Summary
The proximo-distal (PD) axis of the Drosophila leg is thought to be established by the combined gradients of two secreted morphogens, Wingless (Wg) and Decapentaplegic (Dpp). According to this model, high [Wg+Dpp] activates Distalless (Dll) and represses dachshund (dac) in the distal cells of the leg disc while intermediate [Wg+Dpp] activates dac in medial tissue. To test this model we identified and characterized a dac cis-regulatory element (dac RE) that recapitulates the dac’s medial expression domain during leg development. Counter to the gradient model, we find that Wg and Dpp do not act in a graded manner to activate RE. Instead, dac RE is activated directly by Dll and repressed distally by a combination of factors including the homeodomain protein Bar. Thus, medial leg fates are established via a regulatory cascade in which Wg+Dpp activate Dll and then Dll directly activates dac, with Wg+Dpp as less critical, permissive inputs.
Introduction
Animals use multiple mechanisms to establish unique cell types within developing tissues. One well-characterized mechanism depends on morphogens, molecules that trigger distinct responses in responding cells in a concentration-dependent manner. For example, Sonic hedgehog (Shh), secreted from the ventral-most cells of the vertebrate neural tube, exists as a ventral-to-dorsal concentration gradient that in turn establishes five discreet cellular domains along the dorso-ventral (DV) axis (Jessell, 2000). Positional information can also arise from cascades of cross-regulating transcription factors. In the early Drosophila embryo, for example, a network of interacting segmentation genes provides positional information to the pre-blastoderm nuclei along the antero-posterior (AP) axis (Schroeder et al., 2004). In many cases, both mechanisms – morphogen gradients and transcription factor networks – work in concert with each other. Once Shh regulates the expression of an initial set of transcription factors in the neural tube, cross-regulation is required to fully define cellular fates (Briscoe et al., 2000). Analogously, the segmentation gene network in the fly requires an initial asymmetric input that is provided, at least in part, by an anterior-to-posterior gradient of the morphogen Bicoid (Ephrussi and St Johnston, 2004). These and other examples suggest that biological systems often use both mechanisms to generate positional information in developing tissues.
Compared to the vertebrate neural tube and the early Drosophila embryo, the formation of animal appendages requires an additional layer of complexity. In addition to having AP and DV axes, appendages also have a proximo-distal (PD) axis, which forms orthogonally to the two main body axes. Unlike the AP and DV axes, the PD axis is established de novo for each appendage, during embryogenesis. Classical grafting experiments carried out in the cockroach provided important insights into how the PD axis is initiated (French, 1978, 1980). Juxtaposition of nonadjacent leg fragments (e.g. dorsal next to ventral) lead to the formation of supernumerary legs with new PD axes. At the time, the formation of these supernumerary legs was interpreted as resulting from the juxtaposition of different positional values followed by extensive tissue growth to fill in the missing positional values. More recent experiments carried out in Drosophila established that a new PD axis in the leg could be generated by the juxtaposition of two populations of cells, one that expresses the morphogen Wingless (Wg) and one that expresses the morphogen Decapentaplegic (Dpp) (Campbell et al., 1993; Diaz-Benjumea et al., 1994; Lecuit and Cohen, 1997). Moreover, by activating these pathways in a cell-autonomous manner, Lecuit and Cohen (1997) demonstrated that Wg and Dpp have the ability to induce a new PD axis directly, without the induction of another non-autonomous signal. Because Wg and Dpp are expressed in ventral and dorsal sectors, respectively, of developing insect legs (Figure 1A), these observations provided a molecular explanation for the cockroach grafting experiments: the juxtaposition of non-adjacent leg fragments likely resulted in new juxtapositions of Wg and Dpp-expressing cells, which in turn led to the production of a new PD axis.
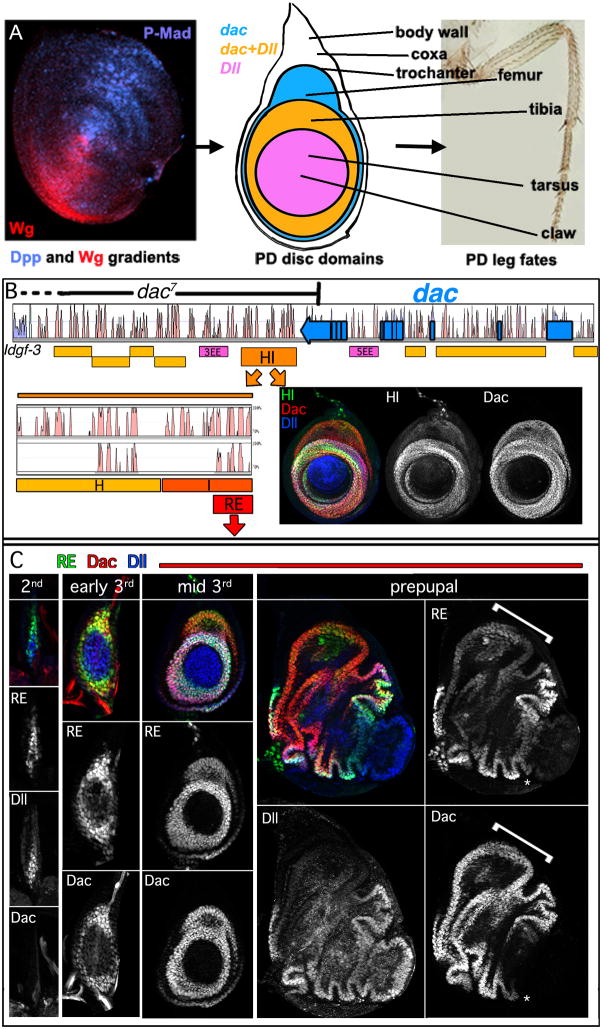
Figure 1. Identification of the Dac Ring Enhancer (RE).
(A) Left: Wg and Dpp gradients in the leg disc shown by staining for Wg (red) and the activated form of the downstream effector of Dpp signaling, phospho-Mad (blue). The middle and right panels show a schematic of a 3rd instar disc and the corresponding proximodistal fates in the adult leg.
(B) Vista plot alignment of D. melanogaster dac locus compared to D. pseudobscura (dac coding region shown in blue). Yellow and orange boxes represent cloned regions tested for the ability to drive reporter gene expression. Pink boxes represent enhancers active in the eye identified previously (Pappu et al., 2005). dac7 is a deletion allele that begins in dac’s last exon, and extends 3′ to the gene, but its 3′ endpoint has not been mapped (Pappu et al., 2005). HI was further subdivided based upon sequence homology (Vista alignments show D. melanogaster HI compared to D. pseudobscura (top) and D. virilis (bottom)).
(C) Leg discs stained for dac RE-lacZ (green), Dac (red), and Dll (blue). lacZ expression was first apparent in the 2nd instar, slightly before Dac protein was detectable. dac RE maintains a ringed pattern throughout development. RE expression is weaker in the Dac-only domain (bracket), compared to the Dac+Dll domain. dac RE is active at high levels at its distal edge where Dac protein is only weakly detected (asterisk).
In addition to initiating PD axis formation, Wg and Dpp have also been proposed to establish positional information along the PD axis of the fly leg in a concentration-dependent manner (Lecuit and Cohen, 1997). In support of this idea, which we refer to here as the gradient model, two readouts of the PD axis were examined. Distalless (Dll), which encodes a homeodomain transcription factor, is expressed in cells that will give rise to the distal-most portion of the leg, from the tip of the tarsus to the middle of the tibia (Figure 1A). In contrast, dachshund (dac), which also encodes a transcription factor, is expressed in a medial domain of the leg’s PD axis, from the tibia to the trochanter (Figure 1A). According to the gradient model, high concentrations of Wg+Dpp activate Dll and repress dac, intermediate concentrations of Wg+Dpp activate dac but not Dll, and lower levels activate neither gene (Lecuit and Cohen, 1997). Three experiments were carried out to support the idea that dac is responsive to lower levels of these signals compared to Dll. First, although both Dll and dac could be induced in cells in which the Dpp pathway was activated cell-autonomously, the outcome depended on the distance the cells were from the endogenous source of Wg: Dll was induced in clones closer to the source of Wg compared to dac induction, and neither readout was induced in clones located far from the source of Wg. Analogously, when the Wg pathway was activated cell autonomously, dac was induced in cells farther from the endogenous source of Dpp compared to Dll induction. Third, when a secreted form of Wg was expressed in dorsal clones (close to the endogenous source of Dpp) it resulted in a nested pattern of PD gene expression, with Dll expressed closest to the source of Wg and dac expressed further from the source (Lecuit and Cohen, 1997). Together, these results suggested that dac and Dll are induced by different levels of Wg and Dpp signaling. However, the expression patterns of Wg and Dpp in the growing leg imaginal disc – the progenitor of the adult leg – raise several questions about how this would happen in molecular terms: because Wg and Dpp are expressed in ventral and dorsal sectors of the leg disc, respectively (Figure 1A), it is unclear how cells might read and integrate a combined Wg+Dpp gradient.
One way to test the gradient model is to dissect the mechanisms that activate Dll and dac during leg development. Dll expression in the leg discs is mediated at least in part by two separable cis-regulatory elements (Estella et al., 2008; McKay et al., 2009). In stage 14 embryos and leg imaginal discs, the ‘leg-trigger’ (LT) enhancer is specifically activated in cells that receive high levels of Wg and Dpp signaling. Consequently, on its own, LT is only active in a small number of cells in the center of the leg disc, close to where the Wg and Dpp expression sectors come into contact. In contrast, when LT is in cis with the Dll maintenance (M) element, which includes the native Dll promoter, an accurate Dll expression pattern is generated. These observations led to the ‘trigger-maintenance’ model, which posits that Dll is first activated by LT or LT-like elements in a small number of cells, and then maintained by the M element in these cells and their progeny (Estella et al., 2008). Notably, due to subsequent growth and maintenance of the Dll-expression domain, this mechanism does not require Dll to interpret graded Wg or Dpp inputs to be accurately expressed. Only high Wg and high Dpp inputs are required to initially activate Dll and, once activated, Wg and Dpp inputs are no longer required. Direct binding of the downstream transcription factors in the Wg and Dpp pathways (Pangolin, Pan, Mothers against Dpp, Mad, and Brinker, Brk) are required for LT activity (Estella et al., 2008). LT activity also requires Sp1, a ventral selector gene that ensures that Wg and Dpp only activate Dll in ventral leg tissue, but not dorsal tissue such as the wing (Estella and Mann, 2010). Together, these results suggest that a gradient mechanism is not required for generating the Dll expression pattern during leg development.
Although the gradient model may not apply to Dll activation, it could still account for the establishment of medial fates along the PD axis, where dac is activated. Here, we test this possibility by characterizing a dac cis-regulatory element that is active in a medial domain along the PD axis. We show that Wg and Dpp inputs play a surprisingly minor role in the direct activation of this element. Consistent with lineage tracing studies showing that the entire dac expression domain is derived from Dll-expressing cells (McKay et al., 2009), we find that direct Dll input is essential for the activation of this element and dac expression. Together, these and other data suggest that dac is activated by a regulatory cascade whereby Wg and Dpp activate Dll, which in turn activates dac. According to this revised model, gradients of Wg and Dpp activities are not required for Dll or dac activation or, therefore, the establishment of positional information along the PD axis. Instead, the combination of Wg and Dpp initiates a regulatory cascade that, coupled to the growth of the leg disc, establishes the PD axis.
Results
Identification of a dac enhancer element
dac7 is a deletion allele of dac that removes DNA 3′ to dac beginning in its last exon (Pappu et al., 2005) (Figure 1B). dac7 completely removes dac expression and function in the leg (see below), but only partially removes dac function in the eye (Pappu et al., 2005), suggesting that cis regulatory elements essential for leg expression are deleted by this allele. Therefore, we searched the DNA deleted by dac7 for a cis-regulatory element that is able to drive expression in medial leg fates along the PD axis during leg development (Fig. 1B). We focused our attention on sequences between dac and the neighboring gene, Idgf-3, in part because of the binding of insulator proteins close to Idgf-3 (Negre et al., 2010). We discovered a 3.6 kb fragment, dac HI, that was able to drive reporter gene expression in a medial leg ring that is very similar to the pattern of Dac protein (Fig. 1B). dac HI was further dissected to a 567 bp fragment that we call the dac Ring Enhancer (RE), which retained strong activity in the medial leg and is well conserved in multiple Drosophila species (Fig. 1B,C). We also looked for, but failed to find, additional leg elements in the dac introns (Figure 1B). In sum, although we cannot exclude that there is an additional element distal to Idgf-3, these results suggest that dac RE is essential for dac expression and function in the leg.
Reporter gene expression driven by dac RE was robust when dac is first activated in the 2nd instar and continued through the remainder of leg development. In the mid to late 3rd instar, reporter gene expression became weaker in the dorsal, Dll-non-expressing, region of the dac expression domain, which gives rise to the femur (referred to here as the ‘Dac-only domain’. Additionally, although Dac protein levels were relatively weak in the 1st tarsal segment, which is the distal-most extent of its domain, expression driven by RE remained strong. dac RE also drove expression in a medial ring in the antenna, but was not active in any of the other imaginal discs (data not shown).
dac RE regulation is similar to dac
Based on previous genetic experiments, dac RE should be regulated by Wg and Dpp in two ways: 1) High levels of Wg and Dpp signaling, which are normally present in the region of the leg disc that will give rise to the distal tarsal segments, should repress RE and 2) intermediate levels of both signals should activate RE. Consistently, in dorsal clones expressing an activated form of Armadillo (ArmΔN), which mimics high Wg activity, both dac and dac RE were repressed, while more lateral ArmΔN clones activated dac and dac RE (Fig. 2A). As previously suggested (Lecuit and Cohen, 1997), the different behavior of these clones is likely due to how close they are to the source of Dpp. Analogously, clones overexpressing a constitutively active Dpp co-receptor, Thickveins (TkvQD), repressed both dac and dac RE in ventral regions of the disc, where endogenous Wg signaling is strong, while more lateral TkvQD clones activated dac and dac RE (Fig. 2B).
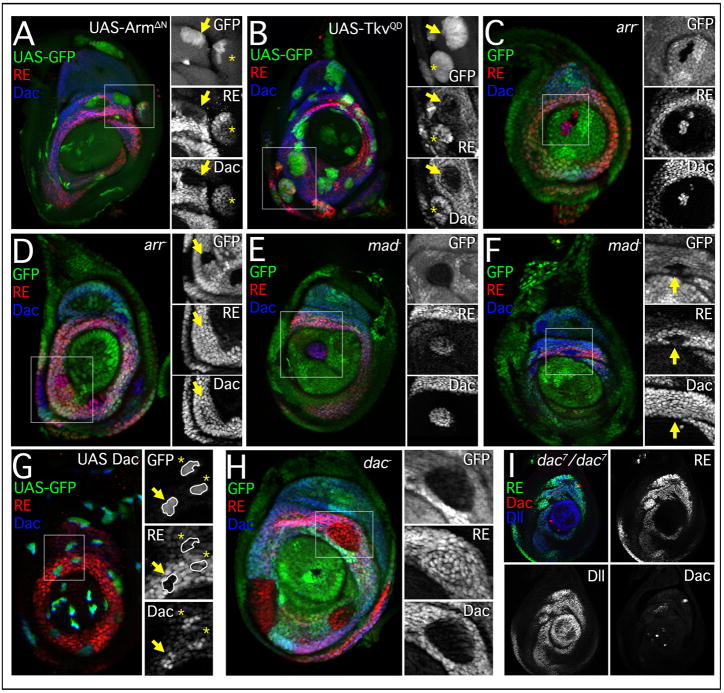
Figure 2. dac RE is regulated in a manner similar to dac.
(A–H) Leg discs stained for RE-lacZ (red), Dac (blue), and Gal4 expressing clones (A, D, G, marked by GFP) or mutant clones (B, C, E, F, H; marked by the absence of GFP). For this and subsequent panels, smaller images show the individual staining patterns of the boxed regions. All clones were examined after growth for 48 hrs at 25°C unless otherwise indicated. arr− and mad− clones were generated in the early 2nd instar.
(A) Clones expressing activated Arm, ArmΔN, create new domains of dac and dac RE activation in lateral tissue (asterisks) and repress both dac and dac RE in medial tissue where high levels of Dpp signaling are present (arrow).
(B) Clones expressing an activated form of the Dpp receptor, TkvQD, repress both dac and dac RE medially (arrow) while activating both dac and dac RE in proximal clones (asterisk).
(C) Distal clones mutant for arrow derepress both dac and dac RE.
(D) Medial clones mutant for arrow have no effect on either dac or dac RE (arrow).
(E) Distal clones mutant for mad derepress both dac and dac RE.
(F) Medial clones mutant for mad have no effect on Dac protein but dac RE activity is absent.
(G) UAS-Dac expression clones. The extent of dac RE repression is variable (compare arrow and asterisks). Discs stained 24 hrs post heatshock.
(H) dac null clones.
(I) dac7/7 mutant disc stained for dac RE-lacZ (green), Dac (red), and Dll (blue). In dac7 mutant discs dac RE activity is still present in a ring-like pattern, while Dac protein is virtually undetectable.
See also Figure S1.
Loss of function analysis of the Wg and Dpp pathways supports the idea that dac RE interprets these signals similarly to dac. Distal clones mutant for the Wg co-receptor, arrow (arr−), derepressed both dac and dac RE, demonstrating that Wg signaling is required to repress dac (Fig. 2C). arr− clones within the dac domain generated after the initiation of dac expression had no effect on dac or dac RE (Fig. 2D). Similarly, distal clones mutant for the transcriptional effector of Dpp signaling, mothers against Dpp (mad), derepressed both dac and dac RE (Fig. 2E). mad− clones within the dac domain also did not affect dac expression (Fig. 2F). However, dac RE activity was lost in some mad− clones (Fig. 2F), indicating a role for continuous Dpp input in maintaining dac RE activity. For both readouts, distal derepression in mad− or arr− clones was only observed when the clones were induced prior to the start of the 3rd instar (72 hrs after egg laying (AEL)); mad− or arr− clones generated in the 3rd instar did not show distal derepression of dac or dac RE-lacZ (Figure S1).
The near-identical behaviours of dac and dac RE-lacZ raised the possibility that dac RE is simply a Dac-responsive autoregulatory element. However, clones overexpressing Dac failed to activate dac RE-lacZ outside the dac domain and showed a reduction in dac RE activity within the dac domain (Fig. 2G). We also tested dac RE activity in clones and leg discs mutant for dac. In both cases dac RE-lacZ expression was unaffected (Fig. 2H,I), demonstrating that dac function is not required for dac RE activity. We also observed that dac mutant clones within the Dac-only domain, where dac RE expression is normally low, resulted in the upregulation of dac RE-lacZ (Figure S1H). Together, these results show that dac RE is not activated by Dac and suggest that dac exhibits negative autoregulation, perhaps to fine-tune its expression levels.
Mutation of multiple binding sites for Wg and Dpp pathway effectors has only a minor effect on dac RE activation
To initially define the direct inputs into dac RE we characterized the in vivo activities of a series of RE deletions (Figure S2). No individual deletion was sufficient to eliminate the ring pattern or to fully derepress distal expression. One deletion, REΔ5, drove ectopic expression in a distal ring at the tarsal/pretarsal boundary and is discussed below. We also tested smaller, 100–200 bp, subfragments of RE and found that none of these were sufficient to reproduce a ring-like expression pattern (Figure S2). Some fragments produced faint arcs of expression in the ventral Wg-expressing domain during late larval stages, suggesting some Wg responsiveness within RE. These results indicate that the control of dac RE activity is distributed along the full element, likely through multiple inputs.
If dac RE is directly activated or repressed in response to the amount of Wg and Dpp signaling, we would expect multiple binding sites for the Wg and/or Dpp transcriptional effectors Pangolin (Pan) and Mad, respectively. Using TargetExplorer (Sosinsky et al., 2003), we found that dac RE has multiple putative binding sites for both transcription factors (Fig. 3A). We performed electrophoretic mobility shift assays (EMSAs) to determine whether or not these candidate sites were able to bind Mad or Pan proteins in vitro (data not shown). Of the seven candidate Pan sites, four showed specific binding to Pan protein that was lost upon mutation of three core nucleotides in the Pan binding site (Figure 3A). Of the seven predicted Mad binding sites, binding to two was lost when the sites were mutated (Figure 3A). As Brinker (Brk) recognizes similar sequences as Mad, we also checked the predicted Mad binding sites for their ability to bind Brk. Four of the seven sequences showed specific binding to Brk that was sequence-specific (Figure 3A).
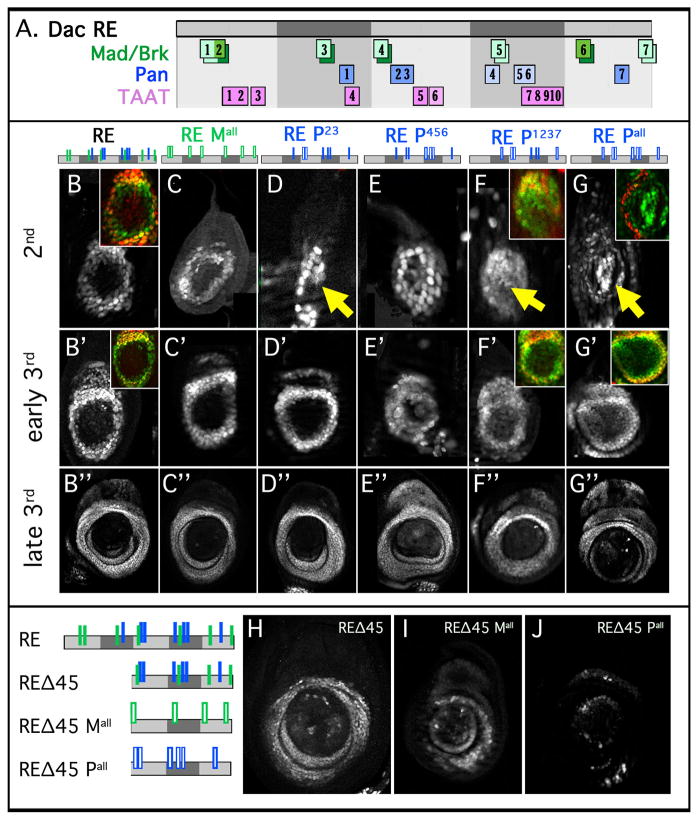
Figure 3. Normal dac RE activation when direct inputs by Mad or Pan are compromised.
(A) Schematic of dac RE showing binding sites for Mad (green), Pan (blue), and TAAT sequences (pink). Binding sites that show specificity in EMSAs have darker shading. Brk binding is shown in boxes dropped down from the Mad boxes.
(B–G) Leg discs at 2nd (B–G), early 3rd (B′–G′), and late 3rd (B″–G″) instar stages stained for the activities of dac RE and mutant versions of dac RE. Mutant elements are schematized above with the mutant sites represented by open bars.
(B) dac RE. The inset shows RE (green) and Dac (red).
(C) dac RE Mall. RE with seven Mad sites mutated.
(D) dac RE P2,3. Mutating two Pan binding sites gives initial expression in some cells distal to the normal Dac domain (arrow).
(E) dac RE P4,5,6. Mutation of the three weak Pan binding sites results in little effect on the RE ring pattern.
(F) dac RE P1,2,3,7. Mutating these four binding sites results in distal expression in the 2nd instar (arrow), in addition to RE’s normal expression domain. A ring-like pattern is eventually formed, but distal expression remains into the 3rd instar. The inset shows RE (green) and Dac (red).
(G) dac RE Pall. Mutation of seven candidate Pan sites results in distal expression in early discs (arrow) but does not significantly affect RE activity in older discs. The inset shows RE (green) and Dac (red).
(H–J) Direct Wg and Dpp inputs are required for expression of a truncated dac RE fragment, REΔ45 (schematics on left).
(H) REΔ45.
(I) REΔ45 fragment with seven Mad binding sites mutated.
(J) REΔ45 fragment with seven Pan binding sites mutated.
We next tested the role of these binding sites in vivo. If intermediate levels of Wg and Dpp inputs activate dac RE in the medial leg, reducing or eliminating these direct inputs should reduce or eliminate dac RE activity during leg development. Single mutants of Mad or Pan sites had no effect on dac RE activity (data not shown). Even when the seven predicted Pan binding sites (RE mPall) or the seven predicted Mad/Brk binding sites (RE Mall) were mutated, enhancer activity was not significantly compromised at the 3rd larval stage (Fig. 3C″,G″). RE Mall drove expression in a largely normal pattern (Fig. 3C″), while RE Pall drove expression in an imperfect ring, with some gaps in the pattern evident in third instar leg discs (Fig. 3G″). Although we cannot rule out that some Pan or Mad binding remains intact in these multiply mutated fragments, these observations do not support models in which dac regulation – both medial activation and distal repression – is sensitive to graded levels of Wg and Dpp inputs.
In contrast to the relatively normal expression patterns these mutant fragments drove in the 3rd instar (Figure 3B″ to G″), some of their activities were not wild type in younger leg discs, e.g. in the 2nd instar (Figure 3B–G). At this stage, RE Mall is expressed like wild type RE, suggesting that Dpp is indirectly required for both distal repression and medial activation (Figure 3B,C). In contrast, when Pan sites were mutated, we observed derepression in distal cells in 2nd instar discs (Figure 3B,D–G). Although the distal derepression driven by these fragments is transient (i.e., is not observed by the 3rd instar), these observations suggest that Pan is directly required for the repression of dac in distal cells when it is first activated in the 2nd instar. Although additional Pan sites not identified by TargetExplorer may also contribute to distal repression, below we provide evidence that other factors are required for repression of dac in the 3rd instar.
Although the above findings are inconsistent with the gradient model, they do not rule out that Mad and/or Pan inputs contribute to dac activation. Support for this idea comes from the analysis of dac RE subfragments, in which Pan or Mad binding sites have been mutated. For example, REΔ45, which is missing subregions 4 and 5, drove a patchy pattern of expression in the medial leg disc (Figure 3H). When all predicted Mad or Pan binding sites were mutated in this compromised context, expression was strongly reduced, particularly when the Pan binding sites were mutated (Figure 3I,J). Together, these observations suggest that Wg and Dpp promote dac RE activity, but that these signaling pathways are not being used in a graded manner to establish positional information along the PD axis.
Dll is essential for dac expression
The above results support a model in which the Wg and Dpp inputs into dac activation are primarily indirect, raising the question of what factor(s) directly activate dac during leg development. Importantly, lineage-tracing experiments demonstrated that the entire dac domain is derived from cells that expressed Dll earlier in development (McKay et al., 2009). Consistent with these observations, Dac protein was first observed in cells that also have low levels of Dll protein (Figure 4A). Dll levels declined in dac-expressing cells until the early 3rd instar when they became undetectable (Figure 4A,B). These observations are consistent with the idea that Dll plays a positive role in dac activation. As a first test of this idea, we examined clones overexpressing Dll, which resulted in an upregulation of both dac and dac RE-lacZ. Upregulation was observed in all parts of the leg disc except in distal regions that normally do not express dac (Figure 4C). Conversely, clones mutant for Dll in the dac domain showed a loss of both dac and dac RE-lacZ expression (Figure 4D). Together, these results demonstrate that Dll is required for the activation of dac.
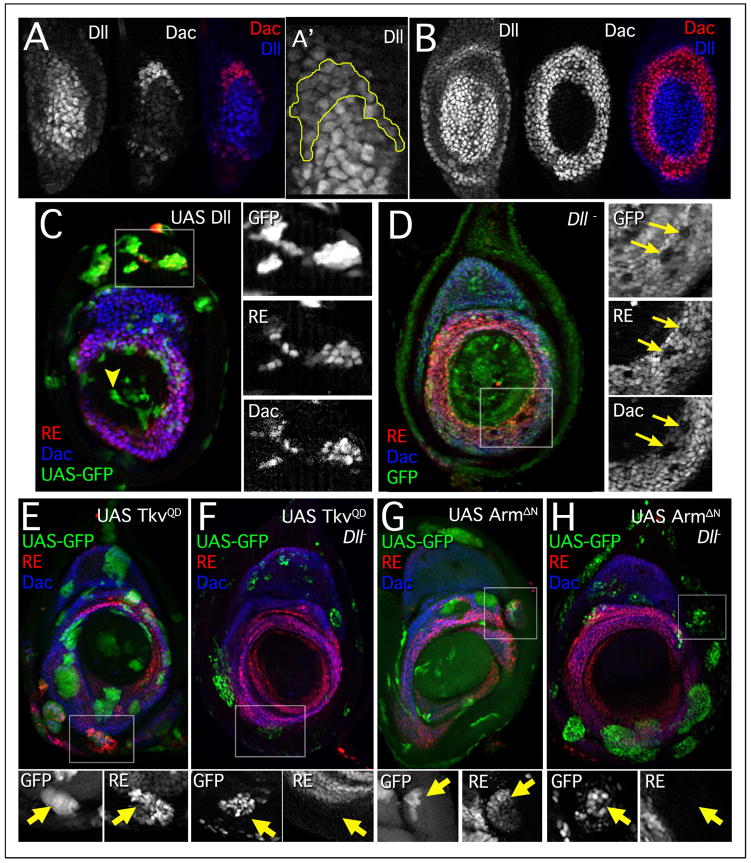
Figure 4. Dll is required for dac and dac RE activation.
(A) Late 2nd instar leg disc stained for Dac (red) and Dll (blue). Dac protein is first observed in cells that have lower levels of Dll, consistent with lineage tracing experiments. A′ shows a closeup of the Dac-expressing cells (outlined) with lower levels of Dll.
(B) Early 3rd instar leg disc showing that Dll (blue) is no longer observed in most of the Dac (red) expressing cells. Dac and Dll have a 1–2 cell overlap at this stage.
(C) Leg disc with clones expressing Dll (green), which activates both dac (blue) and dac RE lacZ (red) in proximal tissue. There is no effect on dac or dac RE activity distally (arrowhead). Larvae heat shocked in early 3rd instar (72–96 hrs AEL) and stained 24 hrs post heat shock.
(D) Dll mutant clones (lack of GFP, green). Larvae were heat shocked in early 3rd instar (72–96 hrs AEL) and stained 24 hrs post heat shock.
(E–H) Clones are marked by GFP+ expression (green) and discs are stained for dac RE lacZ (red) and Dac (blue). Larvae heat shocked between 24–72 hrs AEL, then fixed and stained 48 hrs post heatshock.
(E) UAS-TkvQD clones activate both dac and dac RE lacZ in ventral-proximal tissue.
(F) MARCM clones expressing TkvQD and mutant for Dll do not activate the distal leg program or result in ectopic expression of dac or dac RE lacZ.
(G) UAS-ArmΔN clones express both dac and dac RE lacZ in dorsal-proximal tissue.
(H) MARCM clones expressing ArmΔN and mutant for Dll do not activate dac or dac RE-lacZ.
A central observation in support of the gradient model is that de novo juxtapositions of Wg and Dpp expressing cells activate either Dll or dac in proximal tissues, depending on where in the disc these clones arise (Abu-Shaar and Mann, 1998; Lecuit and Cohen, 1997) (Figure 2). If Dll is an essential activator of dac, we would expect that Wg+Dpp would be unable to activate dac in Dll− cells. To test this prediction we used the MARCM method (Lee and Luo, 2001) to express either TkvQD or ArmΔN in Dll− clones. We found in both cases that Wg or Dpp pathway activation was unable to activate dac or dac RE-lacZ in Dll− clones (Fig. 4E–H). Thus, Dll is essential for dac activation, even at new sites of Wg and Dpp pathway activation.
Dll acts through multiple binding sites to directly activate dac RE
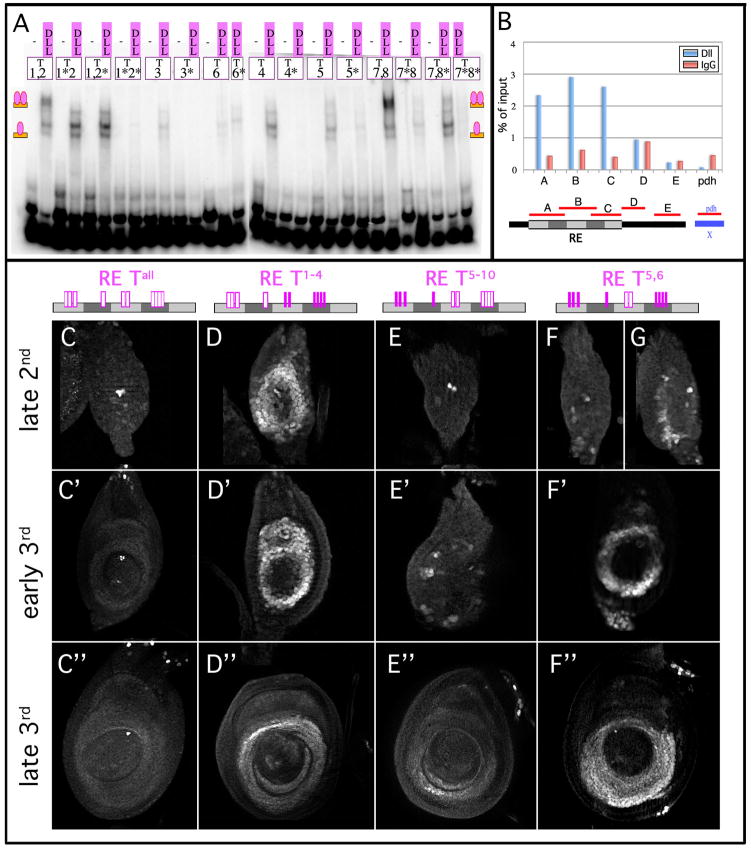
Dll is a homeodomain protein whose predicted target sequence, like many homeodomains, is based around a core TAAT sequence (Berger et al., 2008; Noyes et al., 2008). There are ten TAAT sequences in dac RE (Figure 3A). We tested all ten of these for their ability to bind Dll protein in a series of EMSA experiments. Dll bound specifically to seven of the ten sites (Figures 3A and 5A). To test if Dll is bound to these sites in vivo, in leg discs, we performed Chromosome Imunoprecipitation (ChIP) with 3rd instar leg discs using an anti-Dll antibody. We found that anti-Dll was able to specifically immunoprecipitate dac RE, but not control chromatin, consistent with a direct role for Dll in dac regulation (Fig. 5B).
Figure 5. Dll acts through TAAT sequences in dac RE to directly activate expression.
(A) EMSAs with Dll protein on TAAT-containing oligos from dac RE. All TAAT sites except t6 show specific binding to Dll that is lost upon mutation. Oligos that contain two TAAT sequences (taat1,2 and taat7,8) show a slower mobility band which is due to the occupancy of both sites.
(B) Representative ChIP of 3rd instar leg discs using anti-Dll. Real time PCR of primer sets at dac RE are pulled down specifically relative to IgG controls. Primer sets #1–3 are contained within dac RE, #4 is located just downstream and #5 is located 400 bp further downstream. pdh is a negative control amplicon of the pdh gene on the X chromosome.
(C–G) Leg discs at 2nd (D–G), early 3rd (D′–F′), and late 3rd (D″–F″) instar stained for dac RE reporter genes with mutated TAAT sequences. Mutant dac REs are schematized above with the mutant sites represented by open bars.
(C) dac RE taatall. RE with all 10 TAAT sequences mutated is nearly inactive. Expression is limited to a few distal cells and in a few proximal cells of the trochanter.
(D) dac RE taat1–4. RE with the four 5′ TAAT sequences mutated is expressed in a normal RE ring. In the late 3rd instar there is an ectopic distal ring of expression at the boundary of the pretarsus (see Figure 6).
(E) dac RE taat5–10. Mutation of the six 3′ TAAT sequences results in severely limited expression. By the 3rd instar expression is limited to a few cells in a weak ring-like pattern. In the late 3rd instar there is staining in some ventral medial cells but expression is weak elsewhere. Apparent differences in expression in dorsal and ventral regions of the RE domain are largely due to folds in the discs and slight differences in the focal plane.
(F,G) dac RE taat5,6. Mutation of TAAT sites 5 and 6 results in a delayed expression of the Dac ring. By the 3rd instar expression is largely normal.
See also Table S1.
To test the importance of the Dll binding sites for dac RE activity, we mutated the TAAT core sequences singly and in combination in the context of the full dac RE reporter gene. In contrast to mutation of all predicted Mad or Pan sites, mutation of all ten TAAT sites eliminated activity at all stages of disc development (Fig. 5C). Mutation of the first four TAAT sequences (RE TAAT1–4) did not significantly affect the ring pattern (Fig. 5D), while mutation of the latter six TAAT sites (RE TAAT5–10) resulted in delayed expression and a weaker ring pattern (Fig. 5E). Mutation of the two central TAAT sites (RE TAAT5,6) also significantly delayed the onset of reporter gene expression (Fig. 5F,G), and deletion of these two sites in an already compromised fragment resulted in the near-elimination of RE activity (Figure S2F-H). Interestingly, multimerization of a small portion of RE that contains TAAT sites 5 and 6 was sufficient to drive a ring-like expression pattern in 3rd instar leg discs (4x(3b); Figure S2O). These findings suggest that Dll activates dac RE by binding to multiple TAAT-containing binding sites, and that multiple sites are required for the correct timing and levels of dac RE activity.
Bar directly represses dac RE
The above results argue that Wg and Dpp do not act in a graded fashion to activate dac RE or dac, because eliminating the predicted Pan and Mad binding sites had only a minimal effect on enhancer activity. Notably, by the 3rd instar, mutation of seven Pan and seven Mad binding sites also did not result in significant expression in the distal leg disc (Figure 3B″–G″). These observations raise the question of what factor(s) directly mediate repression in older discs. In addition to activating Dll and dac, Wg and Dpp activate components of the epidermal growth factor receptor (EGFR) pathway, which are important for patterning the tarsal segments. One of the early downstream targets of this pathway is the homeodomain protein, Bar, which is initially expressed in a ring just distal to the dac domain starting in the 3rd instar (Kojima et al., 2000). When this ring of Bar expression initiates it is adjacent to the distal edge of the dac domain. Thus, Bar is a good candidate for maintaining dac’s distal expression boundary in the early 3rd instar.
To test Bar’s ability to repress dac RE, we examined clones overexpressing Bar. Consistent with previous results (Kojima et al., 2000), ectopic Bar was able to repress both dac and dac RE-lacZ (Figure 6A). However, Bar-mediated repression of dac was not observed in all clones, suggesting that it is not sufficient for repression in all contexts (Figure S3). When Bar was ectopically expressed prior to dac, for example using Dll-Gal4, repression was robust, suggesting that the timing of Bar expression is important. Similar resultes were observed in clones expressing an activated form of the EGF Receptor (λtop) (Figure 6B and data not shown). These results suggest that the repression of dac in the 3rd instar is mediated in part by the EGFR pathway through its activation of Bar.
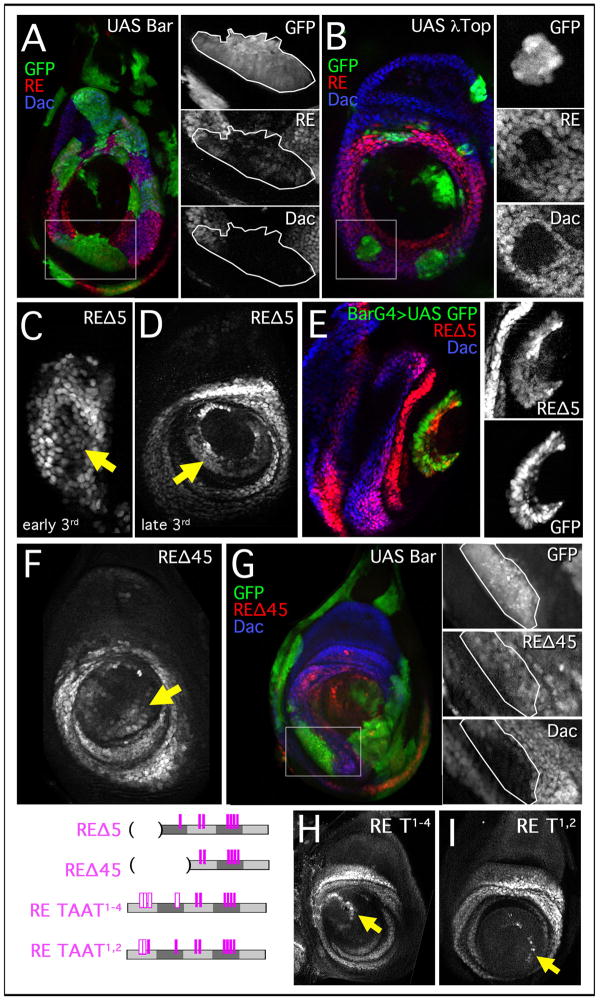
Figure 6. Late stage distal repression elements in dac RE.
(A,B) Leg discs stained for dac RE lacZ (red) and Dac (blue), with Gal4 expressing clones marked by GFP+. Boxed regions are blown up in the right-hand panels.
(A) Ectopic Bar clones. Larvae heat shocked 24–72 hrs AEL and stained 64 hrs post heat shock.
(B) Clones expressing a constitutive EGFR (λTop, green). Larvae heat shocked 24–72 hrs AEL and stained 48 hrs post heat shock.
(C–E) Activity of dac REΔ5 lacZ, a truncated fragment of RE (schematized below).
(C) dac REΔ5 lacZ is expressed in cells distal (arrow) to the normal Dac domain in the early 3rd instar.
(D) By the late 3rd instar there is a prominent ring (arrow) of expression at the 5th tarsal segment at the boundary with the pretarsus.
(E) Late 3rd instar everting leg disc with Bar-Gal4 driving GFP (green) stained for dac REΔ5 lacZ (red) and Dac (blue). The distal ring of REΔ5 is coincident with Bar-Gal4 expression. The distal region of the disc is shown blown up in boxes on right.
(F,G) Activity of dac REΔ45 lacZ, a truncated fragment of RE (schematized below).
(F) dac REΔ45 lacZ is ectopically expressed in a distal ring (arrow).
(G) Clones expressing Bar repress dac (blue) but do not significantly affect dac REΔ45 lacZ expression. Larvae heat shocked 24–72 hrs AEL and stained 64 hrs post heat shock. The boxed region is blown up in the right-hand panels.
(H,I) Expression of lacZ from mutant RE constructs in late 3rd instar discs. Mutant REs are schematized at left with the mutant sites represented by open bars.
(H) dac RE taat1–4. Mutation of the four 5′ TAAT sequences results in a thin ring of expression at the tarsal/pretarsus boundary (arrow).
(I) dac RE taat1,2. Mutating two of the 5′ TAAT sequences results in a partial ring of expression at the tarsal/pretarsus boundary (arrow).
As noted above, one of the dac RE subfragments (REΔ5) drove expression distal to the normal dac domain during the 2nd and early 3rd instar, eventually giving rise to a strong additional distal ring of expression (Figure 6C,D). This ectopic ring of expression coincided with Bar expression (Figure 6E), also supporting a role for Bar in the distal repression of dac. Further, Bar may repress RE through the 5′ end of the element. Consistently, a smaller RE subfragment comprised of just RE’s 3′ half (REΔ45) also drove ectopic expression in a distal ring in the 3rd instar (Figures 6F and 3H). Interestingly, REΔ45 was unaffected in clones that overexpress Bar (Figure 6G).
Like Dll, Bar is a homeodomain protein that is likely to bind DNA binding sites that contain a TAAT core. There are four TAAT sequences missing from REΔ45. Strikingly, when those TAATs are mutated (RE TAAT1–4) there is a similar derepression in a distal ring that coincides with the Bar expression domain (Figure 6H and data not shown). Mutation of just the first two TAAT sites (TAAT1,2) results in a slim ring of distal derepression (Figure 6I). Together, these data provide strong evidence that Bar directly binds to and represses dac RE through multiple TAAT binding sites located in the 5′ portion of the enhancer.
Discussion
Activation of dac in the medial leg by a non-gradient mechanism
The PD axis of the Drosophila leg is initiated by two secreted morphogens, Wg and Dpp. The process begins when these two pathways collaborate to induce the expression of Dll in a small group of ventral cells in each thoracic hemisegment of stage 14 embryos, in part by directly activating the Dll LT enhancer (Estella et al., 2008). Soon after Dll LT is activated, these cells invaginate from the main epithelium, thus establishing the leg imaginal disc. Wg and Dpp continue to be expressed next to the AP compartment boundary in ventral and dorsal cells, respectively, until the end of larval development. As the disc grows during larval stages, Wg and Dpp are secreted and diffuse from the cells that express them, resulting in gradients of both morphogens. These gradients help to pattern the DV axis of the adult leg (Struhl and Basler, 1993).
The question we address here is whether the ventral and dorsal gradients of Wg and Dpp are also used to establish distinct cell fates along the PD axis of the growing leg. To test this idea, we characterized an enhancer element from the dac locus, dac RE, which directs expression in a medial domain along the PD axis. If dac was interpreting intermediate levels of the Wg and Dpp gradients for its activation, two expectations should be fulfilled: 1) binding sites for Pan and Mad should be present in the dac enhancer and 2) reducing the amount of Pan or Mad binding to dac RE should reduce the proximal extent of its activity; in other words, the domain of dac RE activity should shrink as the amount of Pan and Mad input is reduced. However, our data only support the first of these two expectations. Counter to the prediction of the gradient model, eliminating most, if not all, of the Pan or Mad binding sites in dac RE had virtually no effect on its ability to activate this element. Even if some residual Pan or Mad binding remains in these heavily mutated RE reporter genes, the gradient model predicts that they should have had significantly reduced activities. Based on these observations we conclude that this element is not directly integrating intermediate levels of Wg and Dpp activities to be activated.
If Wg and Dpp are not the primary activators of dac, what is? Because lineage-tracing experiments demonstrated that all dac+ cells derive from cells that previously expressed Dll, we tested the idea that Dll is an activator of dac. Both genetic and molecular studies support this idea – Dll− clones in the dac domain lose dac expression; gain-of-function Dll+ clones proximal to the dac domain activate dac; and Dll protein binds to dac RE both in vivo (in leg discs) and in vitro. Further, mutating Dll binding sites in dac RE eliminates its activity. Because ectopic activation of the Wg and Dpp pathway can induce a new PD axis and new sites of dac expression, we asked if this de novo activation of dac also required Dll. We found that Wg or Dpp pathway activation were both incapable of inducing dac or dac RE-lacZ expression when the cells were also mutant for Dll. Together, these data provide strong evidence that Dll is an essential and direct activator of dac and dac RE.
In light of these observations, do Wg and Dpp play any direct role in the activation of dac? Based on the finding that compromised subfragments of dac RE (e.g. dac REΔ45) require Pan and Mad binding sites for their activities, we suggest that they play a permissive role in dac activation. However, the nearly wild type activities of dac RE Pall and dac RE Mall in the 3rd instar imply that this contribution to dac activation is relatively minor compared to Dll.
Distal repression of dac by Wg, EGFR, and Bar
Another question our results raise is, if Dll is an activator of dac, why is dac not activated in the center of the leg disc? Even when Dll is ectopically expressed, it cannot activate dac or dac RE-lacZ in this distal-most domain, although it can activate both of these readouts in proximal cells. We suggest that there are at least two mechanisms that repress dac in the distal leg disc. The first mechanism is due to direct binding of Pan to dac RE. The evidence in support of this idea is that mutating the Pan binding sites (e.g. dac RE Pall) results in distal derepression in 2nd instar discs. The direct repression of dac RE by Pan is noteworthy because it apparently occurs in the presence of active Wg signaling, which is usually associated with transcriptional activation in a mechanism that depends on Arm binding to Pan (Stadeli et al., 2006). However, an alternative DNA binding mode by Pan has been defined that mediates Wg- and Arm-dependent transcriptional repression (Blauwkamp et al., 2008). Although such a mechanism may be operating at dac, we note that Wg signaling cannot be sufficient for dac repression because Wg fails to repress dac in the ventral leg disc, despite high levels of signaling. In the future, it will be interesting to explore this repression mechanism further and to specifically test the idea that Wg signaling collaborates with distally-expressed transcription factors such as Bar to repress dac.
Soon after dac is first activated, the EGFR pathway is triggered in distal cells, due to the expression of EGFR ligands, such as vein, and EGFR pathway proteases, such as rhomboid, that are required for ligand activation (Campbell, 2002; Galindo et al., 2002). Our data suggest that this pathway also contributes to dac repression. First, expressing a constitutively activated form of the EGF receptor (λtop) resulted in the repression of dac and dac RE activity. Second, at least one downstream target of the EGFR pathway, Bar, is required for a subset of the distal repression of dac and dac RE. Our mutagenesis studies of dac RE suggest that Bar mediates repression directly, by binding to multiple homeodomain binding sites in the 5′ end of the enhancer. EGFR signaling results in the patterned expression of several transcription factors in addition to Bar, including aristalless and rotund (Kojima, 2004). Following our evidence for Bar, it is plausible that these, and perhaps other, distally expressed transcription factors also play a role in dac repression. It is also possible that the EGFR pathway directly represses dac, or that there are non-canonical Pan binding sites that also contribute to repression in the 3rd instar.
Interestingly, dac and dac RE-lacZ are derepressed in distal mad− or arr− clones, but only when these clones are induced prior to the start of the 3rd instar. These observations suggest that timing plays a critical role in dac repression. Similarly, the RE Pall mutant reporter gene only shows distal derepression in 2nd instar discs, but not in 3rd instar discs. We suggest that these temporal differences are due to different phases of dac regulation (Figure 7). Specifically, distal dac repression early in development, prior to the activation of the EGFR pathway, depends primarily on Wg. At later stages, other mechanisms come into play. In addition to Bar and other distal transcription factors, the Polycomb group (PcG) of transcriptional repressors may also contribute to the maintenance of dac repression. Consistent with this idea, in flies mutant for the PcG gene pleiohomeotic (pho) dac is derepressed in the distal tip (Kim et al., 2008).
Figure 7. Establishment and elaboration of the PD axis of the leg.
Shown are schematic diagrams of 2nd instar (left), early 3rd instar (middle) and mid 3rd instar leg imaginal discs. In 2nd instar discs, there is a ventral sector of high Wg signaling (light blue) and a dorsal sector of high Dpp signaling (red). Although not shown, these patterns of Wg and Dpp signaling remain the same throughout the remainder of leg development. Only cells in the center of the leg disc (purple) are receiving high inputs for both Wg and Dpp. The combination of high Wg signaling and high Dpp signaling results in the activation of the Dll LT enhancer element and the repression of the dac RE enhancer element. As the disc grows, some Dll-expressing cells move out of this dac repression domain, allowing Dll to activate dac (Dac+Dll domain; orange). Once dac is activated, it may repress Dll, thus contributing to the establishment of the initial Dac-only domain (blue). By the early 3rd instar, the three primary gene expression domains (Dll-only, Dac+Dll, and Dac-only) become fixed by a maintenance mechanism that is independent of Wg and Dpp signaling. For Dll, this maintenance mechanism involves autoregulation mediated by the M element; for dac, autoregulation may also be involved, but this is not yet known. Also during the early 3rd instar, the EGFR pathway is activated in distal cells, leading to the expression of Bar as well as other downstream transcription factors. Bar continues to repress dac in distal cells, thus helping to maintain the Dll-only domain. High levels of Wg and Dpp signaling, still limited to the center of the disc (purple), may continue to contribute to the repression of dac in distal cells.
A temporal cascade of regulatory inputs establish medial fates during leg development
These findings support a revised view of PD axis formation in the Drosophila leg (Figure 7). In the 2nd instar leg disc, we propose that the combination of high levels of Wg signaling and high levels of Dpp signaling, which only occurs in cells close to the center of the disc, results in two outputs: 1) activation of Dll, in part via the LT enhancer (Estella et al., 2008) and 2) repression of dac RE. As the disc grows in size, Dll activates dac in all cells that are not receiving high levels of Wg+Dpp signaling. Soon thereafter, in the early 3rd instar, Dll expression is maintained and high Wg+Dpp activities begin to activate the EGFR pathway (Campbell, 2002; Galindo et al., 2002). Genes downstream of the EGFR pathway, such as Bar, continue to repress dac in distal cells through the rest of leg disc development. The fundamental difference between this model and the gradient model is that graded activities of Wg and Dpp are not relevant for the regulation of either dac or Dll. Instead, by proposing that dac is activated by Dll and repressed by high levels of Wg+Dpp signaling, only peak levels of Wg and Dpp pathway activation are relevant to forming the PD axis. In one case (Dll) activation is direct (mediated by Mad and Pan binding to Dll) while in the other case (dac) activation is indirect, mediated by Dll binding to dac RE.
As described so far, this model would predict that the proximal extent of dac expression should coincide with the proximal extent of Dll expression. (For this discussion, we do not include the proximal trochanter ring, which expresses both Dll and dac but is under separate control.) However, this is not the case: in 3rd instar leg discs there is a Dll-non-expressing Dac-only domain that surrounds the Dll expression domain. This Dac-only domain is larger in the dorsal portion of the disc due in part to the absence of Brk expression (Estella and Mann, 2008). Lineage-tracing experiments show that the progenitors of all dac-expressing cells, including the Dac-only domain, expressed Dll (McKay et al., 2009). Thus, Dll is a transient activator of dac in these cells: it must be expressed to initially activate dac, but then is only maintained in a subset of dac-expressing cells. We suggest that the growth of the leg disc, coupled with a transition to autoregulation or a transcriptional maintenance mechanism, accounts for the formation of the Dac-only domain (Figure 7). In the second instar, dac is first activated in Dll-expressing cells that are not also receiving high levels of Wg and Dpp. As cells in the young disc divide, some of the Dll and dac expressing cells will move out of the range of Wg and Dpp signaling that is minimally required for Dll activation. Consequently, some of these cells still express dac but lose Dll expression. In addition, dac may down-regulate Dll in a negative feedback loop (Figure 7). This suggestion is consistent with previous observations showing that small dac− clones in the Dac-only domain derepress Dll (Abu-Shaar and Mann, 1998).
The above regulatory steps will result in an early 3rd instar disc that contains all three primary expression domains: Dll-only, Dac+Dll, and Dac-only. At about the same time, we suggest that the dac and Dll expression status begins to be maintained independently of these earlier regulatory steps, either by autoregulation or by a transcriptional memory system. For Dll, autoregulation plays a critical role beginning in the early 3rd instar (Estella et al., 2008). Notably, dac RE is not activated by Dac, so does not contain an autoregulatory component and, interestingly, dac RE-lacZ is poorly expressed in the Dac-only domain. These observations are consistent with our model because it predicts that dac expression in the Dac-only domain should be more dependent on autoregulation than in the ‘Dac+Dll’ domain, where Dll is constantly available to activate dac. In summary, we suggest that the distinct proximal extents of the dac and Dll domains in the 3rd instar leg disc are a consequence of the dac and Dll expression status at the time when autoregulation/maintenance initiates. In effect, the Wg- and Dpp-independent maintenance phases of dac and Dll expression lock into place the three domains that are initially established in the late 2nd/early 3rd imaginal disc, and maintained throughout the remainder of development.
The role of temporal transcriptional cascades in development
In more general terms, the logic of gene regulation revealed here highlights the importance of developmental timing and tissue growth in eventually generating stable domains of gene expression. Developmental processes such as PD axis formation are dynamic, with cells changing their states in a directional manner over time, typically from a less differentiated state towards a more differentiated state. Such developmental programs are fundamentally distinct from other transcriptional regulatory scenarios that are used to toggle between bistable states, which may be more typical in terminal differentiation programs (Mikeladze-Dvali et al., 2005; Poole and Hobert, 2006). The regulatory steps in Drosophila leg development – from morphogen signaling (Wg+Dpp
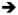 Dll), through a transcription factor cascade and negative feedback (Dll
Dll), through a transcription factor cascade and negative feedback (Dll
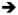 dac, Dac -| Dll), to autoregulation/maintenance (e.g. Dll
dac, Dac -| Dll), to autoregulation/maintenance (e.g. Dll
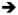 Dll) – represent a series of subcircuits or subroutines that gradually transition from one to another as the tissue grows (Figure 7). This transition through a series of subcircuits is also seen in other dynamic developmental processes such as specification of cell types in the vertebrate neural tube and in the early Drosophila embryo (Ben-Tabou de-Leon and Davidson, 2006; Kutejova et al., 2009; von Dassow et al., 2000). The progression through a series of transient, inherently unstable states may be typical for developmental programs in which cells are continually refining their fates over time and as tissues grow in size.
Dll) – represent a series of subcircuits or subroutines that gradually transition from one to another as the tissue grows (Figure 7). This transition through a series of subcircuits is also seen in other dynamic developmental processes such as specification of cell types in the vertebrate neural tube and in the early Drosophila embryo (Ben-Tabou de-Leon and Davidson, 2006; Kutejova et al., 2009; von Dassow et al., 2000). The progression through a series of transient, inherently unstable states may be typical for developmental programs in which cells are continually refining their fates over time and as tissues grow in size.
Experimental procedures
Transgenes
The initial dac reporter fragments, as well as HI, RE, REΔ5, and REΔ45, were inserted into a standard P-element nuc-lacZ vector (Estella et al. 2008) for which multiple transformants were surveyed for position effects. In addition, all RE reporter genes were inserted into an attB-nuc-lacZ vector modified from (Bischof et al., 2007) with an hsp43 promoter. All attB constructs were inserted into the second chromosome at 51D attP site (gift of K. Basler) via phi-C31 mediated trangenesis (Bischof et al., 2007). Expression at 51D was comparable to the representative first generation standard P-element transformants. MARCM and mutant clone analyses used a P-element RE reporter inserted in the 3rd chromosome. Putative dac regulatory fragments were selected based on sequence conservation to other Drosophilids (Vista Genome Browser, UCSC) and cloned by PCR (dac RE was cloned with primers: CCAACTGAAAAAGGAGCAGCTTTC and ACAAAATTTATACGCCAGATG; all other primer sequences and details are available upon request). 4x(3b) was synthesized as a single oligo with the sequence, TCCAATAATAAAGTTAAATCGATAATTGAGGTCA, repeated four times. dac RE deletion constructs were formed by fusing two PCR fragments made with primers tagged with Spe I digest sites. We used Target Explorer (Sosinsky et al., 2003) to locate potential binding sites for Mad, Brk and Pan, by generating matrices for each protein based upon published binding sites. Mad/Brk matrices were built around a MGCCGCCGM consensus sequence. Pan matrices used the canonical Pan site SCTTTGW and HMG-domain site WTTGWW. Mutant dac RE constructs were generated by PCR. TAATs were mutated to TGGG; for Mad/Brk binding sites the CGC or GCC core was mutated to ATA; for Pan binding sites the AAA or TAA core was mutated to CCC or GGG. The binding sequences are listed in Table S1.
Immunostaining
Imaginal discs were prepared and stained using standard procedures. The primary antibodies used were: rabbit anti-β-Gal (Cappell), mouse anti-Wg & mouse anti-Dac (Developmental Studies Hybridoma Bank; dshb.biology.uiowa.edu), guinea pig anti-P-Mad (gift of E. Laufer and T. Jessell), and guinea pig anti-Dll.
Protein Purification and EMSAs
GST-Mad MH1+L (Xu et al., 1998), GST-dTCF HMG (Lee and Frasch, 2000), GST-Brk 1–100 (gift of C. Rushlow), His-Dll (Estella et al., 2008) were produced and purified by standard procedures (Amersham-Pharmacia or QIAGEN). Protein concentrations were measured by Bradford assay and confirmed by SDS-PAGE and Coomassie blue analysis. Electrophoretic mobility shift assays (EMSAs) were performed as previously described (Gebelein et al., 2004). The amount of protein used in each EMSA was 25 pmol for Brk, 60 pmol for Mad, 40 pmol for dTcf, and 15 pmol for Dll.
Chromatin Immunoprecipitations
ChIP assays were carried out using a previously described protocol (Estella et al., 2008).
Fly Genetics
For gain-of-function experiments, we used the strain yw hsFLP122; tub > y+ > Gal4 UAS-GFP and the following UAS transgenes: UAS-tkvQD (Abu-Shaar and Mann, 1998), UAS-ArmΔN (Pai et al., 1997), UAS-Dac (Shen and Mardon, 1997), UAS-Bar (Sato et al., 1999), UAS-Dll (Dong et al., 2000) and UAS-λTop4.2 (Queenan et al., 1997). Flip-out clones were originated by heat shocking larvae aged 24–48 hrs for 7–10 min at 37°C. Larvae were dissected following 48hrs growth at 25°c unless otherwise indicated. For loss-of-function clones, we used the following genotypes: mad1–2 FRT40A/ubi-GFP FRT40A, yw hsFLP122; FRT42D arr2/FRT42D ubi-GFP, yw hsFLP122; FRT42D Dllsa1/FRT42D ubi-GFP, yw hsFLP122; dac3 FRT40A/ubi-GFP y+ FRT40A. Clones were generated by a 1 hour heat shock at 37°C. Larvae were dissected 48 hrs post heats shock unless otherwise indicated. The dac7 allele is a 3′ deletion that encompasses the RE element and has virtually no Dac expression in the leg (Pappu et al., 2005). To monitor Bar expression we used yw Bar-Gal4; FM7c (Kyoto Stock Center) with dac REΔ5; UAS-GFP. We used the MARCM technique (Lee and Luo, 1999) to express UAS-TkvQD or UAS-ArmΔN in Dll− cells using: FRT42D DllSa1; UAS-ArmΔN (over compound SM6^TM6B), FRT42D DllSa1; UAS-TkvQD (SM6^TM6B), yw hsFLP122 tub-Gal4 UAS-GFP; FRT42D tub-Gal80; dac RE-nucLacZ, yw hsFLP122 tub-Gal4 UAS-GFP; tub-Gal80 FRT40A; dac RE-nucLacZ.
Supplementary Material
Acknowledgments
We thank K. Basler, T. Jessell, E. Laufer, G. Mardon, G. Rubin, and G. Struhl for reagents and James Briscoe, Claude Desplan, Carlos Estella, and Roumen Voutev for comments on the manuscript. We thank Wen Zhang for generating the transgenic lines. This work was supported by an NIH grant GM058575 awarded to R.S.M. and an NRSA fellowship awarded to M.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Shaar M, Mann RS. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development. 1998;125:3821–3830. doi: 10.1242/dev.125.19.3821. [DOI] [PubMed] [Google Scholar]
- Ben-Tabou de-Leon S, Davidson EH. Deciphering the underlying mechanism of specification and differentiation: the sea urchin gene regulatory network. Sci STKE. 2006:pe47. doi: 10.1126/stke.3612006pe47. [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauwkamp TA, Chang MV, Cadigan KM. Novel TCF-binding sites specify transcriptional repression by Wnt signalling. EMBO J. 2008;27:1436–1446. doi: 10.1038/emboj.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Campbell G. Distalization of the Drosophila leg by graded EGF-receptor activity. Nature. 2002;418:781–785. doi: 10.1038/nature00971. [DOI] [PubMed] [Google Scholar]
- Campbell G, Weaver T, Tomlinson A. Axis specification in the developing Drosophila appendage: the role of wingless, decapentaplegic, and the homeobox gene aristaless. Cell. 1993;74:1113–1123. doi: 10.1016/0092-8674(93)90732-6. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Cohen B, Cohen SM. Cell interaction between compartments establishes the proximal-distal axis of Drosophila legs. Nature. 1994;372:175–179. doi: 10.1038/372175a0. [DOI] [PubMed] [Google Scholar]
- Dong PD, Chu J, Panganiban G. Coexpression of the homeobox genes Distal-less and homothorax determines Drosophila antennal identity. Development. 2000;127:209–216. doi: 10.1242/dev.127.2.209. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, St Johnston D. Seeing is believing: the bicoid morphogen gradient matures. Cell. 2004;116:143–152. doi: 10.1016/s0092-8674(04)00037-6. [DOI] [PubMed] [Google Scholar]
- Estella C, Mann RS. Logic of Wg and Dpp induction of distal and medial fates in the Drosophila leg. Development. 2008;135:627–636. doi: 10.1242/dev.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estella C, Mann RS. Non-redundant selector and growth-promoting functions of two sister genes, buttonhead and Sp1, in Drosophila leg development. PLoS Genet. 2010;6:e1001001. doi: 10.1371/journal.pgen.1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estella C, McKay DJ, Mann RS. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev Cell. 2008;14:86–96. doi: 10.1016/j.devcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French V. Intercalary regeneration around the circumference of the cockroach leg. J Embryol Exp Morphol. 1978;47:53–84. [PubMed] [Google Scholar]
- French V. Positional information around the segments of the cockroach leg. J Embryol Exp Morphol. 1980;59:281–313. [PubMed] [Google Scholar]
- Galindo MI, Bishop SA, Greig S, Couso JP. Leg patterning driven by proximal-distal interactions and EGFR signaling. Science. 2002;297:256–259. doi: 10.1126/science.1072311. [DOI] [PubMed] [Google Scholar]
- Gebelein B, McKay DJ, Mann RS. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature. 2004;431:653–659. doi: 10.1038/nature02946. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kim SN, Jung KI, Chung HM, Kim SH, Jeon SH. The pleiohomeotic gene is required for maintaining expression of genes functioning in ventral appendage formation in Drosophila melanogaster. Dev Biol. 2008;319:121–129. doi: 10.1016/j.ydbio.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Kojima T. The mechanism of Drosophila leg development along the proximodistal axis. Dev Growth Differ. 2004;46:115–129. doi: 10.1111/j.1440-169X.2004.00735.x. [DOI] [PubMed] [Google Scholar]
- Kojima T, Sato M, Saigo K. Formation and specification of distal leg segments in Drosophila by dual Bar homeobox genes, BarH1 and BarH2. Development. 2000;127:769–778. doi: 10.1242/dev.127.4.769. [DOI] [PubMed] [Google Scholar]
- Kutejova E, Briscoe J, Kicheva A. Temporal dynamics of patterning by morphogen gradients. Curr Opin Genet Dev. 2009;19:315–322. doi: 10.1016/j.gde.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Cohen SM. Proximal-distal axis formation in the Drosophila leg. Nature. 1997;388:139–145. doi: 10.1038/40563. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- McKay DJ, Estella C, Mann RS. The origins of the Drosophila leg revealed by the cis-regulatory architecture of the Distalless gene. Development. 2009;136:61–71. doi: 10.1242/dev.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen YW, Cohen S, Desplan C. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–787. doi: 10.1016/j.cell.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development. 1997;124:2255–2266. doi: 10.1242/dev.124.11.2255. [DOI] [PubMed] [Google Scholar]
- Pappu KS, Ostrin EJ, Middlebrooks BW, Sili BT, Chen R, Atkins MR, Gibbs R, Mardon G. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005;132:2895–2905. doi: 10.1242/dev.01869. [DOI] [PubMed] [Google Scholar]
- Poole RJ, Hobert O. Early embryonic programming of neuronal left/right asymmetry in C. elegans. Curr Biol. 2006;16:2279–2292. doi: 10.1016/j.cub.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Queenan AM, Ghabrial A, Schupbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- Sato M, Kojima T, Michiue T, Saigo K. Bar homeobox genes are latitudinal prepattern genes in the developing Drosophila notum whose expression is regulated by the concerted functions of decapentaplegic and wingless. Development. 1999;126:1457–1466. doi: 10.1242/dev.126.7.1457. [DOI] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Sosinsky A, Bonin CP, Mann RS, Honig B. Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 2003;31:3589–3592. doi: 10.1093/nar/gkg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadeli R, Hoffmans R, Basler K. Transcription under the control of nuclear Arm/beta-catenin. Curr Biol. 2006;16:R378–385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Xu X, Yin Z, Hudson JB, Ferguson EL, Frasch M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.