but not with BaMV mCherry-TGBp3 (Fig. S3 A). Thus, the sorting signals from different Potexvirus may have distinct features, but nevertheless are conserved in promoting ER tubule localization.
The sorting signal triggers oligomerization and partitioning of TGBp3 into highly curved ER tubules
Our previous studies established that TGBp3 undergoes self-interaction and oligomerization in yeast cells (Lee et al., 2010). Because the TGBp3 sorting mutants showed diffuse ER localization (Fig. 2), we asked whether their defects might be caused by changes in biochemical characteristics. We fused various TGBp3 mutants with a protein A (PA) tag and tested their association with wild-type or mutant TGBp3 fused with a GFP tag. A PA pull-down assay revealed that the self-interactions of the I33A mutant were greatly reduced compared with that of the wild type (Fig. 5 A). The I33A mutant bound better with the wild type than with itself (Fig. 5 A). However, the G44A mutation did not have a significant effect on self-interaction or interaction with the wild-type TGBp3 (Fig. 5 A). Gel filtration analyses showed that wild-type TGBp3 was mostly fractionated with a size larger than half a megadalton. The I34A mutant, which had a minor sorting defect, fractionated mostly into a larger size, with only a small portion below 67 kD, which may represent a monomer and/or dimer; however, all of the other sorting mutants fractionated only into a smaller size (Fig. 5 B). Intriguingly, the G44A mutant fractionated into a range between that of the wild type and the other sorting mutants, which suggests a different extent of oligomerization defect (Fig. 5 B). The separation of various TGBp3 proteins on gel filtration was not caused by the detergent Triton X-100, which forms large micelles, because similar results were obtained with DDM and CHAPS, which form moderate and much smaller micelles, respectively (unpublished data). Collectively, these results demonstrate that oligomerization defects correlate well with TGBp3 localization defects.
Figure 5.
Oligomerization facilitates the partitioning of TGBp3 to the cell periphery. (A) Yeast cells (SEY6210) expressing the proteins indicated were lysed, and the lysate (input) was subjected to pull-down by IgG Sepharose. Samples were analyzed by anti-GFP and anti-PA immunoblots. (B) Yeast cells (SEY6210) expressing various GFP-TGBp3 mutants were size-fractionated on a Superose 6 column. Fractions were analyzed by immunoblotting with anti-GFP antibody (left), and the results were quantified and plotted (right). The elution of molecular weight standards is indicated. (C) Same as B except that a nontagged TGBp3 was overexpressed in cells expressing I33A or G44A GFP-TGBp3. (C, bottom) The plots compare the fractionation profiles for I33A and G44A GFP-TGBp3 in B and C. (D) Various GFP-TGBp3 mutants were transformed with empty vector (−) or a vector overexpressing nontagged TGBp3 (+). The cells were imaged by confocal microscopy (top) and quantified (bottom). Bar, 5 µm.
If oligomerization into the megadalton size is required for TGBp3 partitioning into cortical ER tubules, forcing oligomerization of TGBp3 sorting mutants into larger complexes may partially rescue their sorting defect. We tested this hypothesis by overexpressing wild-type TGBp3 in cells containing I33A or G44A GFP-TGBp3, given that the wild-type TGBp3 interacts with I33A and G44A TGBp3 as suggested in Fig. 5 A. Gel filtration analysis showed that a substantial amount of I33A and G44A GFP-TGBp3 partitioned into the megadalton fractions in the presence of the nontagged TGBp3 (Fig. 5 C). Furthermore, addition of nontagged TGBp3, but not empty vector, enabled GFP-TGBp3 mutants to form peripheral puncta more efficiently (Fig. 5 D). Together, these results support a model in which the sorting signal triggers TGBp3 oligomerization to a highly ordered structure that functions in turn to sort TGBp3 into microdomains in cortical ER tubules.
TGBp3 sorting mutants are localized to ER sheets in plants
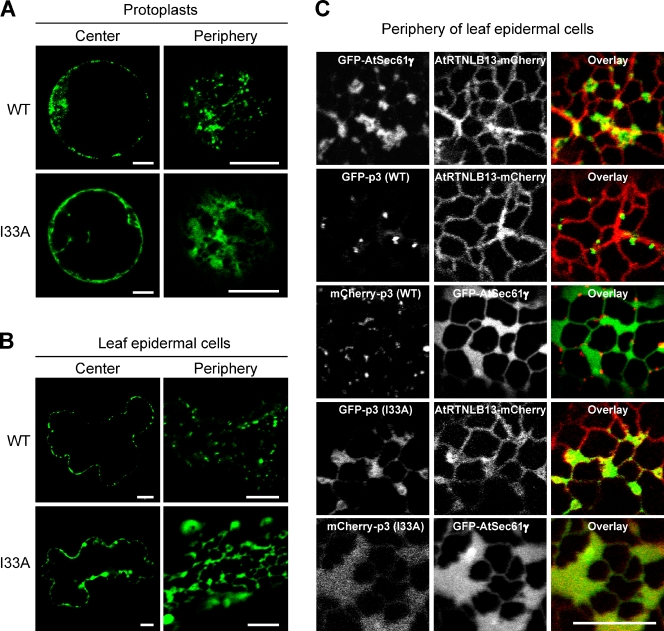
Next, we examined the localization of TGBp3 mutants in plants. Wild-type TGBp3 formed discrete puncta near the periphery of Nicotiana benthamiana protoplasts, whereas the I33A, I40A, I42A, and G44A mutants exhibited a diffuse ER localization along with occasional aggregated structures (Figs. 6 A and S4). The I35A mutant tended to aggregate more in the protoplasts for unknown reasons (Fig. S4). A peripheral view of wild-type GFP-TGBp3 expressed in the leaf epidermal cells of N. benthamiana revealed multiple puncta that exhibited a high mobility (Fig. 6 B and Video 2), whereas the I33A mutant appeared as patches that may represent ER sheets near the cell periphery (Fig. 6 B and Video 3).
Figure 6.
Wild-type but not sorting-defective TGBp3 forms puncta at highly curved ER tubules in N. benthamiana. (A) Protoplasts of N. benthamiana expressing the wild-type or I33A form of GFP-TGBp3 were imaged by confocal microscopy at 8 h after transformation. The center and peripheral sections are shown. (B) Same as A, except that leaf epidermal cells of N. benthamiana were imaged at 6 h after transformation. (C) N. benthamiana epidermal cells expressing various TGBp3 and ER markers (AtRTNLB13, the ER tubule marker; AtSec61γ, the ER sheet marker) were imaged by confocal microscopy. Bars, 10 µm.
To mark ER structures, we showed that the Arabidopsis thaliana AtSec61γ formed patches that were distinct from the network structures labeled by the A. thaliana reticulon isoform AtRTNLB13 (Tolley et al., 2008; Sparkes et al., 2009) when expressed in N. benthamiana (Fig. 6 C). Indeed, wild-type TGBp3 puncta were localized within the highly curved AtRTNLB13-containing tubules. In contrast, the I33A mCherry-TGBp3 mutant colocalized with GFP-AtSec61γ, which is indicative of ER sheets (Fig. 6 C). Thus, the localization of TGBp3 mutants in plants was consistent with our observations in yeast.
TGBp3 sorting mutants impair viral cell-to-cell movement
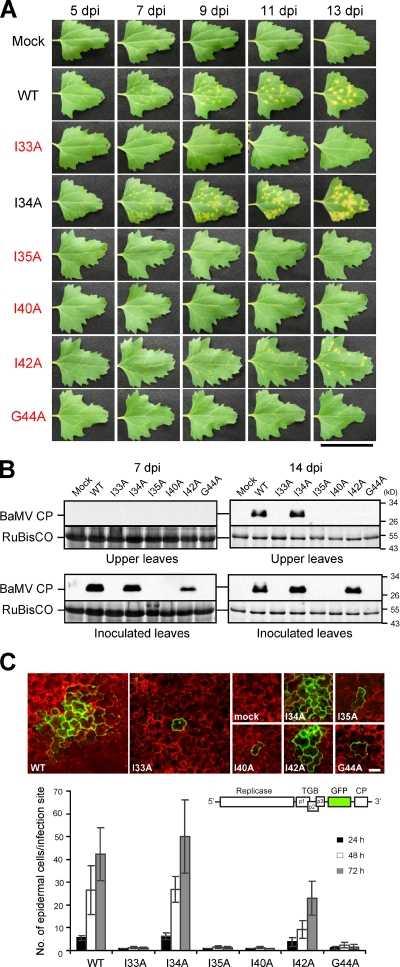
We next asked whether localization of TGBp3 to ER tubules is important for viral pathogenesis. For this purpose, we inoculated a common weed, Chenopodium quinoa, which supports local BaMV spread and elicits an extensive yellow chlorotic lesion phenotype. In this movement assay, wild-type BaMV spread and formed easily visible lesions, whereas mutant viruses containing the TGBp3 sorting mutants had greatly reduced lesion sizes or failed to develop lesions (Fig. 7 A). Consistent with the cytological (Fig. S4) and biochemical (Fig. 5) results, the I34A mutant behaved differently from those with mutations at the adjacent residues I33A and I35A (Fig. 7 A). Measurement of the necrotic lesion expansion rate also showed that the I34A mutant is similar to the wild type (unpublished data). In yeast, this mutant is partially defective in localization (Fig. 2 B); however, the defect can be easily rescued by TGBp2 expression in yeast (unpublished data), which indicates that the mutation has only moderately debilitating effects.
Figure 7.
TGBp3 sorting mutants are defective in viral cell-to-cell movement. (A) Leaves of C. quinoa inoculated with various BaMV viruses were photographed on the days post-inoculation (dpi) as indicated. The critical residues for TGBp3 localization are marked in red. Bar, 5 cm. (B) Various viruses were used to inoculate N. benthamiana plants. The inoculated and upper non-inoculated leaves were harvested and lysed at 7 and 14 dpi. Samples were subjected to immunoblot analysis with anti-BaMV CP antibody. Coomassie blue staining for the plant RuBisCO protein served as a loading control. (C) Various BaMV-GFP viruses harboring the TGBp3 mutants indicated were used to inoculate C. quinoa. At 24, 48, and 72 h after inoculation, at least 15 foci on each inoculated leaf were imaged by confocal microscopy. Images show foci that were selected by random at 48 h after inoculation with mock or the indicated viruses. The number of epidermal cells in each infection focus were counted, and data are plotted as mean ± SD (error bars). The scheme of BaMV-GFP reporter virus is depicted above the plot. The red fluorescence was from autofluorescence of plant chloroplasts. Bar, 50 µm.
Unlike C. quinoa, the tobacco host, N. benthamiana, fails to develop chlorotic lesions, but instead supports systemic infections and develops a systemic mosaic phenotype after BaMV inoculation (Yeh et al., 1999). Upon inoculations with Potexvirus lacking TGBp3, the infections are restricted to the initially infected cells without readily detectable amounts of virus in inoculated leaves (Cruz et al., 1998; Lough et al., 2000). We found that wild-type and I34A viruses produced a similar level of viral CP in the total cell lysates prepared from inoculated and upper non-inoculated leaves of N. benthamiana (Fig. 7 B), which indicates that the viruses were able to spread. For the I42A mutant, CP was also detectable, but only in the inoculated leaves (Fig. 7 B), which implies that the I42A virus was partially defective in cell-to-cell spread and failed to move systemically. Consistent with the results in C. quinoa, viral CP signals were not detected after infection with the I33A, I35A, I40A, and G44A viruses (Fig. 7 B). To exclude defects in replication, we confirmed that the TGBp3 sorting mutants did not affect viral replication at the cellular level in inoculated N. benthamiana protoplasts as examined by Northern blot analysis (Fig. S5). Thus, we conclude that the TGBp3 sorting signal is essential for viral pathogenesis.
To directly assay viral cell-to-cell movement, we constructed a variant BaMV clone that expressed GFP under an additional viral subgenomic promoter that was added after the TGBp3 ORF (termed BaMV-GFP hereafter). Infection with the BaMV-GFP clone elicited lesions in C. quinoa (unpublished data), indicating that the GFP insertion does not compromise movement substantially. Therefore, the spread of GFP fluorescence could be used to provide a direct measurement of viral infection and movement. By confocal microscopy, we found that the GFP fluorescence appeared in the initial infection foci and a few cells around the foci in C. quinoa leaves by 24 h after inoculation with wild-type and I34A BaMV-GFP, which indicates that viral cell-to-cell movement had been initiated. In contrast, leaves inoculated with the BaMV-GFP harboring TGBp3 sorting mutants, such as I33A, had restricted movement of GFP fluorescence from the initial infection foci at 24, 48, and 72 h after inoculation (Fig. 7 C), as expected of defects in cell-to-cell movement. Quantification results showed that all of the sorting-defective mutants were restricted or reduced in cell-to-cell spread as compared with the wild-type reporter virus (Fig. 7 C). Together, our data show that the TGBp3 sorting signal is crucial for cell-to-cell movement of Potexvirus.
Interactions of potexviral TGB and CP proteins in vivo provide a model for cell-to-cell movement
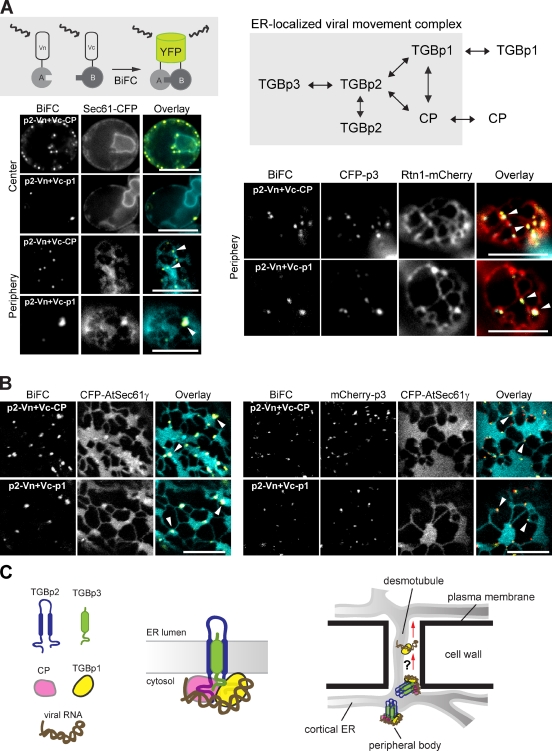
Our results are in agreement with others that TGBp3 is required for targeting its viral partner TGBp2 to the plasmodesmata (Schepetilnikov et al., 2005, 2008). To transport the viral genome properly, a TGBp2–TGBp3 complex has been speculated to bind the viral RNA directly or through interactions with other viral proteins that contain RNA-binding ability (Solovyev et al., 2000; Verchot-Lubicz et al., 2007). Most interactions among the potexviral TGB proteins have been studied by yeast two-hybrid analyses, which may have limitations in identifying positive interactions for membrane-associated movement proteins (Samuels et al., 2007). Thus, we used a bimolecular fluorescence complementation (BiFC) assay, which is considered one of the most sensitive tools to unravel protein–protein interactions in terms of their sites of action (Sung and Huh, 2007), and tested all of the BaMV proteins for potential BiFC interactions in yeast (Fig. 8 A).
Figure 8.
Viral RNA-binding proteins, TGBp1 and CP, interact with TGBp2 in the ER. (A, top left) The scheme of a BiFC assay. Vn tag was fused to a protein of interest termed A, and Vc tag was fused to protein B. The formation of a complex between A and B brings Vn and Vc in close enough proximity to fold into a fluorescent YFP (Venus). (A, top right) Positive interactions among BaMV proteins by BiFC. (A, bottom left) Images of center or periphery of yeast cells (CWY4128) expressing Sec61-CFP and BiFC constructs indicated by fluorescence microscopy. White arrowheads denote the BiFC-positive signals on ER sheets. (A, bottom right) Peripheral views of yeast cells (CWY4093) expressing the Rtn1-mCherry, CFP-TGBp3, and BiFC constructs indicated were imaged by fluorescence microscopy. White arrowheads denote the BiFC-positive signals on ER tubules. Bars, 5 µm. (B, left) Peripheral views of N. benthamiana cells expressing the CFP-AtSec61γ and BiFC constructs indicated were imaged by confocal microscopy. White arrowheads denote the BiFC-positive signals on ER sheets. (B, right) Peripheral views of N. benthamiana cells expressing the mCherry-TGBp3, CFP-AtSec61γ, and BiFC constructs indicated were imaged by confocal microscopy. White arrowheads denote the BiFC-positive signals on ER tubules. Bars, 10 µm. (C) A model for the potexviral RNP complex. TGBp3 forms a stoichiometric complex with TGBp2, which interacts with two RNA-binding proteins (TGBp1 and CP) in the ER. The sorting signal in TGBp3 directs the targeting of the RNP to highly curved ER structures near plasmodesmata for viral cell-to-cell movement. Whether the viral RNA is delivered to the neighboring cells with RNP or only with TGBp1 is unclear.
Besides demonstrating interaction of TGBp2 and TGBp3 (unpublished data), we found that two RNA-binding proteins, CP and TGBp1, exhibited strong BiFC signals with TGBp2, which indicates that they interacted directly with TGBp2 (Fig. 8 A). In addition, TGBp1 and CP showed self-interaction and interaction with each other. In contrast, all of the controls showed very weak if any fluorescent signals (unpublished data). BaMV CP is localized to the cytoplasm and nucleus, whereas TGBp1 often aggregates and shows rod-like structures in the cytoplasm (unpublished data). When Sec61-CFP was used as an ER sheet marker, we found that the BiFC interactions of TGBp2-CP and TGBp2-TGBp1 were largely associated with the ER sheets (Fig. 8 A), which is consistent with the localization of TGBp2 (Fig. 2 C). Importantly, coexpression of CFP-TGBp3 translocated the BiFC signals to CFP-TGBp3 puncta within ER tubules labeled by Rtn1-mCherry (Fig. 8 A). Furthermore, BiFC interactions of TGBp2-CP and TGBp2-TGBp1 in N. benthamiana appeared at cortical ER sheets marked with CFP-AtSec61γ (Fig. 8 B), which is consistent with our yeast results. In addition, mCherry-TGBp3 targeted the BiFC signals to ER tubules and curved ER regions at the borders of the ER sheets in plants (Fig. 8 B). Thus, CP and TGBp1 are likely cargoes that associate with the TGBp2-TGBp3 movement module by interacting with TGBp2 in the ER. Together, these data lead us to propose the existence of an ER-localized potexviral RNP complex comprised of TGBp1, TGBp2, TGBp3, CP, and viral RNA that functions in cell-to-cell movement (Fig. 8 C).
Discussion
The mechanisms by which viral movement proteins sequester viral contents, either virion or RNP, and translocate them through plasmodesmata to the adjacent plant cells are not resolved. Viral movement proteins and some unknown auxiliary factors likely work in concert during these processes. Emerging evidence suggests that many viral movement proteins are associated with the plant ER, which implies that molecules involved in the basic structure and function of the ER may play pivotal roles during plant virus movement. The potexviral nonstructural protein TGBp3 is a unique protein for studying the mechanisms underlying virus movement. This small protein serves as a membrane anchor to recruit its viral partner TGBp2 along the ER to form puncta near the cell cortex (Morozov and Solovyev, 2003). The most intriguing feature of TGBp3 is that the peripheral puncta formed by TGBp3 in both yeast and plants localized exclusively to highly curved tubules of the cortical ER (Fig. 2 A; Lee et al., 2010).
In this study, we identified a novel sorting signal in TGBp3 that sorts proteins into highly curved ER tubules. The hydrophobic residues within the cytoplasmic region of TGBp3 are important for partitioning the protein into microdomains of cortical ER tubules in yeast (Fig. 2). Interestingly, this portion of TGBp3 is necessary and sufficient for the delivery of unrelated transmembrane domains to ER tubules (Fig. 3), which supports it containing a functional sorting signal. Structural prediction implies a hairpin fold in the sorting signal region (Fig. 4). Sequence alignment results and protein domain swapping experiments further suggested that other Potexvirus likely also use this type of sorting signal (Figs. 4 and S3). Biochemical analysis indicated that the signal is involved in protein oligomerization and that the degree of oligomerization is associated with TGBp3 sorting (Fig. 5).
TGBp3 is localized within tubules marked by the ER-shaping protein Rtn1 and Yop1 (Lee et al., 2010). These proteins shape the ER through at least two mechanisms: membrane wedging by the hydrophobic residues that form hairpins and scaffolding by their interactions to form immobile oligomers in the ER (Voeltz et al., 2006; Hu et al., 2008; Shibata et al., 2008). Our analyses revealed that TGBp3 oligomerization mediated by the sorting signal forces the protein to be stabilized in the curved ER subdomains. The putative hairpin structure and the oligomerization-inducing ability may be similar to that of the ER-shaping proteins, although unlike those molecules, TGBp3 cannot shape the ER (unpublished data). In addition, our yeast two-hybrid and protein pull-down analyses revealed that TGBp3 did not interact with yeast Rtn1 and Yop1 (unpublished data), which is consistent with the result that TGBp3 puncta appear to be localized within but are not perfectly colocalized with the tubules labeled by Rtn1 in the 3D reconstruction model (Figs. 2 A and S1). These results, together with the finding that TGBp3 expression did not change the cortical ER network and other ER-related functions in yeast (unpublished data), led us to conclude that the biochemical characteristics that are similar to those of the ER-shaping proteins allow TGBp3 to remain in close association with the highly curved ER subdomains.
The ER is a single organelle with similar functions and structures in diverse organisms (Voeltz et al., 2002; Shibata et al., 2006). The perinuclear ER of metazoans surrounds the nucleus and extends into the peripheral ER sheets, which gradually turn into tubules with three-way junctions near the cell periphery (Terasaki and Jaffe, 1991). In yeast, the peripheral ER is mostly at the cell cortex, termed the cortical ER, with a few tubules connected to the perinuclear ER (Prinz et al., 2000). Although plant systems are more complex than yeast in many ways, the ER sheet and tubule organization is a conserved feature (Sparkes et al., 2009). We showed that, consistent with the conserved ER organization in both yeast and plant systems, wild-type TGBp3 is localized to ER tubules, and the sorting-defective TGBp3 mutants are distributed to ER sheets (Fig. 6). The curved ER largely associating with the cell periphery (Voeltz et al., 2002) explains at least in part why the viral TGBp3 is often found near the cell cortex. The ER tubules may provide an environment for TGBp3 to fold or assemble properly for its function. However, why and how the curved environment contributes to the motility of TGBp3 in plants remains unclear. Although actin and microtubule cytoskeletons are known to be involved in plant ER maintenance and dynamics, they are probably not involved in the dynamic movement of TGBp3 because the TGBp3 peripheral body formation was not affected in plants on treatment with actin and microtubule depolymerization agents (Schepetilnikov et al., 2008).
The localization studies in yeast and plant systems provide the foundation for investigating the functional significance of the sorting signal in TGBp3. By using sorting-defective TGBp3 mutant viruses, we demonstrated that the sorting signal and its ability to oligomerize are important for viral pathogenesis (Fig. 7). In addition, use of the BaMV-GFP reporter viruses, which allowed for direct measurement of cell-to-cell movement, clearly revealed that viruses with sorting-defective TGBp3 were restricted to single cells, which is indicative of strong defects in viral spread (Fig. 7 C). Thus, the sorting signal and its role in partitioning TGBp3 to the highly curved ER subdomain are crucial for TGBp3 function. Furthermore, our BiFC results revealed that TGBp2 interacts with two RNA-binding proteins of Potexvirus (Fig. 8), so TGBp3 likely also moves other viral components along the ER through complex formation with TGBp2. TGBp1 and CP are required for the cell-to-cell movement of Potexvirus (Cruz et al., 1998; Lough et al., 2000). TGBp1 is the only potexviral movement protein able to expand the size exclusion limits of plasmodesmata and may move together with viral RNA to the adjacent cells (Howard et al., 2004). Thus, we propose that TGBp2, which sequesters the viral RNA through interaction with TGBp1 and CP and forms RNP, is targeted by TGBp3 to microdomains in highly curved tubules of the cortical ER (Fig. 8 C). The movement-competent RNP may then move via the ER network toward the tubular ER in plasmodesmata (desmotubules; Fig. 8 C). The model explains why the sorting-defective mutants are defective in viral pathogenesis, given that they are not efficiently moved to the cell periphery, which is enriched in ER tubules, and cannot cross the cell boundary through desmotubules within the plasmodesmata.
In eukaryotic cells, tubules in the cortical ER network are generated and maintained by the ER-shaping proteins of the reticulon/Rtn1 and DP1/Yop1 families (Voeltz et al., 2006). However, little is known about the function of the reticulon-like molecules in plants, despite the existence of at least 20 homologues in A. thaliana (Sparkes et al., 2009). Desmotubules within the plasmodesmata are highly dynamic and are continuous with the cortical ER network in plants (Faulkner et al., 2008). Given that desmotubules undergo frequent remodeling (Tilney et al., 1991; Faulkner et al., 2008; Maule, 2008), it is possible that the ER-shaping proteins may be involved in plasmodesmata biogenesis. However, there is no evidence yet to support the presence of ER-shaping proteins in the plasmodesmata (Maule, 2008). Although we did not find a direct link between TGBp3 and the ER-shaping proteins, we cannot exclude that TGBp3 may couple with plasmodesmata biogenesis machinery or the reticulon-like molecules to move or deposit viral contents to adjacent cells (i.e., cell-to-cell movement). In support of this notion, various viral movement proteins are known to localize exclusively in the central cavities of the branched secondary plasmodesmata (Ding et al., 1992a; Lucas, 2006). Furthermore, our data show that sorting signals are likely present in other Potexvirus TGBp3 and that the sorting signal from different viral species selects for its own kind for oligomerization (Fig. S3), which implies that each sorting signal has a unique structural feature. This feature may ensure that TGBp3 selects for the right viral components in the cells that are infected with more than one type of virus.
In this study, we show that yeast can be used as a model system to identify cellular machinery associated with viral proteins for dissecting plant viral mechanisms. Most importantly, our findings reveal the importance of curved ER tubules for the function of a viral movement protein. Some cellular proteins with selective localization to tubular ER networks might also require the curved environment to exert their functions. This type of molecule, along with ER morphogenesis proteins, may have a function in cortical ER arrangements in general and plasmodesmata biogenesis in plant development.
Materials and methods
Strains, plasmids, and reagents
We used yeast strain backgrounds SEY6210 (MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2–801 suc2-Δ9) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) in this study. CWY2765 (BY4742 RTN1-GFP::HIS), CWY2754 (BY4742 RTN1-mCherry::HIS), CWY4093 (BY4742 RTN1-mCherry::KAN), and CWY4128 (BY4742 SEC61-CFP::KAN) were generated by use of a PCR-based transformation method (Longtine et al., 1998). The protein sequences of the hybrid constructs are summarized in Table S1. Restriction and modifying enzymes were obtained from Thermo Fisher Scientific. The IgG Sepharose 6 Fast Flow was purchased from GE Healthcare, Complete EDTA-free protease inhibitor mixture was obtained from Roche, and Cellulas RS, Macerozyme R10, and plant tissue culture reagents were from Duchefa. All other reagents were from Sigma-Aldrich unless otherwise mentioned. Oligonucleotides were synthesized by Invitrogen. Anti-PA antibody was from Jackson ImmunoResearch Laboratories, Inc. Anti-GFP antiserum (a gift of Y.-C. Chang, National Taiwan University, Taipei, Taiwan) was affinity-purified in our laboratory. Anti-CP antibody was raised in rabbits with GST-BaMV CP fusion protein used as an antigen. The wild-type BaMV infectious clones pBS2-8 and pCass-BaMV were gifts of N.-S. Lin (Academia Sinica, Taipei, Taiwan). The pBaMV-GFP infectious clone was made by duplicating the subgenomic promoter of CP with the addition of cloning sites where GFP was then inserted. All DNA sequencing was performed by the core laboratory at Academia Sinica.
Fluorescence microscopy
Yeast cells were grown in SC medium (0.67% yeast nitrogen base, amino acids, and 2% glucose). For 3D reconstruction of TGBp3 and Rtn1 localization in yeast, cells were grown to mid-log phase and attached to a slide coated with 200 µg/ml concanavalin A for microscopy. Z-section images of 100 slices spaced at 0.05 µm were captured by the DeltaVision system (Applied Precision) with a 100× objective lens (NA 1.4) and a CoolSNAPTM HQ2 charge-coupled device camera (Photometrics). Deconvolution was processed by use of SoftWoRx, followed by 3D reconstruction with use of Imaris 6.4 (Bitplane). To image yeast ER, we used an inverted microscope (PALM laser capture microdissection system; Carl Zeiss, Inc.) equipped with an EC Plan-Neofluar 100×/1.30 NA oil M27 lens and 47HE (CFP), 52 (GFP), 46HE (YFP), and 43 (mCherry) filter sets (all from Carl Zeiss, Inc.) for fluorescence microscopy. Images were acquired with an AxioCam MR3 camera and AxioVision software (Carl Zeiss, Inc.). For confocal microscopy, a microscope (LSM 510 Meta; Carl Zeiss, Inc.) equipped with a Plan-Apochromat 100×/1.4 NA oil lens was used for imaging yeast, and lenses of LCI Plan-Neofluar 63×/1.3 NA immersion, Plan-Apochromat 20×/0.8 NA, and Plan-Apochromat 10×/0.45 NA were used for imaging plants. Images were captured by use of LSM 510 v3.2 software (Carl Zeiss, Inc.) with filters for GFP (488 nm laser, BP500-530), mCherry (561 nm laser, BP575-630), YFP (514 nm laser, BP520-555), and CFP (458 nm laser, BP465-510). Images were processed by use of Photoshop (Adobe).
Gel filtration assay
Yeast spheroplast preparation and lysis were performed as described previously (Lee et al., 2010). The filtered supernatant was subjected to gel filtration analysis with use of the AKTA explorer FPLC system (GE Healthcare) and the Superose 6 HR 10/30 column under the control of UNICORN 4.11 software. PSS200 (20 mM Pipes, pH 6.8, 200 mM sorbitol, 100 mM KOAc, 50 mM KCl, and 1 mM MgCl2) containing 1% Triton X-100 was used as the running buffer. Fractions were collected every 0.5 ml and precipitated immediately with TCA to a final concentration of 10%. Protein samples were resuspended in MURB (50 mM sodium phosphate, 25 mM MES, pH 7.0, 1% SDS, 3 M urea, and 5% β-mercaptoethanol) and analyzed by 12.5% SDS-PAGE followed by immunoblotting with anti-GFP antibody.
PA pull-down assay
Cells were lysed by glass beads in PSS200 buffer supplemented with protease inhibitors, 1 mM PMSF, and 1% Triton X-100. The supernatant after centrifugation at 13,000 g for 10 min at 4°C was incubated with 50 µl IgG Sepharose 6 Fast-Flow beads (GE Healthcare) at 4°C for 2.5 h. After being washed six times in PSS200 with 1% Triton X-100 and 1 mM PMSF, the beads were then mixed with 100 µl MURB and boiled for 6 min to release bound proteins. Samples were analyzed by 12.5% SDS-PAGE followed by immunoblotting with anti-GFP and anti-PA antibodies.
Protein expression in protoplast and leaf epidermal cells of N. benthamiana
Protoplasts were isolated form N. benthamiana suspension cells treated with the protoplast buffer (1.2% Cellulase, 0.6% Macerozyme R-10, 10 mM CaCl2, 12 mM NaOAc, and 11% mannitol, pH 5.7) with mild shaking in the dark, followed by flotation centrifugations as described previously (Lee et al., 2008). For direct DNA uptake, 10 µg p35S-GFP-TGBp3 DNA was incubated with 100 µl protoplasts and 110 µl PEG solution, followed by addition of a 1-ml incubation buffer (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7; Lee et al., 2008). To express GFP-TGBp3 in leaf epidermal cells, 2.5 µg DNA was used to transform a 5-wk-old N. benthamiana leaf by particle bombardment (Lee et al., 2010). After transformation, protoplasts and leaves were incubated at 25°C. Images were acquired with use of a confocal microscope (LSM 510 Meta; Carl Zeiss, Inc.).
Virus replication assay
Protoplasts were isolated from 6-wk-old N. benthamiana leaves as described previously (Lee et al., 2010). In vitro transcription was performed in a 30-µl reaction containing T7 RNA polymerase and 1.2 µg pBS2-8 linearized with SacI. Protoplast inoculation and viral RNA purification were performed as described previously (Donald and Jackson, 1994), with minor modifications. In brief, ∼6 × 105 freshly prepared protoplasts and 28 µl BaMV in vitro transcripts were used for each inoculation. After 24 h, total RNA was isolated in extraction buffer (200 mM ammonium carbonate, pH 9, 2% SDS, 2 mM EDTA, and 200 µg/ml bentonite), followed by Northern blot analysis. A DIG-labeled DNA probe that recognizes BaMV CP sequences was prepared by a PCR reaction with pBS2-8 used as the template, and the detection procedure followed the manufacturer’s instructions (Roche).
Virus inoculation assay
N. benthamiana and C. quinoa plants were grown in a greenhouse at 27°C with a 16/8 h light/dark cycle. For virus inoculation, 1 µg wild-type or mutant pCass-BaMV DNA, with the viral RNA expressed under a 35S promoter, was used to inoculate 4-wk-old N. benthamiana or 5-wk-old C. quinoa leaves by a Cellite-dusted method. The lesions on inoculated C. quinoa leaves and a reference ruler were recorded daily by use of a digital camera, and photos were processed by ImageJ to measure the lesion size. To detect CP accumulation in N. benthamiana, inoculated leaves and upper noninoculated leaves were collected and ground into fine powder in liquid nitrogen. Every 1 g of leaves was mixed with 2 ml HK buffer (50 mM Hepes and 150 mM KCl, pH 7.4) and centrifuged at 5,000 rpm for 5 min to remove cell debris. The supernatants were precipitated by 10% TCA, washed twice with acetone, and resuspended in MURB for immunoblotting with anti-CP antibody. For the detection of BaMV-GFP spread, 2 µg of infectious pBaMV-GFP DNA was used to inoculate leaves of C. quinoa. The infection foci were imaged at 24, 48, and 72 h after inoculation with use of a confocal microscope (LSM 510 Meta). To quantify virus movement, the number of GFP-positive epidermal cells in 15 independent infection foci selected at random was counted.
BiFC assay
To test viral protein interactions by BiFC in yeast, we modified BiFC vectors that were provided by W.-K. Huh (Seoul University, Seoul, Korea; Sung and Huh, 2007). We generated various Vn and Vc constructs that were in frame with the 5′ or 3′ end of the BaMV ORFs. These constructs were transformed into yeast in combination with each other and the controls (Vn or Vc alone). By microscopy, positive BiFC interactions showed YFP fluorescence, in contrast to controls, which showed no or much reduced YFP fluorescence. For the BiFC experiments in N. benthamiana, we used 0.125 µg p35S-TGBp2-Vn, 0.125 µg p35S-Vc-TGBp1, 0.42 µg p35S-Vc-CP, 1.25 µg p35S-mCherry-TGBp3, and 2.5 µg p35S-CFP-AtSec61γ for transformation with particle bombardment to minimize backgrounds caused by protein overexpression. Transformed leaves were incubated at 25°C for 8–10 h and then visualized by confocal microscopy (LSM 510 Meta).
Online supplemental material
Fig. S1 shows the results of two experiments in which TGBp3 was localized to junctions and bended tubules labeled by Rtn1-GFP. Fig. S2 shows that TGBp3 sorting mutants were stably expressed and interacted with TGBp2 in yeast cells. Fig. S3 shows that TGBp3 from different Potexvirus formed distinct peripheral puncta. Fig. S4 shows the localization of TGBp3 sorting-defective mutants in plant protoplasts and leaf epidermal cells. Fig. S5 shows that viruses harboring wild-type or sorting-defective TGBp3 exhibited comparable replication rates. Video 1 shows the 3D reconstruction model of Rtn1-GFP and mCherry-TGBp3 localization in yeast. Video 2 shows a time-lapse movie of a N. benthamiana leaf epidermal cell expressing GFP-TGBp3 (wild-type). Video 3 shows a time-lapse movie of a N. benthamiana leaf epidermal cell expressing GFP-TGBp3 (I33A). Table S1 lists the sequences of hybrid proteins used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006023/DC1.
Acknowledgments
We give special thanks to Dr. Rey-Huei Chen (Institute of Molecular Biology, Academia Sinica) for sharing lab resources and providing invaluable scientific input, as well as critical reading of the manuscript. We are grateful to Drs. Wen-Chi Hu for technical assistance with virus inoculation assays, Ya-Chun Chang (National Taiwan University) for providing GFP antiserum, Dr. Na-Sheng Lin (Institute of Plant and Microbial Biology, Academia Sinica) for providing the BaMV clones, Lin-Yun Kuang (Institute of Plant and Microbial Biology, Academia Sinica) for help with particle bombardment, Dr. Won-Ki Huh (Seoul University) for the BiFC vectors, Dr. Roger Tsien (University of California, San Diego, La Jolla, California) for use of mCherry, and Shu-Chen Shen (Scientific Instrument Center, Academia Sinica) for technical assistance in confocal microscopy. We thank Sue-Ping Lee (Institute of Molecular Biology, Academia Sinica), Mei-Jane Fang (Institute of Plant and Microbial Biology, Academia Sinica), and members of the R.-H Chen laboratory, especially Li-Chuang Tseng and You-Liang Cheng for technical assistance in imaging; Dr. Tien-Hsien Chang (Genomics Research Center, Academia Sinica) for advise on writing; and Dr. Andy Jackson (University of California, Berkeley, Berkeley, California) for help with editing and improving the manuscript.
This research was supported by an internal research fund from IPMB, Academia Sinica, Taiwan. The authors declare no conflict of interest.
C.-W. Wang and C.-H. Wu designed research. C.-H. Wu, S.-C. Lee and C.-W. Wang performed experiments and analyzed data. C.-W. Wang wrote the paper.
Footnotes
Abbreviations used in this paper:
- BaMV
- Bamboo mosaic potexvirus
- BiFC
- bimolecular fluorescence complementation
- CP
- capsid protein
- ORF
- open reading frame
- PA
- protein A
- PVX
- Potato virus X
- TGB
- triple-gene-block
References
- Bayne E.H., Rakitina D.V., Morozov S.Y., Baulcombe D.C. 2005. Cell-to-cell movement of potato potexvirus X is dependent on suppression of RNA silencing. Plant J. 44:471–482 10.1111/j.1365-313X.2005.02539.x [DOI] [PubMed] [Google Scholar]
- Boyko V., Ferralli J., Ashby J., Schellenbaum P., Heinlein M. 2000. Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 2:826–832 10.1038/35041072 [DOI] [PubMed] [Google Scholar]
- Chapman S., Hills G., Watts J., Baulcombe D. 1992. Mutational analysis of the coat protein gene of potato virus X: effects on virion morphology and viral pathogenicity. Virology. 191:223–230 10.1016/0042-6822(92)90183-P [DOI] [PubMed] [Google Scholar]
- Christensen N., Tilsner J., Bell K., Hammann P., Parton R., Lacomme C., Oparka K. 2009. The 5′ cap of tobacco mosaic virus (TMV) is required for virion attachment to the actin/endoplasmic reticulum network during early infection. Traffic. 10:536–551 10.1111/j.1600-0854.2009.00889.x [DOI] [PubMed] [Google Scholar]
- Cruz S.S., Roberts A.G., Prior D.A., Chapman S., Oparka K.J. 1998. Cell-to-cell and phloem-mediated transport of potato virus X. The role of virions. Plant Cell. 10:495–510 10.1105/tpc.10.4.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Haudenshield J.S., Hull R.J., Wolf S., Beachy R.N., Lucas W.J. 1992a. Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell. 4:915–928 10.1105/tpc.4.8.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Turgeon R., Parthasarathy M.V. 1992b. Substructure of freeze-substituted plasmodesmata. Protoplasma. 169:28–41 10.1007/BF01343367 [DOI] [Google Scholar]
- Donald R.G., Jackson A.O. 1994. The barley stripe mosaic virus gamma b gene encodes a multifunctional cysteine-rich protein that affects pathogenesis. Plant Cell. 6:1593–1606 10.1105/tpc.6.11.1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers K., Kollmann R. 2001. Primary and secondary plasmodesmata: structure, origin, and functioning. Protoplasma. 216:1–30 10.1007/BF02680127 [DOI] [PubMed] [Google Scholar]
- Faulkner C., Akman O.E., Bell K., Jeffree C., Oparka K. 2008. Peeking into pit fields: a multiple twinning model of secondary plasmodesmata formation in tobacco. Plant Cell. 20:1504–1518 10.1105/tpc.107.056903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R.L., Beck D.L., Guilford P.J., Voot D.M., Van Dolleweerd C.J., Andersen M.T. 1992. The coat protein of white clover mosaic potexvirus has a role in facilitating cell-to-cell transport in plants. Virology. 191:480–484 10.1016/0042-6822(92)90215-B [DOI] [PubMed] [Google Scholar]
- Gillespie T., Boevink P., Haupt S., Roberts A.G., Toth R., Valentine T., Chapman S., Oparka K.J. 2002. Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of tobacco mosaic virus. Plant Cell. 14:1207–1222 10.1105/tpc.002303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden B.L., Ramakrishnan V., White S.W. 1993. Ribosomal protein L6: structural evidence of gene duplication from a primitive RNA binding protein. EMBO J. 12:4901–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber U.F., Way M. 2006. A superhighway to virus infection. Cell. 124:741–754 10.1016/j.cell.2006.02.018 [DOI] [PubMed] [Google Scholar]
- Hofmann C., Sambade A., Heinlein M. 2007. Plasmodesmata and intercellular transport of viral RNA. Biochem. Soc. Trans. 35:142–145 10.1042/BST0350142 [DOI] [PubMed] [Google Scholar]
- Holt C.A., Beachy R.N. 1991. In vivo complementation of infectious transcripts from mutant tobacco mosaic virus cDNAs in transgenic plants. Virology. 181:109–117 10.1016/0042-6822(91)90475-Q [DOI] [PubMed] [Google Scholar]
- Howard A.R., Heppler M.L., Ju H.J., Krishnamurthy K., Payton M.E., Verchot-Lubicz J. 2004. Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology. 328:185–197 10.1016/j.virol.2004.06.039 [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., Prinz W.A. 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 319:1247–1250 10.1126/science.1153634 [DOI] [PubMed] [Google Scholar]
- Ju H.J., Samuels T.D., Wang Y.S., Blancaflor E., Payton M., Mitra R., Krishnamurthy K., Nelson R.S., Verchot-Lubicz J. 2005. The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol. 138:1877–1895 10.1104/pp.105.066019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J.E. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Kiselyova O.I., Yaminsky I.V., Karpova O.V., Rodionova N.P., Kozlovsky S.V., Arkhipenko M.V., Atabekov J.G. 2003. AFM study of potato virus X disassembly induced by movement protein. J. Mol. Biol. 332:321–325 10.1016/S0022-2836(03)00835-0 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy K., Heppler M., Mitra R., Blancaflor E., Payton M., Nelson R.S., Verchot-Lubicz J. 2003. The Potato virus X TGBp3 protein associates with the ER network for virus cell-to-cell movement. Virology. 309:135–151 10.1016/S0042-6822(02)00102-2 [DOI] [PubMed] [Google Scholar]
- Lee L.Y., Fang M.J., Kuang L.Y., Gelvin S.B. 2008. Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods. 4:24 10.1186/1746-4811-4-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Wu C.H., Wang C.W. 2010. Traffic of a viral movement protein complex to the highly curved tubules of the cortical endoplasmic reticulum. Traffic. 11:912–930 10.1111/j.1600-0854.2010.01064.x [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lough T.J., Netzler N.E., Emerson S.J., Sutherland P., Carr F., Beck D.L., Lucas W.J., Forster R.L. 2000. Cell-to-cell movement of potexviruses: evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant Microbe Interact. 13:962–974 10.1094/MPMI.2000.13.9.962 [DOI] [PubMed] [Google Scholar]
- Lucas W.J. 2006. Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology. 344:169–184 10.1016/j.virol.2005.09.026 [DOI] [PubMed] [Google Scholar]
- Más P., Beachy R.N. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945–958 10.1083/jcb.147.5.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule A.J. 2008. Plasmodesmata: structure, function and biogenesis. Curr. Opin. Plant Biol. 11:680–686 10.1016/j.pbi.2008.08.002 [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. 2000. The PSIPRED protein structure prediction server. Bioinformatics. 16:404–405 10.1093/bioinformatics/16.4.404 [DOI] [PubMed] [Google Scholar]
- Mitra R., Krishnamurthy K., Blancaflor E., Payton M., Nelson R.S., Verchot-Lubicz J. 2003. The potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral cell-to-cell movement. Virology. 312:35–48 10.1016/S0042-6822(03)00180-6 [DOI] [PubMed] [Google Scholar]
- Morozov S.Y., Solovyev A.G. 2003. Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84:1351–1366 10.1099/vir.0.18922-0 [DOI] [PubMed] [Google Scholar]
- Morozov S.Y., Solovyev A.G., Kalinina N.O., Fedorkin O.N., Samuilova O.V., Schiemann J., Atabekov J.G. 1999. Evidence for two nonoverlapping functional domains in the potato virus X 25K movement protein. Virology. 260:55–63 10.1006/viro.1999.9788 [DOI] [PubMed] [Google Scholar]
- Niehl A., Heinlein M. 2011. Cellular pathways for viral transport through plasmodesmata. Protoplasma. 248:75–99 10.1007/s00709-010-0246-1 [DOI] [PubMed] [Google Scholar]
- Nishiguchi M., Motoyoshi F., Oshima N. 1978. Behaviour of a temperature sensitive strain of tobacco mosaic virus in tomato leaves and protoplasts. J. Gen. Virol. 39:53–61 10.1099/0022-1317-39-1-53 [DOI] [Google Scholar]
- Oparka K.J. 2004. Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci. 9:33–41 10.1016/j.tplants.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Overall R.L., Blackman L.M. 1996. A model of the macromolecular structure of plasmodesmata. Trends Plant Sci. 1:307–311 10.1016/S1360-1385(96)88177-0 [DOI] [Google Scholar]
- Pelkmans L., Kartenbeck J., Helenius A. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473–483 10.1038/35074539 [DOI] [PubMed] [Google Scholar]
- Prinz W.A., Grzyb L., Veenhuis M., Kahana J.A., Silver P.A., Rapoport T.A. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150:461–474 10.1083/jcb.150.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambade A., Brandner K., Hofmann C., Seemanpillai M., Mutterer J., Heinlein M. 2008. Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata. Traffic. 9:2073–2088 10.1111/j.1600-0854.2008.00824.x [DOI] [PubMed] [Google Scholar]
- Samuels T.D., Ju H.J., Ye C.M., Motes C.M., Blancaflor E.B., Verchot-Lubicz J. 2007. Subcellular targeting and interactions among the Potato virus X TGB proteins. Virology. 367:375–389 10.1016/j.virol.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Sanderfoot A.A., Lazarowitz S.G. 1996. Getting it together in plant virus movement: cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 6:353–358 10.1016/0962-8924(96)10031-3 [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M.V., Manske U., Solovyev A.G., Zamyatnin A.A., Jr, Schiemann J., Morozov S.Y. 2005. The hydrophobic segment of Potato virus X TGBp3 is a major determinant of the protein intracellular trafficking. J. Gen. Virol. 86:2379–2391 10.1099/vir.0.80865-0 [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M.V., Solovyev A.G., Gorshkova E.N., Schiemann J., Prokhnevsky A.I., Dolja V.V., Morozov S.Y. 2008. Intracellular targeting of a hordeiviral membrane-spanning movement protein: sequence requirements and involvement of an unconventional mechanism. J. Virol. 82:1284–1293 10.1128/JVI.01164-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senshu H., Ozeki J., Komatsu K., Hashimoto M., Hatada K., Aoyama M., Kagiwada S., Yamaji Y., Namba S. 2009. Variability in the level of RNA silencing suppression caused by triple gene block protein 1 (TGBp1) from various potexviruses during infection. J. Gen. Virol. 90:1014–1024 10.1099/vir.0.008243-0 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voeltz G.K., Rapoport T.A. 2006. Rough sheets and smooth tubules. Cell. 126:435–439 10.1016/j.cell.2006.07.019 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., Voeltz G.K. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283:18892–18904 10.1074/jbc.M800986200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieczkarski S.B., Whittaker G.R. 2002. Dissecting virus entry via endocytosis. J. Gen. Virol. 83:1535–1545 [DOI] [PubMed] [Google Scholar]
- Smith A.E., Helenius A. 2004. How viruses enter animal cells. Science. 304:237–242 10.1126/science.1094823 [DOI] [PubMed] [Google Scholar]
- Solovyev A.G., Stroganova T.A., Zamyatnin A.A., Jr, Fedorkin O.N., Schiemann J., Morozov S.Y. 2000. Subcellular sorting of small membrane-associated triple gene block proteins: TGBp3-assisted targeting of TGBp2. Virology. 269:113–127 10.1006/viro.2000.0200 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Frigerio L., Tolley N., Hawes C. 2009. The plant endoplasmic reticulum: a cell-wide web. Biochem. J. 423:145–155 10.1042/BJ20091113 [DOI] [PubMed] [Google Scholar]
- Staehelin L.A. 1997. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 11:1151–1165 10.1046/j.1365-313X.1997.11061151.x [DOI] [PubMed] [Google Scholar]
- Sung M.K., Huh W.K. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 24:767–775 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- Terasaki M., Jaffe L.A. 1991. Organization of the sea urchin egg endoplasmic reticulum and its reorganization at fertilization. J. Cell Biol. 114:929–940 10.1083/jcb.114.5.929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L.G., Cooke T.J., Connelly P.S., Tilney M.S. 1991. The structure of plasmodesmata as revealed by plasmolysis, detergent extraction, and protease digestion. J. Cell Biol. 112:739–747 10.1083/jcb.112.4.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley N., Sparkes I.A., Hunter P.R., Craddock C.P., Nuttall J., Roberts L.M., Hawes C., Pedrazzini E., Frigerio L. 2008. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic. 9:94–102 10.1111/j.1600-0854.2007.00670.x [DOI] [PubMed] [Google Scholar]
- Verchot-Lubicz J., Ye C.M., Bamunusinghe D. 2007. Molecular biology of potexviruses: recent advances. J. Gen. Virol. 88:1643–1655 10.1099/vir.0.82667-0 [DOI] [PubMed] [Google Scholar]
- Verchot-Lubicz J., Torrance L., Solovyev A.G., Morozov S.Y., Jackson A.O., Gilmer D. 2010. Varied movement strategies employed by triple gene block-encoding viruses. Mol. Plant Microbe Interact. 23:1231–1247 10.1094/MPMI-04-10-0086 [DOI] [PubMed] [Google Scholar]
- Voeltz G.K., Rolls M.M., Rapoport T.A. 2002. Structural organization of the endoplasmic reticulum. EMBO Rep. 3:944–950 10.1093/embo-reports/kvf202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Waigmann E., Ueki S., Trutnyeva K., Citovsky V. 2004. The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit. Rev. Plant Sci. 23:195–250 10.1080/07352680490452807 [DOI] [Google Scholar]
- Wolf S., Deom C.M., Beachy R.N., Lucas W.J. 1989. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science. 246:377–379 10.1126/science.246.4928.377 [DOI] [PubMed] [Google Scholar]
- Wright K.M., Wood N.T., Roberts A.G., Chapman S., Boevink P., Mackenzie K.M., Oparka K.J. 2007. Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: evidence from FRAP. Traffic. 8:21–31 10.1111/j.1600-0854.2006.00510.x [DOI] [PubMed] [Google Scholar]
- Yeh T.Y., Lin B.Y., Chang Y.C., Hsu Y.H., Lin N.S. 1999. A defective RNA associated with bamboo mosaic virus and the possible common mechanisms for RNA recombination in potexviruses. Virus Genes. 18:121–128 10.1023/A:1008008400653 [DOI] [PubMed] [Google Scholar]