FRAP reveals that a stable PAR boundary requires balancing diffusive flux of PAR proteins between domains with spatial differences in PAR protein membrane affinities.
Abstract
Polarization of cells by PAR proteins requires the segregation of antagonistic sets of proteins into two mutually exclusive membrane-associated domains. Understanding how nanometer scale interactions between individual PAR proteins allow spatial organization across cellular length scales requires determining the kinetic properties of PAR proteins and how they are modified in space. We find that PAR-2 and PAR-6, which localize to opposing PAR domains, undergo exchange between well mixed cytoplasmic populations and laterally diffusing membrane-associated states. Domain maintenance does not involve diffusion barriers, lateral sorting, or active transport. Rather, both PAR proteins are free to diffuse between domains, giving rise to a continuous boundary flux because of lateral diffusion of molecules down the concentration gradients that exist across the embryo. Our results suggest that the equalizing effects of lateral diffusion are countered by actin-independent differences in the effective membrane affinities of PAR proteins between the two domains, which likely depend on the ability of each PAR species to locally modulate the membrane affinity of opposing PAR species within its domain. We propose that the stably polarized embryo reflects a dynamic steady state in which molecules undergo continuous diffusion between regions of net association and dissociation.
Introduction
The PAR polarity pathway is broadly conserved and essential for many processes in metazoan development, including directed cell motility, asymmetric cell division, and the establishment of tissue architecture. It relies on two sets of antagonistic PAR (partitioning defective) proteins: one composed of PAR-3 (Bazooka), PAR-6, and atypical PKC (aPKC), and another characterized variably by PAR-1, LGL (Lethal Giant Larvae), and/or PAR-2 (Kemphues et al., 1988; Etemad-Moghadam et al., 1995; Boyd et al., 1996; Watts et al., 1996; Tabuse et al., 1998; Wodarz et al., 1999; Betschinger et al., 2003; Hoege et al., 2010). In polarized cells, these two sets of proteins are segregated such that the cell membrane is partitioned into stable, mutually exclusive membrane domains that define the polarity axis. Although progress has been made in identifying the molecular players and many of the interactions, the mechanisms by which these proteins maintain their asymmetric distributions within cells remain elusive.
To generate asymmetric distributions of molecules, cells must possess mechanisms to enrich molecules at defined locations, thereby countering the entropic forces that would otherwise tend to equalize protein concentrations across the cell. Budding yeast, for example, use a variety of such mechanisms, including diffusion barriers (Barral et al., 2000; Takizawa et al., 2000), active transport (Wedlich-Soldner et al., 2003), recruitment to preexisting landmarks or scaffolds (Chant et al., 1995), and self-organizing pattern-forming processes (Goryachev and Pokhilko, 2008; Kozubowski et al., 2008). Developing a physical picture for generating and maintaining cellular asymmetry requires detailed knowledge of the kinetic behaviors and mobilities of the proteins involved and how these are regulated in space.
The initial asymmetric stem cell–like division of the Caenorhabditis elegans embryo is an attractive system for studying PAR polarity (Cowan and Hyman, 2004; Munro and Bowerman, 2009). PAR polarity is essential for this asymmetric division and is established de novo in the single-cell embryo in a reproducible fashion after completion of meiosis II. As in other systems, PAR proteins segregate into two mutually exclusive domains, here anterior and posterior, separated by a stable boundary at midcell. The formation of a stable PAR boundary depends on mutual antagonism between the two sets of PAR proteins (Benton and St Johnston, 2003; Betschinger et al., 2003; Tanentzapf and Tepass, 2003; Chalmers et al., 2005; Hao et al., 2006). If the function of either set of PAR proteins is disrupted, the other set of proteins fail to be confined within their appropriate domain and instead spread throughout the embryo (Etemad-Moghadam et al., 1995; Boyd et al., 1996; Watts et al., 1996; Cuenca et al., 2003). Consistent with this notion of competitive inhibition, mutants in par-2 can often be rescued, at least partially, by depletion of PAR-3, PAR-6, or aPKC (Watts et al., 1996; Labbé et al., 2006). However, it is unclear how this antagonism maintains the asymmetric distribution of PAR proteins in the absence of persistent spatial cues with which to define the domain boundaries.
The actomyosin cortical meshwork has emerged as a strong candidate for playing an organizational role in polarity in a variety of systems. Notably, it is often polarized coincidently with PAR proteins and has the ability to provide persistent spatial landmarks within cells (Munro et al., 2004). Consistent with this picture, much evidence points to a critical role for actin in both the formation and maintenance of PAR domains (Hill and Strome, 1988; Guo and Kemphues, 1996; Shelton et al., 1999; Severson and Bowerman, 2003; Munro et al., 2004; Duncan et al., 2005; Harris and Peifer, 2005; Alford et al., 2009; Liu et al., 2010). During the initial polarity establishment phase, polarized actin flows, driven by the activity of the myosin motor NMY-2, are thought to segregate PAR-3, PAR-6, and aPKC into the anterior, allowing PAR-1, PAR-2, and LGL to associate with the membrane in the posterior (Cheeks et al., 2004; Munro et al., 2004; Beatty et al., 2010; Hoege et al., 2010). Disruption of the actomyosin cortex has also been reported to destabilize PAR domains even after they have been formed (Severson and Bowerman, 2003; Liu et al., 2010), leading to a model in which domains are stabilized by the actomyosin cortex, presumably through feedback between actin and/or myosin and the PAR proteins (Munro, 2006; Tostevin and Howard, 2008). However, several studies have questioned this model, noting that polarity is resistant to short pulses of cytochalasin, and embryos with defects in actomyosin contractility are nonetheless able to undergo some degree of polarization, including the generation of small, rudimentary PAR domains under certain conditions (Hill and Strome, 1988; Rose et al., 1995; Guo and Kemphues, 1996; Shelton et al., 1999; Severson and Bowerman, 2003; Schonegg et al., 2007; Zonies et al., 2010). These observations, combined with recent reports that myosin may not be required for PAR polarity in other systems (Chalmers et al., 2005; Atwood and Prehoda, 2009), suggest that the ability of PAR proteins to form domains can be uncoupled from actomyosin contractile asymmetries. Given these divergent observations, the basic question of how the embryo physically sorts and confines PAR proteins within the appropriate domains remains unanswered.
Addressing the mechanism of PAR protein segregation has been hampered by a lack of quantitative information regarding the mobility of PAR proteins within cells. Even basic questions remain unanswered. For example, is their motion restricted by cytoskeletal elements and/or localized binding sites, or are they relatively free to diffuse when associated with the membrane? Although there is evidence that PAR proteins turn over rapidly within small regions of the cortex (Cheeks et al., 2004; Petrásek et al., 2008), various behaviors could underlie this recovery, including active transport, diffusion, or exchange between membrane and cytoplasmic pools. Each of these behaviors places constraints on models for how PAR proteins can be segregated within the membrane.
Here, we examine the kinetic behavior of two proteins essential for PAR polarity in C. elegans, the anterior protein PAR-6 and the posterior protein PAR-2. Our findings indicate that both proteins are free to undergo extensive diffusion along the membrane and suggest that the equalizing effects of lateral diffusion of PAR proteins on the membrane must be countered by an actin-independent spatial bias in membrane association.
Results
Lateral diffusion of membrane-associated PAR proteins
The kinetic behavior of a protein associated with a cell membrane can in principle result from diffusion, directed motion, and/or exchange between membrane-bound and cytoplasmic states. By measuring the spatial characteristics of FRAP, one can detect and distinguish these various types of mobility (Sprague and McNally, 2005). The initial polarity establishment phase in C. elegans embryos is characterized by a contractile actomyosin cortex, anterior-directed flows of cortical cytoplasm, and the formation of two PAR domains of roughly equal size. During the subsequent maintenance phase, the motion of the actomyosin cortex is down-regulated, and the PAR domains are stably maintained until cytokinesis. The stable distributions of PAR proteins and the lack of cortical motion make the maintenance phase an attractive system for acquiring quantitative measurements of PAR protein dynamics in vivo.
We used GFP fusions to PAR-6 and PAR-2 as proxies for the behavior of the anterior and posterior PAR proteins, respectively. Both complement mutations in their respective genes (unpublished data). Consistent with FRAP experiments performed during the establishment phase (Cheeks et al., 2004), fluorescence recovery during maintenance phase was rapid for both proteins, typically reaching near full fluorescence recovery within 1 min. We did not observe directed motion of bleached spots during maintenance phase, indicating a lack of long-range directional transport of PAR molecules within their domains during this phase, in contrast to the onset phase (Fig. S1). This finding is consistent with the lack of long-range cortical flows in maintenance phase and suggests that recovery is primarily a result of lateral diffusion on the membrane and/or exchange between membrane and cytoplasmic pools.
Lateral diffusion on the membrane can be detected in FRAP experiments by monitoring the spatial characteristics of fluorescence recovery (Liebman and Entine, 1974; Poo and Cone, 1974; Axelrod et al., 1976). In brief, if molecules diffuse laterally during the timescale of the FRAP experiment, recovery will occur first at the edges. If they are only exchanging, recovery will be spatially uniform. To isolate the behavior intrinsic to each PAR protein, we performed FRAP in the absence of mutual antagonism between the two groups of PAR proteins. In other words, we examined GFP–PAR-6 in embryos depleted of PAR-2 and GFP–PAR-2 in embryos depleted of PAR-6. Under these conditions, the GFP fusions are distributed throughout most, if not all, of the embryo membrane. For both proteins, we find that recovery at the edges is more rapid than in the center (Fig. 1, A and B). Although not precluding the existence of membrane–cytoplasmic exchange, this result provides clear evidence that lateral diffusion contributes significantly to the behavior of PAR-2 and PAR-6 on the membrane.
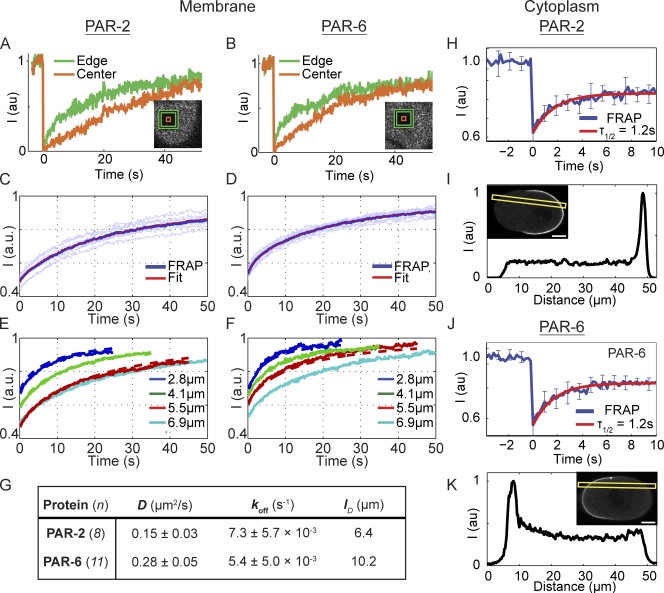
Figure 1.
PAR-2 and PAR-6 kinetics revealed by FRAP. (A and B) A comparison of the recovery at the edges versus the center of bleached boxes on the embryo surface reveals a clear outside-in pattern for both proteins that is characteristic of lateral diffusion. (C and D) Mean, normalized FRAP data (blue line) for a 6.9 × 6.9–µm box were well fit by a diffusion and exchange model (red line) using the measured coefficients in G. Light blue lines show individual replicates. (E and F) Recoveries into boxes of varying size are accurately predicted using the measured coefficients. Note that the larger boxes recover more slowly, consistent with recovery through diffusion. (G) Measured diffusion and binding coefficients for PAR-2 and PAR-6. Mean ± SD is shown. (H) FRAP of PAR-2 in the cytoplasm of embryos exhibited a halftime of recovery of ∼1.2 s for a 9 × 9–µm square bleached at the embryo midplane. Mean ± SD of individual recoveries is plotted (n = 8). (I) No obvious gradient of GFP–PAR-2 was visible when embryos were imaged in a cross section using confocal microscopy. Fluorescence along the A–P axis along the indicated yellow box is shown. (J and K) Same as H and I, but for GFP–PAR-6. Again, cytoplasmic recovery is rapid and no significant gradient is seen. Bars, 10 µm.
To determine the relative contributions of lateral diffusion and membrane–cytoplasmic exchange, we made use of a novel method to simultaneously determine the lateral diffusion coefficient and the rate of exchange, in this case given by the off rate, by bleaching 2D boxes on the membrane (Goehring et al., 2010). FRAP recoveries for both proteins at the membrane were well fit as a combination of lateral diffusion and exchange (Fig. 1, C and D). The near-complete recovery of fluorescence indicates that there is no significant subpopulation of immobile molecules. Fits of recoveries reveal that the rates of detachment from the membrane, koff, for both proteins are similar (Fig. 1 G) and indicate that mean lifetimes for membrane attachment are on the order of 100 s. The lateral diffusion coefficients, D, are also of the same order of magnitude (Fig. 1 G), although PAR-6 diffuses approximately twice as fast. An important test of this lateral diffusion/membrane–cytoplasmic exchange model is the ability of a given set of parameters to accurately predict recoveries into different-sized boxes (Goehring et al., 2010). Because the distance that molecules must diffuse increases with larger boxes, changing the box size will alter the relative contributions of diffusion and exchange and thus the recovery kinetics. We find that the single-set coefficients obtained here accurately predict recoveries into bleach areas of four different sizes (Fig. 1, E and F).
We can use measurements of D and koff to estimate the characteristic diffusion length, lD. This length is the typical distance a PAR molecule can diffuse along the membrane before it falls off and reenters the cytoplasm and thus describes the amount of membrane that a molecule can explore during each membrane association event. It is calculated as
Substituting our measured parameters, we obtain lD = 6.4 µm for PAR-2 and lD = 10.2 µm for PAR-6. Compared with an embryo that is ∼50 µm in length, these diffusion lengths imply that during their lifetime on the membrane, both PAR proteins are capable of exploring a significant fraction of the embryo surface.
An important caveat with these measurements is that our bleach geometry necessarily results in bleaching of the cytoplasmic pool beneath the membrane, which complicates analysis unless care is taken to account for this population. Performing similar experiments in the cytoplasm resulted in recovery timescales on the order of 1 s, at least an order of magnitude faster than what was observed on the cortex (Fig. 1, H and J). Therefore, the cytoplasmic pool is unlikely to be contributing significantly to our measurements of the membrane-associated species. Rather, these results indicate that the cytoplasmic pool undergoes rapid mixing, a picture that is confirmed both by the lack of obvious cytoplasmic gradients of PAR proteins (Fig. 1, I and K) and FCS experiments on cytoplasmic PAR-2 (Petrásek et al., 2008). Thus, we conclude that the intrinsic behavior of both PAR-2 and PAR-6 in the embryo can be characterized as a slow exchange between a rapidly mixing cytoplasmic pool and a membrane-associated state in which PAR proteins undergo significant lateral diffusion.
PAR proteins diffuse across the PAR domain boundary
One striking characteristic of PAR protein distributions observed in fixed embryos is the presence of opposing spatial gradients at the domain interface (Wu and Rose, 2007). Quantification of PAR protein distributions in live embryos simultaneously expressing fluorescent protein fusions to PAR-2 and PAR-6 confirmed the presence of opposing gradients (Fig. 2 A). Such spatially extended gradients can arise because of diffusion of molecules between a source and a sink (Yu et al., 2009). In the absence of mechanisms to prevent diffusion across the anterior–posterior PAR boundary, the concentration differences between the two PAR domains would be expected to drive the lateral diffusion of PAR proteins from regions where they are enriched into regions of the membrane where they are depleted.
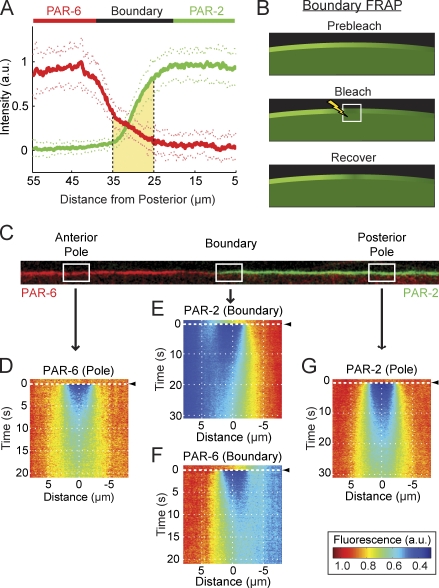
Figure 2.
PAR proteins diffuse across the domain boundary. (A) Quantification of PAR protein distributions during maintenance phase. The levels of PAR-6 (red, left) and PAR-2 (green, right) are relatively homogenous within the anterior and posterior domains, respectively. At midcell, proteins localize to opposing spatial gradients, which overlap (shaded yellow region). Mean ± SD is shown (n = 20 anterior to posterior profiles, 2 each from 10 embryos). Note that distances are represented relative to the posterior pole, with positive distance toward the anterior. (B) A schematic of boundary FRAP. The white box with the lightning bolt indicates the bleached region. (C) An example of a computationally straightened membrane profile from an embryo expressing GFP–PAR-6 (red) and mCherry–PAR-2 (green) in which the boundary gradient and polar regions are indicated. (D–G) For bleaching, embryos expressing GFP–PAR-6 or GFP–PAR-2 were subjected to FRAP within a 4-µm-long box at the approximate positions indicated. Fluorescence profiles were captured along the membrane at each time point, and recovery data were averaged for experiments performed at each position (n > 14). To facilitate visualization, data were assembled into kymographs with time on the y axis. Bleaching occurred at t = 0 s, indicated by black arrowheads. Distance is on the x axis and is expressed relative to the center of the bleached population. At the anterior (D, PAR-6) and posterior poles (G, PAR-2), recovery is symmetric. In contrast, within their respective boundary gradients (E, PAR-2; F, PAR-6), recovery is asymmetric, with recovery from the domain-proximal boundary being much greater than that from the boundary distal to the domain.
To determine whether PAR proteins are free to diffuse down the concentration gradients that exist between the two domains or are instead confined within their respective domains through some form of diffusion barrier, we photobleached a small box within each PAR gradient in a procedure we refer to as boundary FRAP (Fig. 2 B). Recovery occurred with the characteristic outside-in pattern observed in Fig. 1 (A and B), indicating that molecules were entering the bleached region through the edges before spreading into the center (Fig. 2, E and F). In addition, the recovery at the boundary is highly asymmetric; significantly more molecules enter the bleached region from the edge proximal to the respective PAR domain, where fluorescence is higher, than from the edge distal to the domain, where fluorescence is lower. This pattern is consistent with a concentration gradient driving diffusive flux of PAR proteins from their respective domains where concentrations are high into the opposing domains where concentrations are low. Control experiments performed near the center of each of the two PAR domains where concentrations are relatively uniform exhibited the same characteristic outside-in recovery but no asymmetry in recovery (Fig. 2, D and G), consistent with what would be expected given the lack of an underlying concentration gradient in this region.
The diffusion length limits the distance over which a molecule can communicate information along the membrane. Consequently, it should place limits on the length scale or sharpness of the boundary gradient formed by the molecule. Fitting the fluorescence distributions with an error function provides a measure of the boundary gradient decay length (σ) for each protein. Strikingly, the resulting values (σPAR-2 = 7.8 ± 0.1 µm and σPAR-6 = 11.0 ± 0.4 µm, n = 16) are similar to the calculated diffusion lengths, which correctly predict that the PAR-6 gradient is broader than that of PAR-2. Although the precise shape and extent of the boundary gradients will depend on the exact mechanism of protein segregation, the correlation between lD and σ that we observe supports the idea that the characteristic opposing gradients of PAR proteins at midcell are the result of PAR proteins diffusing down their respective concentration gradients within the boundary interface region.
Intriguingly, further examination of the PAR gradients revealed that although both proteins are relatively depleted within the boundary region, consistent with previous postulation of a gap between the domains (Munro et al., 2004), the two gradients clearly overlap one another within this region (Fig. 2 A). This overlap argues against a simple barrier separating the two domains. Instead, combined with the aforementioned results, it suggests a picture in which PAR proteins are not physically compartmentalized but are subject to a constant diffusive flux out of their respective domains into a boundary region. Consequently, both PAR proteins are present together within the boundary region, which, in principle, would permit direct physical interactions between membrane-associated species.
Lateral diffusion is not countered by sorting or transport
So far, we have shown that PAR proteins undergo lateral diffusion between the two PAR domains. Because of the tendency of diffusion to equalize concentrations in space, the maintenance of a stably positioned, steady-state PAR gradient requires some mechanism to compensate for this diffusive flux across the boundary interface. In principle, this could occur in two ways: first, molecules that have diffused down the gradient could be continuously removed, with new molecules being added to the membrane to replace those that are lost. Alternatively, a process of lateral sorting caused by active or passive mechanisms could directly work against the diffusive fluxes across the boundary. Molecules that are subject to such sorting should experience a force at the interface that leads to directional displacement of individual molecules over time. We refer to this displacement as drift and the rate of displacement as the drift velocity.
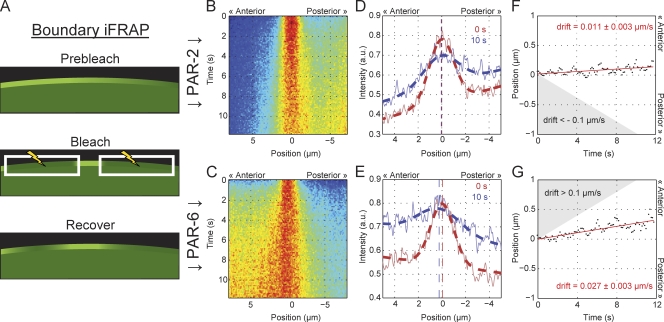
To distinguish between these ideas, we tested whether PAR proteins experience drift within the interface region by performing inverse FRAP (iFRAP; Lippincott-Schwartz et al., 2003). In the reverse of boundary FRAP, we bleached the entire gradient within the frame, with the exception of a small box, leaving a small fluorescent band (Fig. 3 A). This allowed us to follow the fate of an isolated population of molecules within the gradient. For a lateral sorting mechanism, we would expect drift of the unbleached band up the gradient immediately after iFRAP because of directed movement of molecules within the interface region toward their “correct” domain. Because this drift must be sufficient to move the PAR molecules across the interface region (10 µm) during their lifetime on the membrane (koff−1 ≈ 100 s), we estimate this drift velocity must be, at minimum, of the order of 0.1 µm/s. In reality, it must likely be higher given the rates of lateral diffusion for both molecules. In contrast, for asymmetric attachment/detachment, we expect little to no drift at short times, although spatially biased detachment and reattachment will still lead to some distortion in the observed distribution. Kymographs of fluorescence distributions over time indicate very little drift (Fig. 3, B and C). Fitting the fluorescence distribution with a modified Gaussian at each time point allows an estimate of potential drift velocity (Fig. 3, D–G). The resulting fits yield drift velocities of 0.01 µm/s (PAR-2) and 0.03 µm/s (PAR-6), where positive values represent drift toward the anterior. Note that a sorting mechanism would require drift of molecules within the two gradients to occur in opposing directions rather than in the same direction toward the anterior as we see here. This suggests that this residual drift we observe may be the result of a slow displacement of the entire boundary region toward the anterior. Importantly, in neither case do we observe drift up the gradient exceeding the ∼0.1 µm/s that would be expected for a lateral sorting mechanism. Therefore, lateral sorting does not seem to be required for countering the diffusive flux of PAR proteins across the PAR boundary. Rather, our results indicate that diffusive flux is countered by the net removal of molecules in the “wrong” domain and their net association in the correct domain, which most likely is driven by differences in the rates of attachment to the membrane and/or detachment from the membrane between the two domains.
Figure 3.
PAR proteins exhibit minimal drift within the boundary gradients. (A) A schematic of boundary iFRAP. White boxes with lightning bolts indicate bleached regions. (B–G) Embryos expressing GFP–PAR-6 or GFP–PAR-2 were subject to bleaching of the gradient with the exception of a 4-µm-long box, leaving a small unbleached population. As in Fig. 2, fluorescence profiles were captured along the membrane at each time point and assembled to form the kymographs shown. Time is on the y axis. Distance relative to the center of the unbleached population is on the x axis with positive distance toward the anterior. Data are averaged from multiple experiments (n > 11). (B and C) Kymographs reveal that the unbleached fluorescent population spreads outward from the center over time. At later time points, fluorescence returns throughout the gradient as molecules diffuse into the imaged region from unbleached portions of the embryo, and molecules undergo membrane–cytoplasmic exchange. These processes will eventually tend to distort the distribution. (D and E) Fluorescence distributions (solid lines) and the resulting fits (dashed lines) for the distribution of unbleached molecules at 0 s and 10 s after bleach. The fit peaks of the distribution of unbleached molecules are indicated by vertical dashed lines for each time point. (F and G) Plotting the position corresponding to the peak of unbleached distribution over time shows that although some drift is evident, the rates of drift are much below what would be minimally required to counteract diffusive flux. The predicted required levels are indicated here by the shaded regions, which indicate drift velocities that exceed 0.1 µm/s up the respective gradient. Note that the estimated drift velocity (slope of the red line) for PAR-2 and PAR-6 is opposite relative to their respective gradients, indicating that both drift slightly toward the anterior.
Actin depolymerization does not affect PAR proteins during the polarity maintenance phase
How could such asymmetries in attachment and/or detachment occur? One possibility is that an asymmetric cortical actin scaffold could locally stimulate the association of the anterior PAR proteins with the membrane or destabilize the association of posterior PAR proteins in the anterior, a hypothesis that has been proposed in various forms (Cheeks et al., 2004; Tostevin and Howard, 2008). We developed a method to introduce drugs into embryos at defined time points and used it to examine the effects of acute disruption of the actin cytoskeleton on PAR proteins during maintenance phase. Despite effective disruption of the actin cytoskeleton (Fig. 4, A and B), treatment with cytochalasin D (CD) or latrunculin A did not lead to obvious defects in PAR domains during maintenance phase (Fig. 4 C), consistent with previous observations that such treatment does not disrupt polarity (Hill and Strome, 1988). Boundary gradients appear to be similar several minutes after treatment with either drug (Fig. 4 D and Fig. S2). Quantification revealed only minor broadening (CD: PAR-2, 7.9 ± 1.9 µm and PAR-6, 13.0 ± 2.1 µm, n = 6; latrunculin A: PAR-2, 11.0 ± 1.8 µm and PAR-6, 14.9 ± 1.9 µm, n = 8). The kinetic behavior of PAR proteins using FRAP under these conditions was also similar, ruling out major effects of the actomyosin cortex on the lateral diffusion and membrane association of PAR proteins (Fig. 4, E and F). Consistent with an actomyosin-independent association of PAR proteins with the cell membrane, PAR proteins remained stably associated with expanding bleb membranes (Fig. S3 and Video 4). Therefore, PAR proteins do not appear to rely on the actomyosin cortical network to associate with the membrane, and their dynamics on the membrane appear to be largely independent of the actomyosin cortex during the maintenance phase. Consequently, the cortical actin network is unlikely to be providing the required spatial asymmetries in attachment and detachment required for maintenance of the PAR boundary.
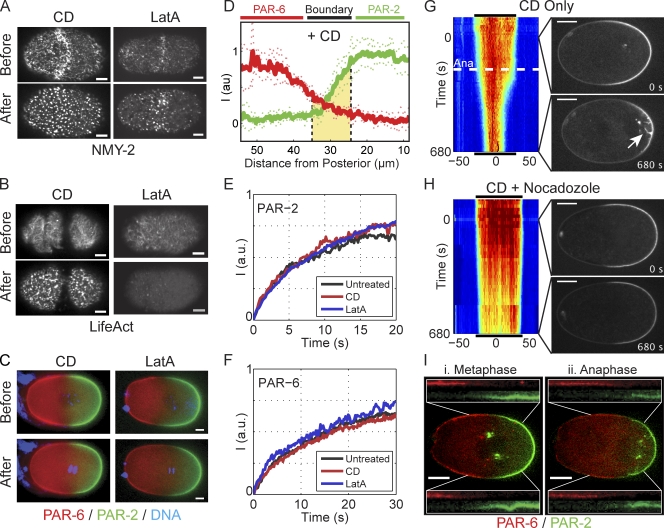
Figure 4.
PAR domain maintenance does not require an intact actin cytoskeleton. (A and B) Treatment of permeable embryos with CD or latrunculin A leads to rapid disruption of the actomyosin cortex as visualized with NMY-2–GFP (A) or LifeAct–GFP (B) using spinning disk confocal microscopy of a cortical plane taken before and 2–3 min after drug treatment. (C) Treatment of permeable embryos expressing fluorescently tagged PAR-2 (green)/PAR-6 (red) with CD or latrunculin A does not lead to loss of PAR domains. Select wide-field images of the embryo midplane are shown (Videos 1 and 2). (D) PAR distributions several minutes after treatment with CD are similar to untreated embryos (compare with Fig. 2 A). Mean ± SD is shown (n = 6 anterior to posterior profiles). Similar measurements for latrunculin A are provided in Fig. S2. (E) The recovery of GFP–PAR-2 during FRAP is similar in embryos left untreated compared with embryos treated with either CD or latrunculin A. Box size was 4.1 × 4.1 µm. In each case, two to four FRAP curves were averaged and normalized to allow comparison (see Materials and methods). (F) Same as E, but for GFP–PAR-6 embryos with a 6.9 × 6.9–µm box size. (G) A kymograph of GFP–PAR-2 in a CD-treated embryo shows that the domain remains relatively stable until anaphase (Ana, dashed white line), when it undergoes a dramatic contraction. Time is relative to nuclear envelope breakdown. Distance is relative to the center of the PAR-2 domain. Select images show a PAR-2 domain before and after anaphase. The PAR-2 invaginations that accompany domain contraction are indicated (white arrow; Video 3). Black lines indicate the extent of the PAR-2 domain at the beginning of time series and are shown above and below the kymograph to facilitate size comparisons. (H) Same as G, but including nocodazole plus CD. Disruption of microtubules eliminates both PAR-2 invaginations and anaphase PAR-2 domain contraction. (I) After anaphase onset, the boundaries of both mCherry–PAR-2 (green) and GFP–PAR-6 (red) shift to the posterior in CD-treated embryos. Images of a CD-treated embryo before (Metaphase) and after anaphase onset (Anaphase) illustrate the posterior migration of both PAR-2 and PAR-6 domain boundaries as a result of invaginations. In the insets, identical 25 × 25–µm regions encompassing the boundary region are taken from images before and after anaphase as indicated, and channels are shown individually to demonstrate the shift of both domain boundaries. Bars: (A–C) 5 µm; (G–I) 10 µm.
These results superficially disagree with two reports that disruption of the actomyosin cortex during maintenance phase destabilizes PAR domains, either by depolymerization of actin filaments using latrunculin (Severson and Bowerman, 2003) or by compromising cortical contractility through the use of a fast-acting mutation in nmy-2 (Liu et al., 2010). Similar to Liu et al. (2010), we observed that as embryos lacking a functional cortex entered anaphase, the PAR-2 domain contracted, accompanied by a corresponding expansion of the anterior PAR domain (Fig. 4, G and I). This contraction coincided with removal of PAR-2 from the cortex to the centrosome through what appeared to be membrane invaginations arising because of microtubule pulling forces on the posterior cortex in embryos lacking a stiff cortical actomyosin meshwork (Redemann et al., 2010). Therefore, anaphase destabilization does not appear to be caused by a failure in the mechanism for maintaining a PAR boundary, but instead results from the aberrant removal of PAR-2 from the cortex and sequestration within cytoplasmic aggregates. Supporting our interpretation, similar accumulations are visible in Severson and Bowerman (2003; Fig. 2) and Liu et al. (2010; Fig. 2 B). As expected, treatment with nocadozole prevents anaphase contraction in CD-treated embryos (Fig. 4 H). These results suggest that once the boundaries of the PAR domains are specified, they do not require input from the actin cytoskeleton beyond resisting the effects of microtubule pulling forces.
Discussion
Here, we have shown that PAR polarity determinants are not only dynamic but exhibit both (a) a boundary flux as membrane-associated proteins diffuse laterally out of their respective domains as a result of the concentration differences that exist across the PAR domain boundaries and (b) an exchange between the membrane-associated state and a rapidly mixing cytoplasmic pool. This dynamic behavior contrasts with the relative stability of PAR domains during the maintenance phase when both the size and position of PAR domains are relatively unchanged over the course of nearly 10 min. We have examined several potential mechanisms to explain the segregation of PAR proteins, including (a) a physical barrier separating the two domains, (b) passive sorting, for example, caused by repulsive or attractive forces such as those that drive the phase separation of oil and water, or (c) active transport on the membrane, such as actin- or microtubule-dependent processes. Our data are inconsistent with the involvement of any of these mechanisms, at least during the polarity maintenance phase. Rather, our results suggest a physical picture in which PAR proteins undergo continuous and what appears to be unopposed diffusion across the PAR boundary interface separating the two domains. In other words, PAR proteins tend to diffuse out of their respective domains where they are enriched and into the opposing domains where they are depleted.
Because there does not appear to be a mechanism to directly oppose diffusion on the membrane, there must be some way for cells to ensure that the membrane association of PAR proteins is biased such that there is net association only within the proper domain and net dissociation in the incorrect domain. It has been proposed that the cortical actomyosin network could provide for such asymmetries in membrane affinity (Cheeks et al., 2004; Tostevin and Howard, 2008). However, results here and elsewhere indicate that such a cytoskeletal scaffold model is unlikely to be the key driving force for maintaining segregated domains (Hill and Strome, 1988; Zonies et al., 2010; this study). In fact, our findings suggest that beyond preventing microtubule-dependent membrane invaginations, the actin cortex plays little role in the membrane association or kinetic behaviors of PAR proteins during the maintenance phase. Therefore, it is likely that the spatial regulation of PAR protein membrane affinity arises instead from the characteristics of the PAR proteins themselves. At the same time, there is clearly a role of the actin cortex outside of the maintenance phase, both in promoting normal establishment of PAR polarity and in the alignment of the PAR boundary during cytokinesis (Munro et al., 2004; Schenk et al., 2010; Zonies et al., 2010), although exactly how actin mechanistically accomplishes these tasks remains somewhat unclear.
What, then, drives the association of PAR proteins with the membrane? Two actin-independent links to the membrane have been reported, the regulation of which could be subject to spatial control. PAR-3 has lipid-binding activity (Wu et al., 2007; Krahn et al., 2010), and PAR-6 is known to bind CDC-42, which is itself membrane associated (Joberty et al., 2000; Lin et al., 2000; Qiu et al., 2000; Gotta et al., 2001). Intriguingly, active CDC-42 localizes predominantly to the anterior during the maintenance phase and is important for the cortical localization of PAR-6 during this phase (Gotta et al., 2001; Aceto et al., 2006; Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006; Kumfer et al., 2010). However, it does not appear to be the sole means of membrane association because it is not essential for initial domain formation, and PAR-6 can associate with the membrane independently of CDC-42 as long as PAR-2 is depleted (Aceto et al., 2006; Schonegg and Hyman, 2006). In the posterior, the mechanisms of membrane association remain unclear. Phosphorylation of two posterior proteins, PAR-2 and LGL, likely by aPKC, appears to be important for displacing them from the membrane and is presumably responsible for excluding them from the anterior domain (Hao et al., 2006; Hoege et al., 2010). Clearly, much remains to be discovered regarding the mechanisms of membrane association of the PAR proteins, how these interactions could be spatially regulated within the cell, and how these processes are impacted by cross talk between PAR proteins.
Based on observations that polarity can be established under conditions lacking cortical flow, it has been suggested that mutual antagonism may be sufficient to drive segregation of PAR proteins under certain conditions (Zonies et al., 2010). Combined with our observations, one can envision an intuitive self-organizing scheme based on competition between the membrane-associated species; high concentrations of PAR-3/PAR-6/aPKC favor detachment of PAR-2 (and presumably other posterior proteins such as LGL) from the membrane in the anterior, whereas high concentrations of PAR-2 favor dissociation of PAR-3/PAR-6/aPKC in the posterior (Fig. 5). Such localized regions of enhanced dissociation effectively serve as sinks, removing PAR proteins that diffuse into the wrong domain. These detached PAR proteins then redistribute throughout the cytoplasm and finally reassociate with the membrane within the boundaries of the correct domain, completing the cycle. During the maintenance phase, all of these processes presumably balance, resulting in a stable domain boundary and a cycling flux of molecules. In other words, the stably polarized embryo comprises a nonequilibrium steady state. This scheme is consistent with both our observation of a diffusive boundary flux and our results suggesting that spatial differences in membrane affinity are responsible for maintaining PAR asymmetry.
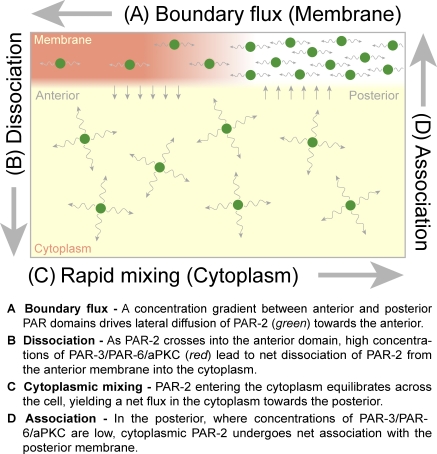
Figure 5.
Creating a stable boundary through diffusive flux and asymmetries in membrane association/dissociation. Because the regions of net association to and dissociation from the membrane occur in different domains, the source and sink are displaced in space, leading to a buildup of concentration gradients across the cell. These concentration gradients drive flux of molecules across the cell both on the membrane (boundary flux) and in the cytoplasm. At steady state, all of these processes balance, the local concentrations of PAR proteins no longer change with time, and the resulting boundary gradient is stable. Here, we show the proposed schematic of this cycle for the posterior PAR protein, PAR-2, with a region of high PAR-3/PAR-6/aPKC serving as a sink in the anterior (red membrane region). A model for PAR-6 would be similar but with a reversed orientation along the A–P axis and a region of high PAR-2 concentrations serving as a sink.
This model envisions interactions between anterior and posterior PAR proteins that are primarily limited to competition between membrane-associated species. Consequently, one would expect that the behavior of a membrane-associated PAR species would only be altered when it enters a region enriched in the opposing PAR complex. Consistent with this picture, FRAP of PAR proteins inside their domains in normal embryos and of PAR proteins in embryos in which the opposing PAR species is depleted by RNAi yields similar results (Fig. S4). Although a direct comparison of the association/dissociation rates for a given PAR protein between the two domains would be informative, the low concentrations of PAR proteins in the wrong domains preclude measurement using FRAP.
Although potentially attractive, such a PAR-intrinsic model raises significant questions. Is the ability of molecules to mutually inhibit one another at the membrane sufficient to ensure mutually exclusive distributions, let alone the formation of spatial patterns? Or is a more complicated pattern of interactions required? More fundamentally, it is unclear what features of the PAR polarity network could provide the required spatial information to define the boundaries of the PAR domains. If one domain is too small, that domain must be favored to allow it to expand at the expense of the other. However, individual PAR proteins are unlikely to know where they are in the cell. Therefore, domain size control would seem to require that the cell senses either the size of the domains or the position of the boundary and adjusts the competition between PAR proteins accordingly, a prospect that does not follow intuitively from a model based solely on mutual antagonism between membrane-associated species.
In the end, understanding cell polarity requires us to confront the central problem of coordinating the mobility and activities of the relevant molecules in space and time. Such spatiotemporal modulation of the transitions between diffusive states appears to be a central theme in polarity, at least within the C. elegans embryo. This is true not only as we show here for PAR proteins, which exchange between a rapidly mixing cytoplasmic state and a more slowly diffusing membrane-associated state, but also for cytoplasmic determinants such as MEX-5, PIE-1, and P granule components, all of which polarize in response to PAR domains through a mechanism that appears to involve asymmetric switching between fast- and slow-diffusing cytoplasmic states (Tenlen et al., 2008; Brangwynne et al., 2009; Daniels et al., 2009). Going forward, increasing insight into the kinetic properties of these molecules, combined with the development of quantitative approaches to understanding the complex dynamic relationships between them, will be critical as we begin to bridge the gap between molecular interactions and the long-range spatiotemporal patterns observed in cells.
Materials and methods
Worm strains and growth conditions
Worm stocks were maintained at 16°C and shifted to 25°C ∼24 h before analysis unless otherwise noted. Embryos were imaged at room temperature (18–22°C). Unless otherwise noted, embryos were dissected in 0.1 M NaCl and 4% sucrose and mounted on agarose pads. RNAi was performed using the feeding method essentially as described previously (Kamath and Ahringer, 2003). All feeding clones were obtained from GeneService except mlc-4, which was described recently (Redemann et al., 2010). Transgenic worms used in this study were JJ1473 (NMY-2–GFP; Nance et al., 2003), TH25 (GFP–PAR-6), TH120 (mCherry–PAR-6 and GFP–PAR-2) and TH129 (GFP–PAR-2 [RNAi resistant]; Schonegg et al., 2007), TH209 (mCherry–PAR-2; Brangwynne et al., 2009), and TH220 (LifeAct–GFP; Redemann et al., 2010). JJ1473 was obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health’s National Center for Research Resources.
Drug treatment using permeabilized embryos
Embryos were made permeable by placing L4 larvae on FO8F8.2(RNAi) plates for 20–24 h at 25°C. Embryos were dissected in Shelton’s growth medium (SGM; Shelton and Bowerman, 1996), mounted in a flow chamber consisting of a glass slide and a coverslip separated by 20.85 ± 0.5–µm polystyrene beads (Bangs Laboratories), and sealed on two sides with VALAP. Permeability was confirmed by adding SGM containing Hoechst. Drugs were introduced by capillary action at the following concentrations: 30 µg/ml nocodazole, 100 µM latrunculin A, and 10–20 µg/ml CD.
Imaging
Wide-field images were captured on a microscope (Axioplan II; Carl Zeiss) using a 63×/1.4 oil Plan Apochromat objective, a camera (Orca ER; Hamamatsu Photonics), and MetaVue (Molecular Devices). Spinning disk confocal images were captured on either an M3 microscope (Carl Zeiss) equipped with a spinning disk head (Yokogawa) using a 63×/1.4 oil Plan Apochromat objective, a 488-nm argon laser (CVI Melles Griot), a camera (Orca ER), and MetaVue or a microscope (IX71; Olympus) equipped with a spinning disk head using a 60×/1.35 oil UPlanSApo objective, 488- and 561-nm lasers (DPSS), a camera (EMCCD; iXon), and ImageIQ (Andor Technology). For still images and gradient length scale measurements, spinning disk images were captured, unbinned, with a 1–2-s exposure. For time-lapse experiments, 200-ms exposures were captured every 15–30 s to minimize photobleaching. For cortical imaging, four z sections spanning 1 µm were captured, and a maximum projection was calculated. Unless otherwise noted, all images were taken at the embryo midplane defined as the focal plane with a maximum cross-sectional area. All maintenance phase measurements were taken during the stable period between the end of nuclear envelope breakdown and anaphase onset.
Image analysis
Spinning disk confocal images were used for all quantitative analysis and image processing performed in Fiji and Matlab (Mathworks). In brief, for membrane fluorescence, a 25-pixel-wide line encompassing the membrane was extracted and computationally straightened. The maximum fluorescence corresponding to an ∼0.8-µm-wide section spanning the membrane was taken as total membrane fluorescence for each point along the line, resulting in a fluorescence profile. For kymographs, the profile at each time was aligned by the approximate center of the PAR-2 domain before assembly. Signal was normalized to peak fluorescence within the kymograph. For gradient analysis, each channel from two-channel mCherry–PAR-2/GFP–PAR-6 images was used individually to generate a profile from the posterior pole to the anterior pole. Profiles were subject to smoothing using a 9-pixel moving mean and fit using an error function of the form
where I(x) is the intensity, b is the edge of the domain defined as the midpoint of the gradient, σ is the characteristic length scale of the gradient, and A and B scale the intensity. Individual fits gave the positions of the edge of the PAR-2 and PAR-6 domains separately. The point midway between these two positions was then defined as the center of the boundary and used as a reference point to align all individual profiles. Profiles were normalized to peak fluorescence and aligned to the reference point before being averaged and refit to obtain a mean value for σ. SDs of individual fits are reported to illustrate the variance of the data.
FRAP
Embryos expressing GFP–PAR-2 or GFP–PAR-6 were depleted for PAR-6 or PAR-2, respectively, by RNAi for 24 h at 25°C. As a result, GFP was typically localized throughout most, if not all, of the embryo membrane during late maintenance phase. Under these conditions, the behavior of the GFP fusions should reflect their intrinsic dynamics in the absence of mutual antagonism. Similar results were obtained within domains in wild-type embryos, suggesting that mutual antagonism does not significantly affect the dynamics of PAR proteins away from the boundary region (Fig. S4).
Embryos were dissected as described in Worm strains and growth conditions, except that coverslips were sealed with VALAP to reduce evaporation and coverslip drift. FRAP was performed and analyzed as previously described (Goehring et al., 2010). In brief, 128 × 128–pixel images were captured at 0.188-s intervals using a 488-nm laser line at 1% power with the pinhole closed down to 200 nm, yielding a back-projected pinhole diameter of ∼490 nm to limit the depth of the imaging region of interest. This yields a theoretical region of interest of ∼0.5 × 0.5 × 1.3 µm. A 50 × 50–pixel box was bleached using a 405-nm DPSS and 488-nm argon laser line at full power for 0.094 s (one scan iteration). The size of the bleach region was altered using the built-in zoom function at 12, 15, 20, and 30×. To minimize the effects of noise, three to five individual FRAP traces were averaged to form a replicate. Replicates were individually fit, and the mean and SD of replicates were reported. For fitting, we incorporated a one-frame bleach offset for PAR-2 or a five-frame bleach offset for PAR-6. Bleaching of the membrane using this bleach geometry necessarily results in bleaching of the cytoplasm underlying the membrane. However, because recovery in the cytoplasm is much faster than recovery at the membrane, this offset allows cytoplasmic fluorescence to equilibrate, which minimizes the effects of fluorescence recovery in the cytoplasm on our measurements (Goehring et al., 2010). The relatively large error in the koff measurements is most likely caused by the fact that, even at this size range, diffusion contributes more to recovery than exchange. Larger bleach areas, which would be more influenced by exchange, were not possible given the size limitations of the embryo.
For FRAP experiments in drug-treated embryos, FRAP was performed on FO8F8.2(RNAi) embryos in SGM. For each experiment, multiple FRAP traces were obtained using either a 4.1 × 4.1– (PAR-2) or 6.9 × 6.9–µm (PAR-6) box within the central region of the PAR-2 or PAR-6 domain. Individual traces were averaged and normalized to allow comparison (PAR-2: untreated, n = 2; CD, n = 2; latrunculin A, n = 5; PAR-6: untreated, n = 7; CD, n = 4; latrunculin A, n = 4). The drug was allowed to incubate for several minutes before FRAP to ensure disruption of the actin cytoskeleton, and embryos were examined for a lack of any cytokinesis furrow ingression to confirm disruption.
Boundary FRAP
Experiments were performed essentially as described in the previous section, with the following changes. Instead of focusing on the cortex, the focal plane was placed in the center of the embryo to generate a section through the center of the embryo such that the imaging and bleaching lasers are aligned parallel to the plane of the membrane. Because we are only interested in diffusion aligned with the gradient, we maximized the confocal depth in z by increasing the pinhole to 650 nm, reducing the problem to an essentially 1D geometry. Images were captured in a 256 × 64–pixel region at 0.13-s intervals. A 4 × 15–pixel region was bleached in the membrane, which yielded a ∼4-µm-wide bleached stripe centered in the gradient. After image capture, a 30-pixel-wide line containing the membrane was extracted, and the intensity of the top five pixels at each position in x was summed to give a fluorescence profile within each frame. Profiles were normalized to fluorescence within a control region of the membrane during the prebleach period. The center of the bleached region was identified by fitting a Gaussian profile to the mean profile of the first three frames. Because of significant noise in individual experiments, profiles from multiple experiments were aligned and averaged to generate the kymographs shown. To reduce the influence of cortical motion that could bias measurements, we performed experiments under a condition of mild MLC-4 depletion in which polarity was established with normal timing. Importantly, we obtained similar results in experiments performed in wild-type embryos if we verified that no cortical motion occurred during FRAP. However, because of the frequent occurrence of transient cortical motion in wild-type embryos, it was difficult to obtain a large sample size.
Boundary iFRAP
For iFRAP, experiments were performed as in boundary FRAP to generate a mean kymograph, except that rather than bleaching a small region of the membrane within the gradient, the entire membrane within the imaging frame was bleached with the exception of a small 4-µm-wide strip. We then fit the mean profile at each time with the function
where mx + b captures the local recovery of the underlying gradient as molecules reassociate with the cortex, and
is a modified Gaussian that captures the distribution of the unbleached population. µ gives the center of mass of this population, with Δµ/t giving the drift velocity. Again, all experiments were performed under conditions of mild MLC-4 depletion as described in the previous paragraph. One could argue that by depleting MLC-4, we are eliminating the very active transport processes that we are trying to measure. However, under these conditions, and even under disruption of the actomyosin cortex (Fig. 4, C and D), PAR proteins remain stably segregated, indicating that although this flow may affect PAR protein distributions, it is not an essential component for maintaining the segregated state that is our focus here.
Online supplemental material
Fig. S1 shows line FRAP experiments to test for the presence of directional flows. Fig. S2 shows quantification of PAR protein distributions in latrunculin A–treated embryos. Fig. S3 shows localization of PAR proteins to membrane blebs in pod-1(RNAi) embryos. Fig. S4 examines FRAP of PAR proteins in wild-type embryos versus embryos in which the opposing species is depleted by RNAi. Videos 1 and 2 show permeable embryos expressing GFP–PAR-2 and mCherrry–PAR-6 that are treated with CD or latrunculin A, respectively. Video 3 shows GFP–PAR-2 being removed from the posterior membrane by microtubule-dependent invaginations. Video 4 shows the localization of NMY-2, LifeAct, PAR-2, and PAR-6 to membrane blebs in pod-1(RNAi) embryos. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201011094/DC1.
Acknowledgments
We thank the various readers who provided critical comments on this work, A. Pozniakovsky for assistance with molecular cloning, S. Ernst for assistance with microparticle bombardments, and P. Khuc Trong, J. Bois, E. Nicola, and D. Chowdhury for helpful discussions.
N.W. Goehring was supported by the Alexander von Humboldt Foundation and a Marie Curie grant (219286) from the European Commission.
Footnotes
Abbreviations used in this paper:
- aPKC
- atypical PKC
- CD
- cytochalasin D
- iFRAP
- inverse FRAP
- SGM
- Shelton’s growth medium
References
- Aceto D., Beers M., Kemphues K.J. 2006. Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev. Biol. 299:386–397 10.1016/j.ydbio.2006.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford L.M., Ng M.M., Burgess D.R. 2009. Cell polarity emerges at first cleavage in sea urchin embryos. Dev. Biol. 330:12–20 10.1016/j.ydbio.2009.02.039 [DOI] [PubMed] [Google Scholar]
- Atwood S.X., Prehoda K.E. 2009. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. 19:723–729 10.1016/j.cub.2009.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D., Koppel D.E., Schlessinger J., Elson E., Webb W.W. 1976. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16:1055–1069 10.1016/S0006-3495(76)85755-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y., Mermall V., Mooseker M.S., Snyder M. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell. 5:841–851 10.1016/S1097-2765(00)80324-X [DOI] [PubMed] [Google Scholar]
- Beatty A., Morton D., Kemphues K. 2010. The C. elegans homolog of Drosophila Lethal giant larvae functions redundantly with PAR-2 to maintain polarity in the early embryo. Development. 137:3995–4004 10.1242/dev.056028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., St Johnston D. 2003. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 115:691–704 10.1016/S0092-8674(03)00938-3 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A. 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 422:326–330 10.1038/nature01486 [DOI] [PubMed] [Google Scholar]
- Boyd L., Guo S., Levitan D., Stinchcomb D.T., Kemphues K.J. 1996. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 122:3075–3084 [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Chalmers A.D., Pambos M., Mason J., Lang S., Wylie C., Papalopulu N. 2005. aPKC, Crumbs3 and Lgl2 control apicobasal polarity in early vertebrate development. Development. 132:977–986 10.1242/dev.01645 [DOI] [PubMed] [Google Scholar]
- Chant J., Mischke M., Mitchell E., Herskowitz I., Pringle J.R. 1995. Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 129:767–778 10.1083/jcb.129.3.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks R.J., Canman J.C., Gabriel W.N., Meyer N., Strome S., Goldstein B. 2004. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 14:851–862 10.1016/j.cub.2004.05.022 [DOI] [PubMed] [Google Scholar]
- Cowan C.R., Hyman A.A. 2004. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 20:427–453 10.1146/annurev.cellbio.19.111301.113823 [DOI] [PubMed] [Google Scholar]
- Cuenca A.A., Schetter A., Aceto D., Kemphues K., Seydoux G. 2003. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 130:1255–1265 10.1242/dev.00284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels B.R., Perkins E.M., Dobrowsky T.M., Sun S.X., Wirtz D. 2009. Asymmetric enrichment of PIE-1 in the Caenorhabditis elegans zygote mediated by binary counterdiffusion. J. Cell Biol. 184:473–479 10.1083/jcb.200809077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan F.E., Moss S.B., Schultz R.M., Williams C.J. 2005. PAR-3 defines a central subdomain of the cortical actin cap in mouse eggs. Dev. Biol. 280:38–47 10.1016/j.ydbio.2004.12.034 [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B., Guo S., Kemphues K.J. 1995. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 83:743–752 10.1016/0092-8674(95)90187-6 [DOI] [PubMed] [Google Scholar]
- Goehring N.W., Chowdhury D., Hyman A.A., Grill S.W. 2010. FRAP analysis of membrane-associated proteins: lateral diffusion and membrane-cytoplasmic exchange. Biophys. J. 99:2443–2452 10.1016/j.bpj.2010.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryachev A.B., Pokhilko A.V. 2008. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 582:1437–1443 10.1016/j.febslet.2008.03.029 [DOI] [PubMed] [Google Scholar]
- Gotta M., Abraham M.C., Ahringer J. 2001. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 11:482–488 10.1016/S0960-9822(01)00142-7 [DOI] [PubMed] [Google Scholar]
- Guo S., Kemphues K.J. 1996. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature. 382:455–458 10.1038/382455a0 [DOI] [PubMed] [Google Scholar]
- Hao Y., Boyd L., Seydoux G. 2006. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev. Cell. 10:199–208 10.1016/j.devcel.2005.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J.C., Peifer M. 2005. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J. Cell Biol. 170:813–823 10.1083/jcb.200505127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D.P., Strome S. 1988. An analysis of the role of microfilaments in the establishment and maintenance of asymmetry in Caenorhabditis elegans zygotes. Dev. Biol. 125:75–84 10.1016/0012-1606(88)90060-7 [DOI] [PubMed] [Google Scholar]
- Hoege C., Constantinescu A.-T., Schwager A., Goehring N.W., Kumar P., Hyman A.A. 2010. LGL can partition the cortex of one-cell Caenorhabditis elegans embryos into two domains. Curr. Biol. 20:1296–1303 10.1016/j.cub.2010.05.061 [DOI] [PubMed] [Google Scholar]
- Joberty G., Petersen C., Gao L., Macara I.G. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531–539 10.1038/35019573 [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Ahringer J. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Kemphues K.J., Priess J.R., Morton D.G., Cheng N.S. 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 52:311–320 10.1016/S0092-8674(88)80024-2 [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Saito K., Johnson J.M., Howell A.S., Zyla T.R., Lew D.J. 2008. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr. Biol. 18:1719–1726 10.1016/j.cub.2008.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn M.P., Klopfenstein D.R., Fischer N., Wodarz A. 2010. Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr. Biol. 20:636–642 10.1016/j.cub.2010.01.065 [DOI] [PubMed] [Google Scholar]
- Kumfer K.T., Cook S.J., Squirrell J.M., Eliceiri K.W., Peel N., O’Connell K.F., White J.G. 2010. CGEF-1 and CHIN-1 regulate CDC-42 activity during asymmetric division in the Caenorhabditis elegans embryo. Mol. Biol. Cell. 21:266–277 10.1091/mbc.E09-01-0060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé J.-C., Pacquelet A., Marty T., Gotta M. 2006. A genomewide screen for suppressors of par-2 uncovers potential regulators of PAR protein-dependent cell polarity in Caenorhabditis elegans. Genetics. 174:285–295 10.1534/genetics.106.060517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman P.A., Entine G. 1974. Lateral diffusion of visual pigment in photoreceptor disk membranes. Science. 185:457–459 10.1126/science.185.4149.457 [DOI] [PubMed] [Google Scholar]
- Lin D., Edwards A.S., Fawcett J.P., Mbamalu G., Scott J.D., Pawson T. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540–547 10.1038/35019592 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Altan-Bonnet N., Patterson G.H. 2003. Photobleaching and photoactivation: following protein dynamics in living cells. Nat. Cell Biol. Suppl:S7–S14 [PubMed] [Google Scholar]
- Liu J., Maduzia L.L., Shirayama M., Mello C.C. 2010. NMY-2 maintains cellular asymmetry and cell boundaries, and promotes a SRC-dependent asymmetric cell division. Dev. Biol. 339:366–373 10.1016/j.ydbio.2009.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F., Sugimoto A. 2006. Sequential functioning of the ECT-2 RhoGEF, RHO-1 and CDC-42 establishes cell polarity in Caenorhabditis elegans embryos. Nat. Cell Biol. 8:978–985 10.1038/ncb1459 [DOI] [PubMed] [Google Scholar]
- Munro E.M. 2006. PAR proteins and the cytoskeleton: a marriage of equals. Curr. Opin. Cell Biol. 18:86–94 10.1016/j.ceb.2005.12.007 [DOI] [PubMed] [Google Scholar]
- Munro E., Bowerman B. 2009. Cellular symmetry breaking during Caenorhabditis elegans development. Cold Spring Harb. Perspect. Biol. 1:a003400 10.1101/cshperspect.a003400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E., Nance J., Priess J.R. 2004. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell. 7:413–424 10.1016/j.devcel.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Nance J., Munro E.M., Priess J.R. 2003. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development. 130:5339–5350 10.1242/dev.00735 [DOI] [PubMed] [Google Scholar]
- Petrásek Z., Hoege C., Mashaghi A., Ohrt T., Hyman A.A., Schwille P. 2008. Characterization of protein dynamics in asymmetric cell division by scanning fluorescence correlation spectroscopy. Biophys. J. 95:5476–5486 10.1529/biophysj.108.135152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo M., Cone R.A. 1974. Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature. 247:438–441 10.1038/247438a0 [DOI] [PubMed] [Google Scholar]
- Qiu R.G., Abo A., Steven Martin G. 2000. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCzeta signaling and cell transformation. Curr. Biol. 10:697–707 10.1016/S0960-9822(00)00535-2 [DOI] [PubMed] [Google Scholar]
- Redemann S., Pecreaux J., Goehring N.W., Khairy K., Stelzer E.H.K., Hyman A.A., Howard J. 2010. Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS ONE. 5:e12301 10.1371/journal.pone.0012301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L.S., Lamb M.L., Hird S.N., Kemphues K.J. 1995. Pseudocleavage is dispensable for polarity and development in C. elegans embryos. Dev. Biol. 168:479–489 10.1006/dbio.1995.1096 [DOI] [PubMed] [Google Scholar]
- Schenk C., Bringmann H., Hyman A.A., Cowan C.R. 2010. Cortical domain correction repositions the polarity boundary to match the cytokinesis furrow in C. elegans embryos. Development. 137:1743–1753 10.1242/dev.040436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonegg S., Hyman A.A. 2006. CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Development. 133:3507–3516 10.1242/dev.02527 [DOI] [PubMed] [Google Scholar]
- Schonegg S., Constantinescu A.T., Hoege C., Hyman A.A. 2007. The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc. Natl. Acad. Sci. USA. 104:14976–14981 10.1073/pnas.0706941104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A.F., Bowerman B. 2003. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J. Cell Biol. 161:21–26 10.1083/jcb.200210171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton C.A., Bowerman B. 1996. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development. 122:2043–2050 [DOI] [PubMed] [Google Scholar]
- Shelton C.A., Carter J.C., Ellis G.C., Bowerman B. 1999. The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J. Cell Biol. 146:439–451 10.1083/jcb.146.2.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague B.L., McNally J.G. 2005. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 15:84–91 10.1016/j.tcb.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Tabuse Y., Izumi Y., Piano F., Kemphues K.J., Miwa J., Ohno S. 1998. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 125:3607–3614 [DOI] [PubMed] [Google Scholar]
- Takizawa P.A., DeRisi J.L., Wilhelm J.E., Vale R.D. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 290:341–344 10.1126/science.290.5490.341 [DOI] [PubMed] [Google Scholar]
- Tanentzapf G., Tepass U. 2003. Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5:46–52 10.1038/ncb896 [DOI] [PubMed] [Google Scholar]
- Tenlen J.R., Molk J.N., London N., Page B.D., Priess J.R. 2008. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development. 135:3665–3675 10.1242/dev.027060 [DOI] [PubMed] [Google Scholar]
- Tostevin F., Howard M. 2008. Modeling the establishment of PAR protein polarity in the one-cell C. elegans embryo. Biophys. J. 95:4512–4522 10.1529/biophysj.108.136416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J.L., Etemad-Moghadam B., Guo S., Boyd L., Draper B.W., Mello C.C., Priess J.R., Kemphues K.J. 1996. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 122:3133–3140 [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Altschuler S., Wu L., Li R. 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 299:1231–1235 10.1126/science.1080944 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U., Knust E. 1999. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 402:544–547 10.1038/990128 [DOI] [PubMed] [Google Scholar]
- Wu J.-C., Rose L.S. 2007. PAR-3 and PAR-1 inhibit LET-99 localization to generate a cortical band important for spindle positioning in Caenorhabditis elegans embryos. Mol. Biol. Cell. 18:4470–4482 10.1091/mbc.E07-02-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Feng W., Chen J., Chan L.-N., Huang S., Zhang M. 2007. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol. Cell. 28:886–898 10.1016/j.molcel.2007.10.028 [DOI] [PubMed] [Google Scholar]
- Yu S.R., Burkhardt M., Nowak M., Ries J., Petrásek Z., Scholpp S., Schwille P., Brand M. 2009. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 461:533–536 10.1038/nature08391 [DOI] [PubMed] [Google Scholar]
- Zonies S., Motegi F., Hao Y., Seydoux G. 2010. Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR-2. Development. 137:1669–1677 10.1242/dev.045823 [DOI] [PMC free article] [PubMed] [Google Scholar]