Abstract
Objective: Recently, a high frequency of mutations in mitochondrial DNA (mtDNA) has been detected in ovarian cancer. To explore the alterations of proteins in mitochondria in ovarian cancer, a pair of human ovarian carcinoma cell lines (SKOV3/SKOV3.ip1) with different metastatic potentials was examined. Methods: Cancer cells SKOV3.ip1 were derived from the ascitic tumor cells of nude mice bearing a tumor of ovarian cancer cells SKOV3. SKOV3.ip1 exhibited a higher degree of migration potential than its paired cell line SKOV3. The proteins in the mitochondria of these two cells were isolated and separated by 2-D gel electrophoresis. The differently expressed proteins were extracted and identified using matrix assisted laser desorption ionisation/time-of-flight/time-of-flight (MALDI-TOF/TOF), and finally a selected protein candidate was further investigated by immunohistochemistry (IHC) method in nude mice bearing tumor tissues of these two cells. Results: A total of 35 spots with different expressions were identified between the two cells using 2D-polyacrylamide gel electrophoresis (PAGE) approach. Among them, 17 spots were detected only in either SKOV3 or SKOV3.ip1 cells. Eighteen spots expressed different levels, with as much as a three-fold difference between the two cells. Twenty spots were analyzed using MALDI-TOF/TOF, and 11 of them were identified successfully; four were known to be located in mitochondria, including superoxide dismutase 2 (SOD2), fumarate hydratase (FH), mitochondrial ribosomal protein L38 (MRPL38), and mRNA turnover 4 homolog (MRTO4). An increased staining of SOD2 was observed in SKOV3.ip1 over that of SKOV3 in IHC analysis. Conclusions: Our results indicate that the enhanced antioxidation and metabolic potentials of ovarian cancer cells might contribute to their aggressive and metastatic behaviors. The underlying mechanism warrants further study.
Keywords: Ovarian carcinoma, Mitochondria, Invasion, Proteomic, Superoxide dismutase 2 (SOD2)
1. Introduction
Ovarian cancer is characterized by invasion and metastasis. Due to the difficulties in detecting the location of the primary tumor, most patients are diagnosed with advanced disease (stage III/IV), with widespread implantations in the abdominal and pelvic cavities. Furthermore, ovarian cancer has a high recurrence rate; therefore, ovarian cancer is still the leading cause of death from gynecologic malignancies despite important advances in surgery and chemotherapy over the past 20 years (Naora and Montell, 2005). Thus, exploring the mechanism of invasion of ovarian tumors, as well as tumorous molecular biologic alterations, is one key to understanding ovarian cancer. Recently, results of a few studies have suggested that the cancer cells’ aggressive behavior and cancerous prognosis might relate to a mitochondrial functional disorder (Gottlieb and Tomlinson, 2005). Thus, we focused our study on the alterations of mitochondria of ovarian cancer cells.
Our previous study had shown that the abnormal copy number of mitochondrial DNA (mtDNA) was related with the type and grade of ovarian cancer (Wang et al., 2006). It is known that the repair and replication of mtDNA depend mainly on nuclear DNA (nDNA)-encoded genes. Thus, it was our hypothesis that the mitochondrion or its controlling system might be associated with ovarian cancer development and progress. However, whether the global expression alterations of the proteins in mitochondria, most of them encoded by nDNA and target mitochondria, are involved in cancer metastasis is still unknown. In light of these considerations, we used two ovarian cancer cells (SKOV3/SKOV3.ip1) with different migration potentials as a model. A proteomic approach was used to identify differential expressions of proteins in mitochondria between the two cells.
2. Materials and methods
2.1. Cell culture and harvest
SKOV3 cells originating from human ovarian carcinoma tissues were obtained commercially from ATCC (USA); its subseries SKOV3.ip1 cells, derived from ascitic tumor cells of nude mice bearing a tumor of SKOV3 cells, were kindly provided by Prof. You-ji FENG of Fudan University, China, who, in turn, firstly received them from M.D. Anderson (USA). The two cell lines were grown in RPMI 1640 media with 15% fetal bovine serum and 2 mmol/L l-glutamine and maintained in the incubator at 37 °C with 5% CO2. When reaching an 80% growth density, the cells were harvested with 2.5 mg/ml trypsin, and then suspended and centrifuged. The resulting cell pellets each contained ≥2×107 cells and were stored at −80 °C for further use.
2.2. Enrichment and confirmation of mitochondria
Mitochondria enrichment was performed by the use of the mitochondria isolation kit for culture cells (Pierce, Rockford, IL USA) and the process was performed according to the manufacturer’s instructions with minor modification. In detail, prior to the process, the protease inhibitor mixture (Sigma, USA) was added to Reagents A and B. The cells were resuspended in 800 μl Reagent A and mixed by 5 s vortex and incubated on ice for 2 min. After 10 μl Reagent B was added and mixed by 5 s vortex, the cell suspensions were incubated on ice for 5 min and transferred to Dounce tissue grinder. They were then homogenized on ice with 80 strokes until over 80% cells were lysed under a microscope check. The lysed cells were returned to the original tubes, with 800 μl of Reagent C added, and mixed by inverting the tubes several times. The mixtures were centrifuged at 700×g for 10 min at 4 °C. The supernatants were transferred to new tubes and centrifuged again at 3 000×g for 15 min at 4 °C. After the supernatants were removed, the pellets containing mitochondria were added to 500 μl Reagent C and centrifuged at 12 000×g for 15 min at 4 °C. The final pellets containing mitochondria were stored at −80 °C until further use.
The enriched mitochondrial fractions were confirmed under a transmission electron microscope (TEM; JEM-100CX2, JEOL; Department of Histology and Embryology, Peking University Health Science Center, Beijing, China) observation. Briefly, a portion of freshly prepared mitochondria were washed by 2.5% (v/v) glutaric dialdehyde at volume ratio 10:1, and fixed by 1% (v/v) osmic acid with an addition of saturate uranyl acetate. After incubation over night and dehydration by gradient acetone, the mitochondria were displaced by propylene oxide and embedded by cyclo-lipoids 618; sections were made by ultramicrotome and observed under TEM.
2.3. Protein isolation and quantification
The mitochondrial fractions isolated from SKOV3 and SKOV3.ip1 cells were transmitted to the Beijing Protein Innovation to perform protein isolation, quantification, 2-dimensional (2-D) gels electrophoresis, and protein sequencing. Briefly, the mitochondrial pellets were lysed by sonication in lysis buffer on ice for 1 min. The mitochondrial solutions were then incubated overnight at −20 °C in 5× volumes of precipitate buffer. The proteins of mitochondria were precipitated by centrifugation and washed with protein precipitate buffer twice. After drying, the proteins were dissolved by sonication for 5 min in lysis buffer containing 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), 2 mmol/L ethylenediaminetetraacetic acid (EDTA), and 20 mmol/L dithiothreitol (DTT). The lysate was centrifuged at 20 000×g for 30 min. The supernatant was collected and used for the 2-D electrophoresis analysis.
Protein quantification was achieved by modified Bradford measurement. Different concentrations of bovine serum albumin (BSA; Beijing Yuanpinghao Biotech Co., China) were diluted to make a standards curve, and to the interested samples Coomassie brilliant blue G-250 (Amersham Pharmacia, Chalfont St. Giles, UK) was added. The quantification was determined by its relative absorption under 595 nm.
2.4. 2-Dimensional gel electrophoresis
2.4.1. Immobilized pH gradient (IPG) strip rehydration and isoelectric focusing (IEF)
A total of 150 μg of proteins in mitochondria were re-suspended in freshly prepared rehydration buffer, and then dropped in the sample-loading cups at the anode ends of the strips. We carefully located the 18-cm pH 3–10L IPG strips in the electrode plate of the IPGphor apparatus (Amersham Pharmacia). IEF was conditioned with a max voltage at 8 kV, totaling 75 kV/h. The strips were removed and subjected to the second dimension of electrophoresis.
2.4.2. Equilibration
The strips were taken out after electrophoresis, and the oil on the gel surface was absorbed. The DTT and iodoacetamide were added in turn to the equilibrate buffer, and shaken for 15 min. The strips were washed with distilled water and dried up by a filter paper.
2.4.3. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
The equilibrated IPG was loaded carefully on the tops of six pre-cast 12% SDS polyacrylamide gels (Sigma). To be assured of the thorough contact of the IPG with the SDS gels, we sealed them with acrylamide gel, and then put them into the perpendicular plate of elecrophoresis apparatus (ETTAN II, Amersham Pharmacia), which was then loaded in the electrophoresis chamber with a running buffer. SDS-PAGE was carried out under constant power at maxima of voltage and electric current for 6 h until the bromophenol blue electrophoresis reached the lower edge. Then the gels were taken off for further staining and imaging.
2.4.4. Gel staining
After fixation by 40% ethanol and 10% glacial acetic acid, and sensitization by 30% ethanol, sodium acetate, three hydro sodium acetate and sodium thiosulfate, the silver staining was performed by AgNO3 with 37% methanol. The gels were rinsed twice with water, followed by coloration carried out by Na2CO3 with 37% methanol, and stopped by EDTA·Na2·2H2O, and then stored in 1% glacial acetic acid at 4 °C.
2.5. Image analysis, spot cut, and enzymatic digestion
2.5.1. Image analysis
The images of six gels (three from SKOV3 and three from SKOV3.ip1) were scanned by Power Look 2100 xl Gel Image Scanner (Amersham Pharmacia). The color information in the images was discarded using the Photoshop software, then analyzed by ImageMaster 2D Platinum 5.0 software (Amersham Pharmacia). Spot detection and matching between the six gels were performed by using Wizard mode automatically, followed by manual matching. We rearranged the serial numbers of spots, after decreasing the background and normalization of the spot densities against the whole-gel densities. We set up a virtual average gel based on the reference gel, set molecular weight and pI, and then worked out the results in an Excel file. Statistical analysis was performed by independent Student’s t-test. The P values of the intensity changes of each spot among three independent experiments were calculated. The protein spots with P values less than 0.05 were considered as those displaying significant changes between SKOV3 and SKOV3.ip1 cells.
2.5.2. Spot cut and enzymatic digestion
The gel pieces containing the interested spots were carefully cut off from the SDS gels. These gel pieces were washed with 50 µl Milli-Q H2O for 5 min twice, followed by 50 µl acetonitrile (ACN) with 50 mmol/L NH4HCO3 (50%/50%, v/v) for 30 min twice. The supernatants were discarded. When the color of Coomassie blue began to vanish, the gel pieces were dehydrated using 50 µl ACN and dried by vacuum centrifugation. The dried gels were added with 10 mmol/L DTT/25 mmol/L NH4HCO3 and incubated at 56 °C for 1 h. Then they were immerged by 55 mmol/L iodoacetamide (IAM)/25 mmol/L NH4HCO3 and incubated in the dark for 45 min. After that, the gels were washed in sequence with 25 mmol/L NH4HCO3, 50% ACN, and pure ACN for 10 min twice and dried by vacuum centrifugation again. The proteins in the gels were digested with trypsin and enzymolyzed by adding 25 mmol/L NH4HCO3 at 37 °C for 4–6 h. Then the proteins were incubated with 3× volumes of digestion buffer containing 67% ACN in water-bath for 30 min. Finally, the proteins were concentrated by vacuum centrifugation and stored at −80 °C.
2.6. Mass spectrometry (MS)
2.6.1. Sample analysis
The peptide of digest mixture was analyzed using matrix assisted laser desorption ionisation/time-of-flight/time-of-flight (MALDI-TOF/TOF) analyzer (Autoflex, BRUKER, USA). The ionization source of mass spectrometer is a pulsed nitrogen laser of wavelength 337 nm. MS spectra were generated at 25 kV accelerating voltage. The data acquisition was employed by positive ion mode and automatic acquisition pattern.
2.6.2. Data collection and analysis
The mass scan range of the peptide mass fingerprint (PMF) was 700–4 000 Da. The external standard of data system of spectra was calibrated by peptide calibration standard II supplied by BRUKER. The results were searched for in MASCOT from Matrix Science. The proteins with the highest probability based on MOWSE score were reported and also selected by comparing the molecular weight and pI derived from the 2-D gels.
2.7. Immunohistochemistry (IHC)
Tissue sections originating from nude mice bearing tumors of SKOV3 and SKOV3.ip1 cells were obtained after being fixated by formalin and buried by paraffin. Each sample was sectioned at 4-µm intervals for two slides. One was used as negative control, using phosphate buffer solution (PBS) to reply to the primary antibody. After deparaffinization in xylene for 5 min twice, the slides were washed with graded ethanol solutions followed by distilled water, and then incubated in fresh 3% (v/v) hydrogen peroxide for 10 min in room temperature to block the activities of endogenous peroxidase. The slides were washed with PBS thrice and added with 0.01 mol/L sodium citrate buffer. To recover the antigens, the slides were boiled for 12 min and dried at room temperature. Then they were washed again and incubated with 10% (v/v) normal goat serum in PBS for 20 min at room temperature to block nonspecific binding. The slides were incubated with primary antibody (mouse monoclonal to superoxide dismutase 2 (SOD2); Abcam, CB4 OFW, UK) at a 1:200 dilution in PBS at 4 °C overnight. After being washed and incubated for 20 min with polymer helper (freshly prepared), the slides were incubated for 30 min at room temperature with secondary antibodies (multiclonal goat mouse anti-IgG; Santa Cruz, USA), which were labeled with biotin and diluted with PBS to 1:100. After being washed with PBS, the slides were incubated with streptavidin/peroxidase for 20 min at 37 °C and washed with PBS again. Coloration of the slides was then performed by using diaminobenzidine (DAB) for 5 min, ending with adding water. The slides were stained with hematoxylin for 5 s and washed with water followed by 1% (v/v) HCl ethanol. Then the slides were immersed in 1% (v/v) ammonia water for 30 s to change the color to blue. After dehydration in gradient ethanol, the slides were washed with xylene and mounted in neutro gummi for future image analysis.
3. Results
3.1. Successful isolation of mitochondria from SKOV3 and SKOV3.ip1 cells
The appearances of mitochondria separated from SKOV3 and SKOV3.ip1 cells, as described above, were closely observed by TEM. Representative photographs of mitochondria under TEM were shown in Fig. 1. The mitochondria could be identified with the round shape with intact membrane in different sizes. This was different from the granule shape of normal mitochondrion, partly due to limited swelling. The mitochondrial cristae are shown as dense staining among the mitochondrial matrix. The enriched mitochondria were randomly distributed in the matrix that contained a part of the subfraction structure of cytoplasm and the apparent clearing with nucleus.
Fig. 1.
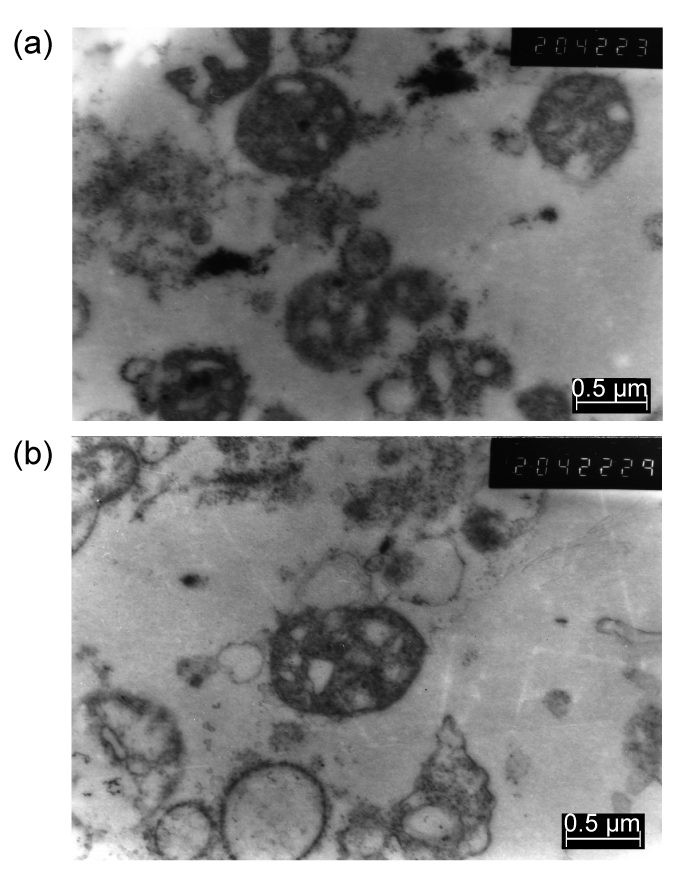
Electron microscope photographs of mitochondria isolated from SKOV3 (a) and SKOV3.ip1 (b) cells
Mitochondria were distributed randomly in the matrix without nucleus
3.2. Separation of proteins in mitochondria by 2-D gel electrophoresis
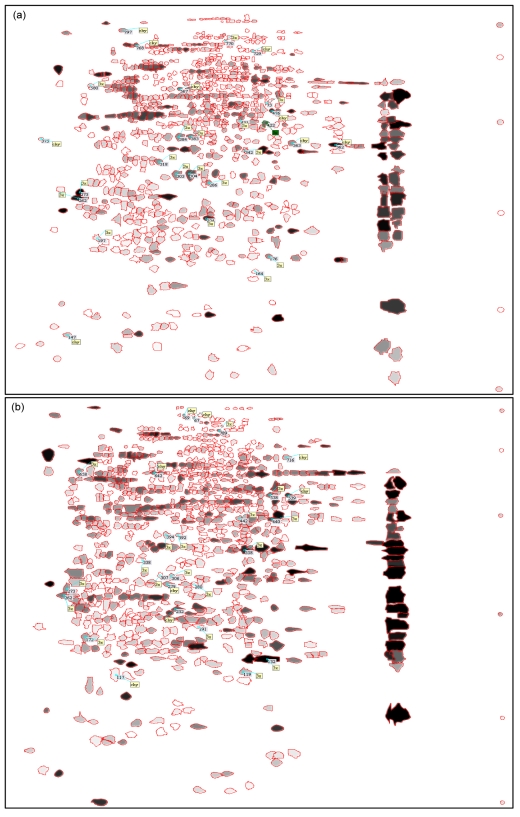
Mitochondria of SKOV3 and SKOV3.ip1 cells were fractionated and the proteins of the fractions were separated by 2-D gel electrophoresis. About 800 spots were detected in the gels. A representative gel image of each cell is shown in Fig. 2. The spots were allotted on the gels with a clean background and no tailing pattern. Each of the three gels from the same cells was synthesized into one gel named gel-syn by analysis software. The gels were analyzed and the spots were filtered. Each valid spot filtered out was assigned one accession number of a corresponding database that was based on its location in the gel, which represented its pI and molecular weight (Fig. 3). The match rates of the three gels from the same cells were all over 85%, indicating that the electrophoresis was reproducible. The distribution of the spots was similar between the two cells. The match rate of protein spots between two cells was 69.9% (Table 1).
Fig. 2.
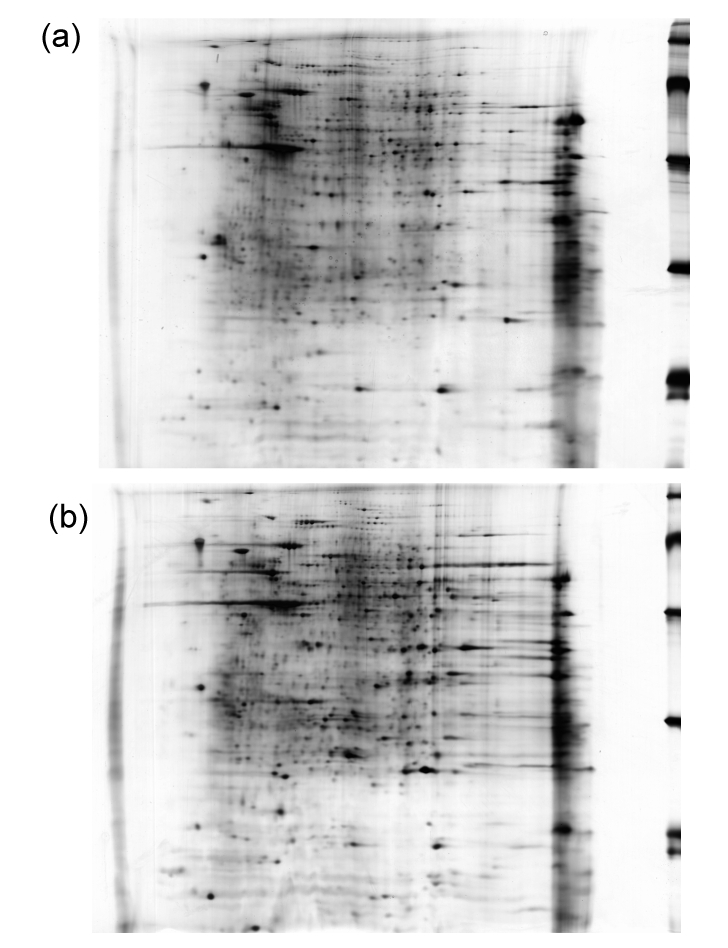
Representative images of 2-D gels of SKOV3 (a) and SKOV3.ip1 (b) cells
Fig. 3.
Images of synthesized 2-D gels of SKOV3 (a) and SKOV3.ip1 (b) cells
Each valid spot was circled by a red line and assigned one accession number of the corresponding database rank by its location in the gel, which represents its pI and molecular weight
Table 1.
Amounts of spots and matched spots and the match rates of six gels
| Cells-gel number | No. of spots | Number of matched spots/match ratio (%) |
Cells-gel number | No. of spots | Number of matched spots/match ratio (%) |
|||||
| SKOV3-1 | SKOV3-2 | SKOV3-3 | SKOV3.ip1-1 | SKOV3.ip1-2 | SKOV3.ip1-3 | SKOV3-syn | ||||
| SKOV3-1 | 810 | SKOV3.ip1-1 | 838 | |||||||
| SKOV3-2 | 839 | 780/94.6% | SKOV3.ip1-2 | 842 | 810/96.4% | |||||
| SKOV3-3 | 744 | 665/85.6% | 699/88.3% | SKOV3.ip1-3 | 840 | 786/93.7% | 780/92.7% | |||
| SKOV3-syn | 811 | 779/96.1% | 811/98.3% | 706/90.8% | SKOV3.ip1-syn | 831 | 831/99.6% | 809/96.7% | 800/95.8% | 574/69.9% |
3.3. Identification of differentially expressed spots
Thirty-five differentially expressed spots were detected. Seventeen spots were found only in one cell line. Among these spots, eight in gels of SKOV3.ip1 cells and nine in gels of SKOV3 cells. In addition, 18 spots were detected with different expression levels over three-fold, nine spots with higher levels in SKOV3 and the others with higher levels in SKOV3.ip1. The IDs, pIs, molecular weights (M w) of the spots detected are all listed in Table 2, and the densities, areas, and volumes of these spots are also listed.
Table 2.
Differential expressed gel spots between SKOV3 and SKOV3.ip1 cells
| Spot | ID | pI | Mw | Density | Area | Volume (%) |
| Three-fold spots | 119 | 7.31 | 24276.00 | 13632.10 | 9.62 | 0.08 |
| 132 | 7.73 | 25618.00 | 45465.30 | 26.07 | 1.62 | |
| 172 | 4.63 | 28116.00 | 13957.00 | 9.57 | 0.26 | |
| 191 | 6.56 | 29398.00 | 6754.37 | 6.24 | 0.04 | |
| 262 | 4.27 | 32996.00 | 21237.90 | 9.14 | 0.19 | |
| 273 | 4.32 | 33704.00 | 18221.00 | 8.84 | 0.15 | |
| 280 | 6.48 | 34197.00 | 4291.43 | 6.19 | 0.03 | |
| 306 | 6.09 | 35164.00 | 11519.20 | 5.50 | 0.04 | |
| 307 | 5.90 | 35249.00 | 4565.97 | 4.24 | 0.02 | |
| 338 | 5.60 | 37113.00 | 5543.63 | 7.21 | 0.04 | |
| 358 | 7.34 | 38324.00 | 32350.00 | 12.13 | 0.35 | |
| 392 | 6.20 | 40300.00 | 8919.30 | 4.20 | 0.03 | |
| 394 | 6.00 | 40374.00 | 10080.50 | 6.01 | 0.04 | |
| 440 | 7.80 | 42482.00 | 28229.50 | 6.52 | 0.17 | |
| 442 | 7.25 | 42611.00 | 13740.80 | 5.70 | 0.32 | |
| 538 | 7.77 | 49131.00 | 23256.20 | 5.18 | 0.10 | |
| 658 | 4.52 | 57216.00 | 18384.00 | 8.72 | 0.18 | |
| 789 | 6.91 | 79590.00 | 25571.70 | 1.58 | 0.04 | |
| Differential spots | 67 | 6.48 | 90064.00 | 14010.70 | 0.92 | 0.01 |
| 69 | 6.31 | 92483.00 | 21038.00 | 0.92 | 0.02 | |
| 117 | 5.15 | 23979.00 | 6091.21 | 17.53 | 0.08 | |
| 232 | 6.16 | 31492.00 | 25637.40 | 8.53 | 0.15 | |
| 279 | 6.02 | 34239.00 | 16318.30 | 7.53 | 0.07 | |
| 539 | 8.08 | 49015.00 | 33059.60 | 4.67 | 0.17 | |
| 642 | 5.80 | 56741.00 | 24336.40 | 4.08 | 0.11 | |
| 716 | 8.05 | 62857.00 | 13434.70 | 6.69 | 0.08 | |
| 147 | 4.16 | 17852.00 | 13148.20 | 6.36 | 0.09 | |
| 362 | 8.86 | 39610.00 | 38133.90 | 10.55 | 0.50 | |
| 363 | 8.10 | 39880.00 | 19782.10 | 7.75 | 0.15 | |
| 375 | 3.69 | 40525.00 | 6460.44 | 3.00 | 0.02 | |
| 476 | 7.74 | 46395.00 | 34413.10 | 3.61 | 0.18 | |
| 567 | 6.12 | 53597.00 | 28007.70 | 2.89 | 0.12 | |
| 729 | 7.41 | 70748.00 | 14317.00 | 2.55 | 0.03 | |
| 768 | 5.35 | 75537.00 | 25957.40 | 3.08 | 0.08 | |
| 797 | 5.14 | 91031.00 | 17079.10 | 4.76 | 0.07 | |
3.4. Identification of eleven proteins by MALDI-MS on the basis of peptide mass matching
Twenty spots with larger differences and larger volumes were selected to be excised from each of three gels. MS analysis was carried out for each excised spot individually. The identification from the MS analysis was based on matching peptides in the database search, together with the theoretical molecular weight and pI values. Ten spots were identified as gene products, listed in Table 3; one was a hypothetical protein. Nine spots failed to be identified from MS analysis, this being attributed mainly to the limitation of the spot volume. Among the eleven spots, seven spots were found expressed at different levels between the two cells, and four spots were expressed only in SKOV3 or SKOV3.ip1. Four of ten proteins identified were located in mitochondria, and the MS analysis is shown in Fig. 4. Four proteins were found in the cytoplasm, one in the endoplasmic reticulum, and the location of the remaining one was unknown.
Table 3.
Differential expressions of gene products identified by MS analysis and a hypothetical protein
| Spot | SKOV3.ip1/SKOV3 | Gene name | Gene | Gene ID | Location |
| 132 | 14.05210 | Manganese-dependent superoxide dismutase | MnSOD/SOD2 | 6648 | Mitochondrion |
| 358 | 5.74082 | mRNA turnover 4 homolog | MRTO4 | 51145 | Mitochondrion |
| 442 | 3.99338 | Mitochondrial ribosomal protein | MRPL38 | 64978 | Mitochondrion |
| 658 | 5.12857 | Heat shock protein gp96 precursor | LOC145811 | 145811 | Endoplasmic reticulum |
| 119 | 3.80444 | Dedicator of cytokinesis 11 | DOCK11 | 139818 | Cytoplasm |
| 172 | 3.15006 | Sperm associated antigen 9 isoform 1 | SPAG9 | 9043 | Cytoplasm |
| 273 | 0.24013 | Dynein, cytoplasmic 1, heavy chain 1, isoform CRA_f | DYNC1H1 | 1778 | Cytoplasm |
| 232 | SKOV3.ip1 | Zinc finger, RAN-binding domain containing 2 | ZRANB2 | 9406 | Unknown |
| 539 | SKOV3.ip1 | Fumarate hydratase | FH | 2271 | Mitochondrion |
| 363 | SKOV3 | Interleukin enhancer binding factor 3 | ILF3 | 3609 | Nucleus or cytoplasm |
| 362 | SKOV3 | Predicted: hypothetical protein |
Fig. 4.
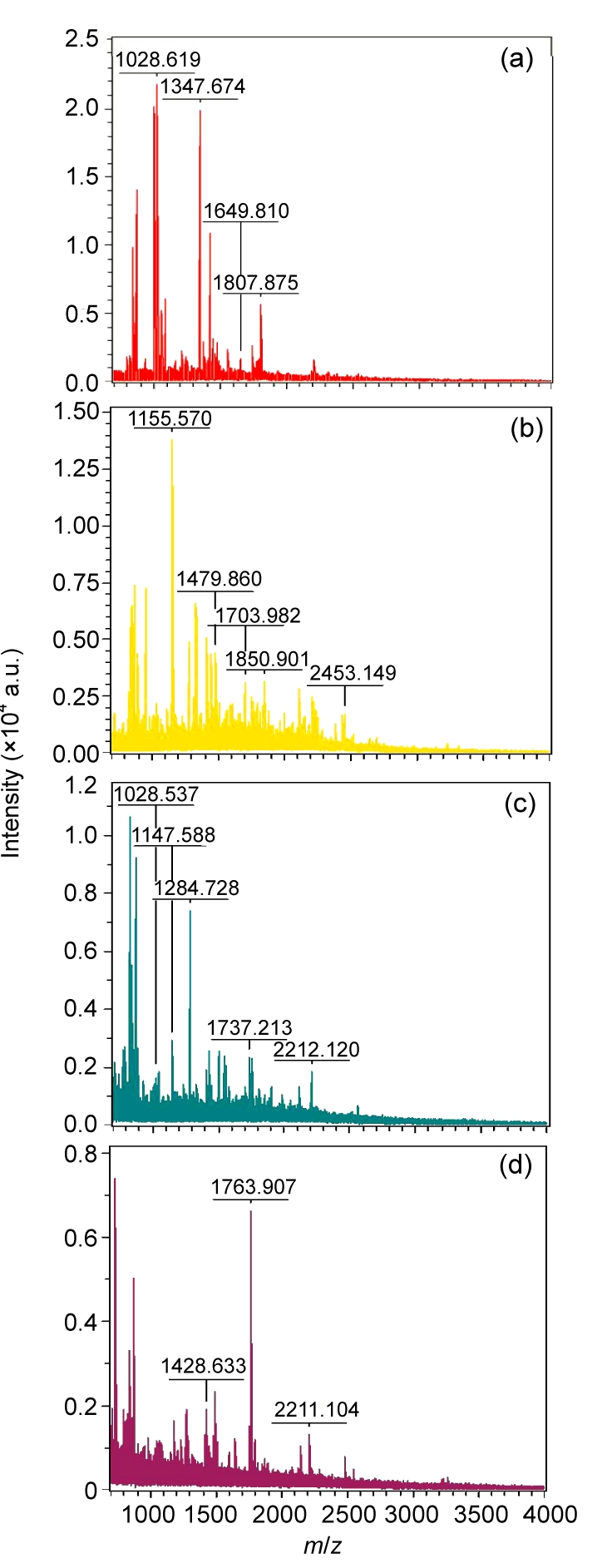
MS patterns of the four identified peptides located in mitochondria
(a) Spot 132, SOD2; (b) Spot 358, MRTO4; (c) Spot 442, MRPL38; (d) Spot 539, FH
3.5. Different expression levels of SOD2 between SKOV3.ip1 and SKOV3 cells
SOD2 was expressed in both SKOV3 and SKOV3.ip1 tumors in IHC analysis. It is obvious that stronger staining was observed in the SKOV3.ip1 tumor compared to the SKOV3 tumor (Fig. 5).
Fig. 5.
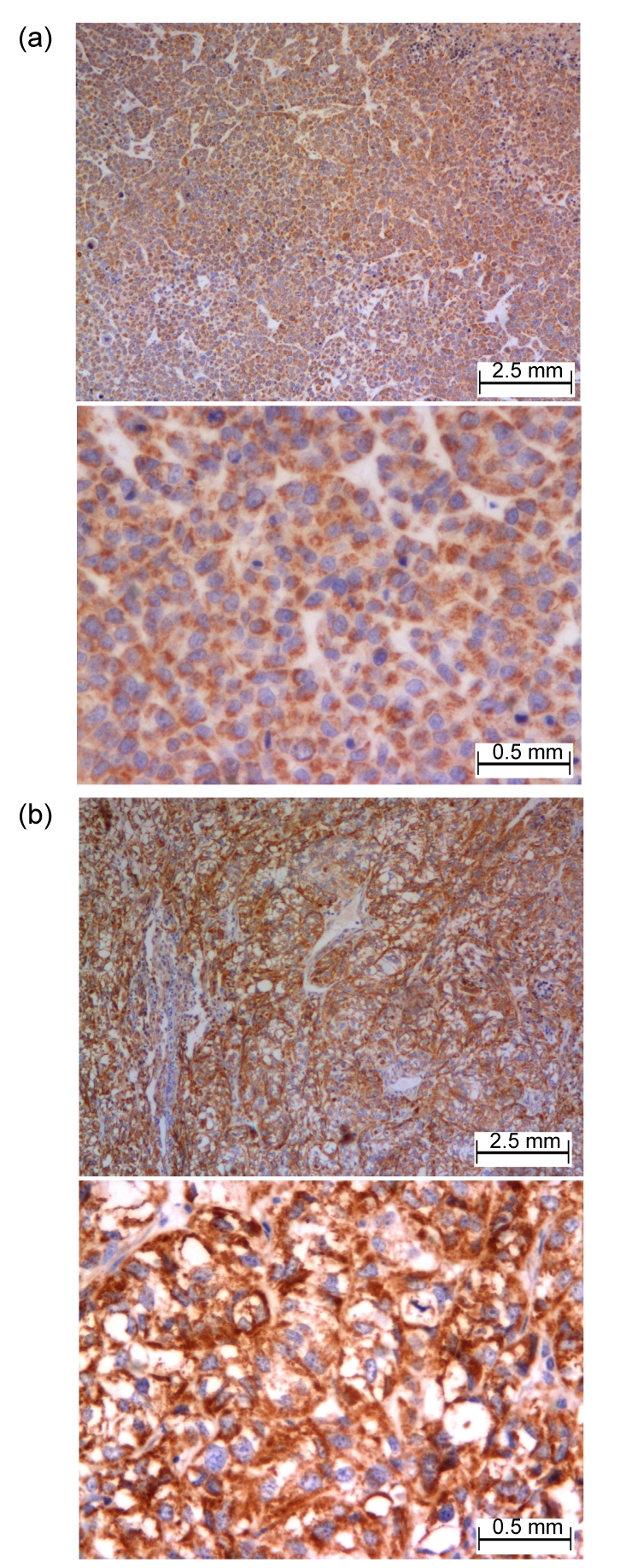
Staining of SOD2 by IHC in SKOV3 (a) and SKOV3.ip1 (b) tumors
The expression level of SOD2 was significantly higher in SKOV3.ip1 than in SKOV3
4. Discussion
We investigated the protein alterations in mitochondria using ovarian cells. Our data show that not only antioxidative ability, but also an energy metabolism disorder, might be relevant to the aggressive behaviors of ovarian cancer cells.
The mitochondrion produces adenosine triphosphate (ATP) needed by cellular metabolism through oxidative phosphorylation (OXPHOS). In addition, mitochondria are also involved in apoptosis (Newmeyer and Ferguson-Miller, 2003). Theoretically, the structural alteration or functional disorder of proteins in mitochondria may result in the activity changes of OXPHOS enzymes, which may lead to the reduced ATP, and therefore influence the signal in the apoptosis pathway and the accumulation of reactive oxygen species (ROS) in cells. The imbalance of metabolism and energy production may contribute to the strength of the invasion and metastasis of tumor cells. In fact, the membrane potential of mitochondrion has been found to be related to the aggressive behaviors of colon-rectum cancer cells (Heerdt et al., 2005).
Based on a previous study, the SKOV3.ip1 was found to exhibit higher degrees of migration and invasive potential than its mother cell line, SKOV3 (Bai et al., 2006). These two cells were an optimal in vitro model to investigate the molecular variations that might relate to the metastasis of ovarian cancer. However, the difference we detected here still represented only the alterations between two cells in vitro, especially the enzymes involved in antioxidation and metabolism, which may be influenced heavily by circumstance. For this reason, we selected manganese-dependent superoxide dismutase (MnSOD or SOD2). This enzyme is the most valuable finding in this study, as a candidate protein for further investigation of its expression in vivo by IHC using the original tissue sections from nude mice bearing SKOV3 and SKOV3.ip1 tumors. A higher expression level of SOD2 was detected in the SKOV3.ip1 tissue than in SKOV3 tissue. This result confirms our findings from the in vivo state.
Using a 2D-PAGE/MS proteomic approach, several proteins identified might relate to ovarian cancer metastasis. Four proteins were known to be located in mitochondria. Among them, SOD2 was the most valuable finding. SOD2 is an antioxidant enzyme located in the mitochondrial matrix. Mitochondria are known as the major sites for reactive oxygen species (ROS) production. The generation of ROS might damage DNA and other large cellular molecules. SOD2 catalyzes the dismutation of superoxide radicals into hydrogen peroxide and oxygen, thus being a defense against oxidative damage induced by ROS. SOD2 deficiency increases the susceptibility of mitochondria to oxidative injury. Different studies have suggested the association of the SOD2 with cancers. For instance, the polymorphism of SOD2 was found to be related to the risk of lung cancer (Zejnilovic et al., 2009). Moreover, the SOD2-depleted cells exhibit a slower growth rate and decreased viability. These cells show a high sensitivity to superoxide, and die through apoptosis (Takada et al., 2009). Thus, it is documented that, compared with normal cell counterparts, SOD2 levels were decreased in neoplastic transformed cells, such as in ovarian cancer (Hu et al., 2005), and the suppression of SOD2 expression by small interfering RNA (siRNA) leads to stimulation of cell proliferation in vitro and more aggressive tumor growth in vivo. Interestingly, in contrast, our findings suggest that decreased SOD2 might be relevant to the aggressive behavior of ovarian cancer cells.
However, similar to our findings, in several recent studies, elevated SOD2 levels have also been reported to be associated with increased frequencies of tumor invasion and metastasis in certain cancers. For example, the severity of invasion was directly correlated with SOD2 expression levels in fibrosarcoma cells and bladder tumor cells (Connor et al., 2007). Expression of SOD2 was consistently increased in high-grade and advanced-stage bladder tumors (Hempel et al., 2009). Activity of SOD2 was significantly higher in advanced-stage tumor or tumor with nodal metastasis in the head and neck squamous cell carcinoma (HNSCC) (Salzman et al., 2007). Elevated SOD2 level was associated with lymph node metastasis in oral squamous cell carcinoma (Ye et al., 2008).
Several studies have suggested the underlying mechanism for the observed up-regulations of SOD2 involved in cancer metastasis. It is known that the matrix degrading protein matrix metalloproteinases (MMPs) play a critical role in the process of stromal invasion and metastasis of cancer cells. Nelson et al. (2003) found that increased SOD2 could increase the DNA-binding activities of MMP transcription factors AP-1 and SP-1. Moreover, SOD2 enhanced MMP-1 promoter activity via the Ras/mitogen-activated protein kinase/extracellular signal-regulated kinase (Ras/MAP/MEK) signaling cascade, and thus elevated MMP production. In addition, enhanced collagen degradation was also observed, as it is combined with overexpression of SOD2 (Nelson et al., 2003). Another interpretation is that the expressions of MMP-9 and vascular endothelial-derived growth factor (VEGF), an ascertained metastasis-related factor by promoting angiogenesis, were shown to be H2O2-dependent. Increased SOD2 expression was found to be accompanied by a significant decrease in catalase (CAT) activity, resulting in a net increase in H2O2 production in cells (Hempel et al., 2009). In addition, the high level of SOD2 led to oxidative inactivation of PTEN (phosphatase and tensin homolog deleted from chromosome 10) and resulted in activation of Akt/PI3K signals, and consequently up-regulated the expression of VEGF (Connor et al., 2005). Furthermore, SOD2 overexpression was associated with a loss of vinculin-positive focal adhesion (Connor et al., 2007). In HeLa cells, the overexpression of SOD2 decreased autophagy and cell death, and blocking SOD2 expression by siRNA increased ROS generation (Chen et al., 2007).
The expression of SOD2 is correlated with the cell response to environmental insults and the many factors involved. The epigenetic silencing, including DNA methylation and histone acetylation, constitutes one mechanism leading to the decreased expression of SOD2 in breast cancer (Hitchler et al., 2006) and some pancreatic cell lines (Hurt et al., 2007). Mutations in the promoter region of the SOD2 gene might activate AP-2-dependent dysregulation of SOD2 expression in cancer cells. The high level of SOD2 observed in aggressive cancer cells may be due in part to the absence of AP-2 transcriptional repression (Xu et al., 2008). Recently, a new control mechanism of the regulation of SOD2 has been revealed. The tumor suppressor protein p53, c-myc, and the life-span determinant protein p66shc promote oxidative stress in mitochondrion. p53 has been found to repress SOD2 expression by either binding directly to the promoter of SOD2 or indirectly suppressing its transcription factor SP1. Thereby, the loss of p53 function could produce the up-regulation of SOD2 and the enhancements of the ability of tumor cells against oxidative damage and resistance to apoptosis (Pani et al., 2009).
Another interesting finding in our study was that fumarate hydratase (fumarase, FH) was also more highly up-regulated in SKOV3.ip1 than in SKOV3. FH is an important Krebs cycle enzyme in mitochondria. It is well documented that FH underlies a tumor susceptibility syndrome, hereditary leiomyomatosis and renal cell cancer (HLRCC). This syndrome is characterized by benign cutaneous and uterine leiomyomas, renal cell carcinomas, and uterine leiomyosarcomas (Stewart et al., 2008). Recently, down-regulation of FH was found in stabilization of transcription factor hypoxia-inducible factor (HIF). The HIF increased the expressions of angiogenesis-regulated genes, such as VEGF, which led to high microvessel density and tumorigenesis. Therefore, FH functions as a tumor suppressor (Sudarshan et al., 2009; Yogev et al., 2010). Since no other enzyme of the Krebs cycle was detected with a significantly different expression in this study, whether or not the increased expression of FH correlated with the migration and invasive potential of ovarian cancer cells was unknown. The underlying mechanism that links FH and aggressive cancer cells appears obscure, and requires further study.
Little attention has been given to the remaining two differently expressed proteins in mitochondria detected in this study. Mitochondrial ribosomal protein L38 (MRPL38), a mitochondrial ribosomal protein, was reported to be four-fold overexpressed in precursor T-cell lymphoblastic lymphoma/leukemia (pre-T LBL) compared to that in thymic tumors (Lin and Aplan, 2007). MRPL38 has been proposed as a candidate gene that might contribute to T-cell leukemogenesis, and a potential therapeutic target for treatment of pre-T LBL. mRNA turnover 4 homolog (MRTO4), a gene related to reproduction, has been reported only in association with spermiogenesis in male mice (Huang et al., 2009). In addition, it was not surprising that no mtDNA-encoded genes were differentially detected between the two cells, since only 13 proteins encoded by mtDNA and only 20 significantly expressed spots were selected from the 2-D gels analyzed by MALDI-MS.
Our results indicate that the alterations of proteins in mitochondria might play an important role in the migration and invasion of ovarian cancer cells. Increases in SOD2 and FH expressions indicated the enhancements of antioxidation and metabolic potentials. The changes in tumor cells might modulate the migration and invasion of tumor cells, contributing to their aggressive and metastatic behaviors. The underlying mechanism warrants further study.
References
- 1.Bai F, Feng J, Cheng Y, Shi J, Yang R, Cui H. Analysis of gene expression patterns of ovarian cancer cell lines with different metastatic potentials. Int J Gynecol Cancer. 2006;16(1):202–209. doi: 10.1111/j.1525-1438.2006.00296.x. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007;120(23):4155–4166. doi: 10.1242/jcs.011163. [DOI] [PubMed] [Google Scholar]
- 3.Connor KM, Subbaram S, Regan KJ, Nelson KK, Mazurkiewicz JE, Bartholomew PJ, Aplin AE, Tai YT, Aguirre-Ghiso J, Flores SC, et al. Mitochondrial H2O2 regulates the angiogenic phenotype via PTEN oxidation. J Biol Chem. 2005;280(17):16916–16924. doi: 10.1074/jbc.M410690200. [DOI] [PubMed] [Google Scholar]
- 4.Connor KM, Hempel N, Nelson KK, Dabiri G, Gamarra A, Belarmino J, van de Water L, Mian BM, Melendez JA. Manganese superoxide dismutase enhances the invasive and migratory activity of tumor cells. Cancer Res. 2007;67(21):10260–10267. doi: 10.1158/0008-5472.CAN-07-1204. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5(11):857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 6.Heerdt BG, Houston MA, Augenlicht LH. The intrinsic mitochondrial membrane potential of colonic carcinoma cells is linked to the probability of tumor progression. Cancer Res. 2005;65(21):9861–9867. doi: 10.1158/0008-5472.CAN-05-2444. [DOI] [PubMed] [Google Scholar]
- 7.Hempel N, Ye H, Abessi B, Mian B, Melendez JA. Altered redox status accompanies progression to metastatic human bladder cancer. Free Radic Biol Med. 2009;46(1):42–50. doi: 10.1016/j.freeradbiomed.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hitchler MJ, Wikainapakul K, Yu L, Powers K, Attatippaholkun W, Domann FE. Epigenetic regulation of manganese superoxide dismutase expression in human breast cancer cells. Epigenetics. 2006;1(4):163–171. doi: 10.4161/epi.1.4.3401. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Rosen DG, Zhou Y, Feng L, Yang G, Liu J, Huang P. Mitochondrial manganese-superoxide dismutase expression in ovarian cancer: role in cell proliferation and response to oxidative stress. J Biol Chem. 2005;280(47):39485–39492. doi: 10.1074/jbc.M503296200. [DOI] [PubMed] [Google Scholar]
- 10.Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ, Li W, Bi J, Zhang LJ, Fu Q, et al. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39(8):786–794. doi: 10.1111/j.1872-034X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- 11.Hurt EM, Thomas SB, Peng B, Farrar WL. Molecular consequences of SOD2 expression in epigenetically silenced pancreatic carcinoma cell lines. Br J Cancer. 2007;97(8):1116–1123. doi: 10.1038/sj.bjc.6604000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin YW, Aplan PD. Gene expression profiling of precursor T-cell lymphoblastic leukemia/lymphoma identifies oncogenic pathways that are potential therapeutic targets. Leukemia. 2007;21(6):1276–1284. doi: 10.1038/sj.leu.2404685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev Cancer. 2005;5(5):355–366. doi: 10.1038/nrc1611. [DOI] [PubMed] [Google Scholar]
- 14.Nelson KK, Ranganathan AC, Mansouri J, Rodriguez AM, Providence KM, Rutter JL, Pumiglia K, Bennett JA, Melendez JA. Elevated SOD2 activity augments matrix metalloproteinase expression: evidence for the involvement of endogenous hydrogen peroxide in regulating metastasis. Clin Cancer Res. 2003;9(1):424–432. [PubMed] [Google Scholar]
- 15.Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112(4):481–490. doi: 10.1016/S0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 16.Pani G, Koch OR, Galeotti T. The p53-p66shc-manganese superoxide dismutase (MnSOD) network: a mitochondrial intrigue to generate reactive oxygen species. Int J Biochem Cell Biol. 2009;41(5):1002–1005. doi: 10.1016/j.biocel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Salzman R, Kankova K, Pacal L, Tomandl J, Horakova Z, Kostrica R. Increased activity of superoxide dismutase in advanced stages of head and neck squamous cell carcinoma with locoregional metastases. Neoplasma. 2007;54(4):321–325. [PubMed] [Google Scholar]
- 18.Stewart L, Glenn GM, Stratton P, Goldstein AM, Merino MJ, Tucker MA, Linehan WM, Toro JR. Association of germline mutations in the fumarate hydratase gene and uterine fibroids in women with hereditary leiomyomatosis and renal cell cancer. Arch Dermatol. 2008;144(12):1584–1592. doi: 10.1001/archdermatol.2008.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudarshan S, Sourbier C, Kong HS, Block K, Valera Romero VA, Yang Y, Galindo C, Mollapour M, Scroggins B, Goode N, et al. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1α stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol. 2009;29(15):4080–4090. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takada S, Inoue E, Tano K, Yoshii H, Abe T, Yoshimura A, Akita M, Tada S, Watanabe M, Seki M, et al. Generation and characterization of cells that can be conditionally depleted of mitochondrial SOD2. Biochem Biophys Res Commun. 2009;379(2):233–238. doi: 10.1016/j.bbrc.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Liu VW, Xue WC, Cheung AN, Ngan HY. Association of decreased mitochondrial DNA content with ovarian cancer progression. Br J Cancer. 2006;95(8):1087–1091. doi: 10.1038/sj.bjc.6603377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Fang F, Dhar SK, Bosch A, St. Clair WH, Kasarskis EJ, St. Clair DK. Mutations in the SOD2 promoter reveal a molecular basis for an activating protein 2-dependent dysregulation of manganese superoxide dismutase expression in cancer cells. Mol Cancer Res. 2008;6(12):1881–1893. doi: 10.1158/1541-7786.MCR-08-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye H, Wang A, Lee BS, Yu T, Sheng S, Peng T, Hu S, Crowe DL, Zhou X. Proteomic based identification of manganese superoxide dismutase 2 (SOD2) as a metastasis marker for oral squamous cell carcinoma. Cancer Genomics Proteomics. 2008;5(2):85–94. [PMC free article] [PubMed] [Google Scholar]
- 24.Yogev O, Yogev O, Singer E, Shaulian E, Goldberg M, Fox TD, Pines O. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 2010;8(3):e1000328. doi: 10.1371/journal.pbio.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zejnilovic J, Akev N, Yilmaz H, Isbir T. Association between manganese superoxide dismutase polymorphism and risk of lung cancer. Cancer Genet Cytogenet. 2009;189(1):1–4. doi: 10.1016/j.cancergencyto.2008.06.017. [DOI] [PubMed] [Google Scholar]