Abstract
First-principles quantum mechanical/molecular mechanical (QM/MM)-free energy (FE) calculations have been performed to examine catalytic mechanism for cocaine esterase (CocE)-catalyzed hydrolysis of (+)-cocaine in comparison with CocE-catalyzed hydrolysis of (−)-cocaine. It has been shown that the acylation of (+)-cocaine consists of nucleophilic attack of hydroxyl group of Ser117 on carbonyl carbon of (+)-cocaine benzoyl ester and the dissociation of (+)-cocaine benzoyl ester. The first reaction step of deacylation of (+)-cocaine, which is identical to that of (−)-cocaine, is rate-determining, indicating that CocE-catalyzed hydrolyses of (+)- and (−)-cocaine have a common rate-determining step. The computational results predict that the catalytic rate constant of CocE against (+)-cocaine should be the same as that of CocE against (−)-cocaine, in contrast with the remarkable difference between human butyrylcholinesterase-catalyzed hydrolyses of (+)- and (−)-cocaine. The prediction has been confirmed by experimental kinetic analysis on CocE-catalyzed hydrolysis of (+)-cocaine in comparison with CocE-catalyzed hydrolysis of (−)-cocaine. The determined common rate-determining step indicates that rational design of a high-activity mutant of CocE should be focused on the first reaction step of the deacylation. Further, the obtained mechanistic insights into the detailed differences in the acylation between the (+)- and (−)-cocaine hydrolyses provide indirect clues for rational design of amino acid mutations that could more favorably stabilize the rate-determining transition state in the deacylation and, thus, improve the catalytic activity of CocE. This study provides a valuable mechanistic base for rational design of an improved esterase for therapeutic treatment of cocaine abuse.
Introduction
Cocaine is recognized as the most reinforcing drug of abuse.1–3 Recent surveys in the United States show that, among the causes of illicit drug related emergency department visits, cocaine was the first on the list.4,5 Disastrous medical and social consequences of cocaine addiction have made the development of an anti-cocaine medication a high priority.6–7 There is still no FDA-approved medication for treatment of cocaine abuse and toxicity.8–12
Cocaine esterase (CocE)13 is the most efficient native enzyme for metabolizing the naturally occurring cocaine yet identified.14 In rodent models, CocE can both prevent and reverse extreme cocaine toxicity,15–16 and even robustly protects rodents from the lethal effects of cocaine.17 Although native CocE is unstable at physiological temperature, CocE mutants designed by a novel computational approach significantly improved its thermostability, increasing the probability of clinical application of this enzyme for therapeutic use against cocaine.18–21
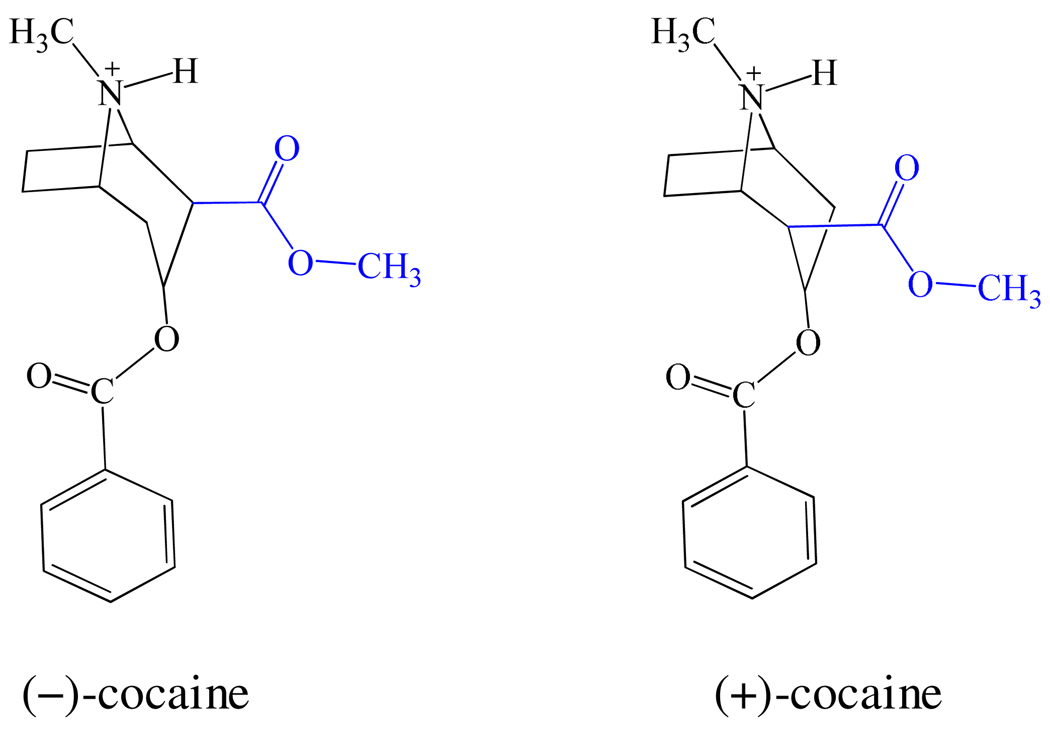
Cocaine has two enantiomers: one is the naturally occurring (−)-cocaine which is biologically active; the other is synthetic and biologically inactive (+)-cocaine. A remarkable difference between (−)- and (+)-cocaine is associated with the relative positions of the methyl ester group (Chart 1). The methyl ester group of (−)-cocaine remains on the same side of the carbonyl of the benzoyl ester, whereas the methyl ester group of (+)-cocaine remains on the opposite side. The structural difference could cause a difference in hydrogen bonding, electrostatic, and van der Waals interactions during the catalytic process and result in a significant difference in free energies of activation. Understanding such mechanistic difference has been proven beneficial to computational design of high-activity mutants of human butyrylcholinesterase (BChE) against (−)-cocaine.9,22–27 In particular, the catalytic efficiency of native human BChE against (−)-cocaine is three-orders-of-magnitude lower than that against (+)-cocaine.23 With an effort to understand the mechanistic differences between the BChE-catalyzed hydrolyses of (+)- and (−)- cocaine, our further computational design followed by wet experimental studies23–24,27–31 have resulted in discovery of various BChE mutants with a considerably improved catalytic efficiency against (−)-cocaine.9,11–12,24,29–31 One of the BChE mutants has a ~2000-fold improved catalytic efficiency against (−)-cocaine compared to wild-type BChE.11
Chart 1.
Structures of (−)-cocaine and (+)-cocaine
Based on the background discussed above, it is important to understand the mechanistic differences between the CocE-catalyzed hydrolyses of (+)- and (−)-cocaine, as their mechanistic differences could be beneficial to the design of high-activity mutants of CocE against cocaine. In a previous study,32 we have elucidated the catalytic mechanism of CocE-catalyzed hydrolysis of (−)-cocaine. However, the detailed catalytic mechanism of CocE-catalyzed hydrolysis of (+)-cocaine remains unknown. In order to understand the mechanistic differences between the (+)- and (−)-cocaine hydrolyses, it is also necessary to uncover and understand the reaction mechanism of CocE-catalyzed (+)-cocaine hydrolysis. For this purpose, the present study was first focused on the detailed mechanism of CocE-catalyzed hydrolysis of (+)-cocaine.
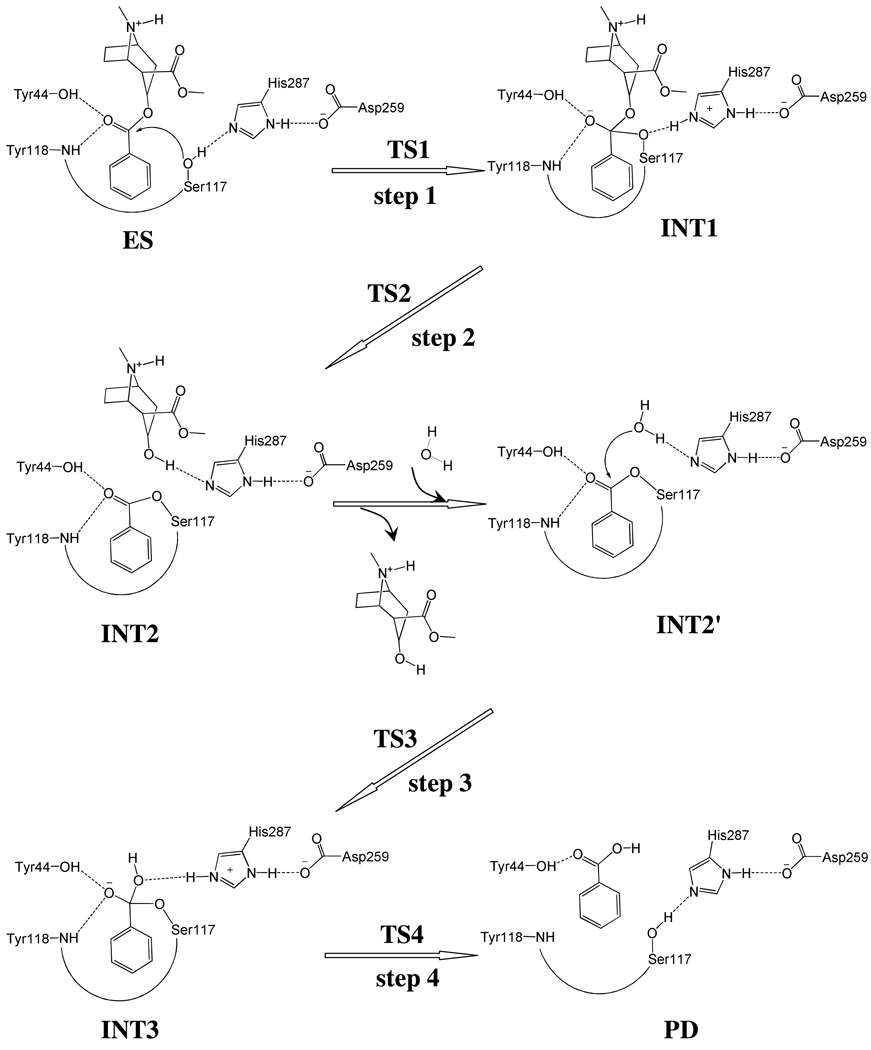
X-ray crystallographic14 and site-directed mutagenesis33 studies have revealed that CocE is a serine carboxylesterase with a catalytic triad formed by Ser117, His287, and Asp259, and with an oxyanion hole formed by the backbone amide of Tyr118 and the hydroxyl group of Tyr44. In light of the mechanistic insights obtained from our recent computational studies on CocE-catalyzed hydrolysis of (−)-cocaine as well as BChE-catalyzed hydrolysis of carboxylic esters (e.g. acetylcholine, butyrylcholine, (+)-cocaine, and (−)-cocaine),11–12,22,26–27,34–36 CocE-catalyzed hydrolysis of (+)-cocaine might undergo a similar pathway as that for CocE-catalyzed hydrolysis of (−)-cocaine consisting of two major stages. The first stage is acylation, leading to formation of a covalent bond between (+)-cocaine and the enzyme and the departure of ecgonine methyl ester of (+)-cocaine. The second stage is deacylation, resulting in the dissociation of the (+)-cocaine benzoyl ester and enzyme, in which a water molecule acts as the nucleophile and the free form of enzyme is restored.
Notably, the (−)- and (+)-cocaine hydrolyses share the same deacylation stage (see Scheme 1) and thus, in the present study, we first focused on the reaction coordinate calculations on the first stage, i.e. acylation, of CocE-catalyzed hydrolysis of (+)-cocaine. Pseudobond first-principles quantum mechanical/molecular mechanical-free energy (QM/MM-FE) approach,37–40 which has been demonstrated to be a powerful tool in simulating a variety of enzymes,11,32,41–52 was employed to uncover the detailed reaction pathway and determine the free energy profile for CocE-catalyzed hydrolysis of (+)-cocaine. The computational simulations were followed by wet experimental tests. The computational data demonstrate that the rate-determining step for CocE-catalyzed hydrolysis of (+)-cocaine should be the same as that for CocE-catalyzed hydrolysis of (−)-cocaine, unlike the reported findings for BChE-catalyzed hydrolyses of (+)-cocaine and (−)-cocaine. The new insights into the catalytic mechanisms of CocE against (+)- and (−)-cocaine have been supported by wet experimental kinetic data.
Scheme 1.
Proposed catalytic mechanism for CocE-catalyzed hydrolysis of (+)-cocaine.
Computational and Experimental Methods
QM/MM-FE Simulation
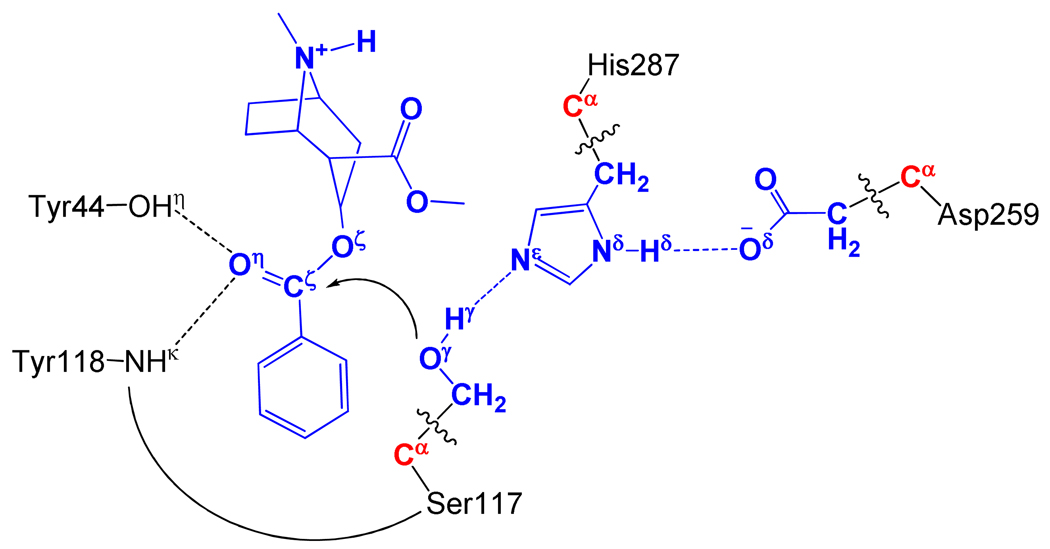
All of the QM/MM calculations were performed by a pseudobond QM/MM method37–38 implemented recently in a revised version11 of Gaussian03 and AMBER8 programs. The QM-MM interface was treated by a pseudobond approach, where a seven-valence-electron atom with an effective core potential is constructed to replace the boundary atom of the environment part and to form a pseudobond with the boundary atom of the active part. The starting structure of the CocE-(+)-cocaine complex was constructed by using the same strategy as used in our previous study on the fundamental mechanism of CocE-catalyzed hydrolysis of (−)-cocaine.32 The coordinates of CocE from previously QM/MM-optimized structure of pre-reactive CocE-(−)-cocaine complex and the structure of (+)-cocaine were used in the molecular docking simulation followed by a ~4 ns MD simulation to understand the detailed binding mode of CocE binding with (+)-cocaine. In QM/MM calculations, all atoms of (+)-cocaine and the side chains of Ser117, His287, and Asp259 were considered as the QM atoms, whereas the other atoms were regarded as MM atoms (Figure 1). The QM/MM calculations were performed using an iterative minimization procedure39 at the B3LYP/6-31*:AMBER level, i.e., the QM calculations were carried out at the B3LYP/6-31G* level, whereas the MM calculations were carried out by using the AMBER force field implemented in the AMBER8 program.53 For QM subsystem, the convergence criterion for geometry optimizations follows the original Gaussian0354 defaults; for MM subsystem, the geometry optimization convergence criterion is the root-mean-square deviation (RMSD) of the energy gradient less than 0.1 kcal·mol−1·Å−1. An iterative restrained optimization procedure39 was then repeatedly applied to different points along the reaction coordinate, resulting in a minimum-energy path. Full QM/MM geometry-optimizations at the B3LYP/6-31G*:AMBER level followed by vibrational frequency analyses were performed to characterize the reactant, intermediates, and transition states. The contribution of the QM subsystem fluctuation to the free energy change was then calculated with the obtained vibrational frequencies using the harmonic approximation. In addition, single-point energy calculations were carried out at the QM/MM(MP2/6-31+G*:AMBER) level for each geometry along the minimum-energy path.
Figure 1.
Division of the QM/MM system for simulating the acylation stage of CocE-catalyzed hydrolysis of (+)-cocaine. Atoms in blue are treated by QM method. Three boundary carbon atoms (Cα, colored in red) are treated with the improved pseudobond parameters.37 All other atoms belong to the MM subsystem.
The free energy changes associated with the QM-MM interaction were then determined by the free energy perturbation (FEP) method39–40 using a revised version32 of the AMBER8 program. The FEP calculations enabled us to more reasonably determine relative free energy changes due to the QM-MM interaction. In the FEP calculations, sampling of the MM subsystem was carried out with the QM subsystem frozen at different states along the reaction path.39 Technically, the final (relative) free energy determined by the QM/MM-FE calculations is the QM part of the QM/MM energy (excluding the Columbic interaction energy between the point charges of the MM atoms and the ESP charges of the QM atoms) plus the relative free energy change determined by the FEP calculations. In FEP calculations, the time step used was 2 fs, and bond lengths involving hydrogen atoms were constrained. In sampling of the MM subsystem by MD simulations, the temperature was maintained at 298.15 K. Each FEP calculation consisted of 50 ps of equilibration and 300 ps of sampling.
The MD simulations and QM/MM-FE calculations were performed on a supercomputer (e.g. IBM X-series Cluster with 340 nodes or 1,360 processors) at University of Kentucky Center for Computational Sciences. The other less-time-consuming modeling and computations were carried out on SGI Fuel workstations and a 34-processor IBM x335 Linux cluster in our own lab.
Expression and Purification of Cocaine Esterase
A potential problem in wet experimental studies on CocE was that the wild-type enzyme is unstable, with a half-life of only ~10 minutes at 37°C. Due to the low thermostability, it was difficult to accurately measure the catalytic activity in wet experiments. Nevertheless, a thermostable version of CocE (i.e. the T172R/G173Q mutant) was developed recently.18 The T172R/G173Q mutations significantly increase the half-life of CocE to ~5 to 6 hours at 37°C without changing its catalytic function because residues #172 and #173 are not in the active site. So, in order to accurately measure the kinetic parameters of the enzyme, we carried out the kinetic characterization using the thermostable version of CocE. The previously prepared CocE cDNA cloned in a bacterial expression vector, pET-22b (+)18 was used to express the enzyme as 6xHis-tagged proteins in Escherichia coli BL-21 (DE3) cells grown at 37°C. Protein expression was induced with 1 mM isopropyl-β-thiogalactopyranoside (Sigma Aldrich) for ~15 h at 18°C. Cells were pelleted, resuspended in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and a protease inhibitor cocktail (34 ug/ml each of L-tosylamido-2-phenylethyl chloromethyl ketone, 1-chloro-3-tosylamido-7-amino-2-heptanone, and phenylmethylsulfonyl fluoride, and 3 µg/ml each of leupeptin and lima bean trypsin inhibitor) and lysed using a French Press (Thermo Fisher Scientific, Waltham, MA). The 6xHis-tagged enzyme was enriched using HisPur™ Cobalt Resin (Thermo Fisher Scientific, Waltham, MA) storge buffers containing 20 mM HEPES, pH 8.0, 2 mM MgCl2, 1 mM EDTA, and 1 mM dithiothreitol. The fractions were concentrated by using an Amicon Ultra-50K centrifuge (Millipore, Billerica, MA). The enzyme concentration was determined using CB-Protein Assay™ Kit (from CALBIOCHEM) with bovine serum albumin as a standard.
Michaelis-Menten Kinetics of Cocaine Esterase
The catalytic activities of the enzyme against (+)- and (−)-cocaine were determined at the same time under the same experimental conditions. The initial rates of the enzymatic hydrolysis of (+)/(−)-cocaine were estimated by following the change in the intrinsic absorbance of (+)/(−)-cocaine at 230 nm with time using a GENios Pro microplate reader (TECAN, Research Triangle Park, NC) with the XFluor software. The initial rates were estimated from the linear portion of the progress curves and spanned no longer than 15 min. The reaction was initiated by adding 100 µl of an enzyme solution (phosphate buffered saline (PBS, pH 7.4) to 100 µl of a cocaine solution (50 ng/ml enzyme, 100 mM phosphate buffer, pH 7.4). Final (+)/(−)-cocaine concentrations were as follows: 100, 50, 20, 15, 10, 7.5, 5, and 2.5 µM. Vmax and KM values were calculated by using Prism 5 (GraphPad Software Inc., San Diego, CA). All of the activity assays were performed at room temperature (~25°C).
Results and Discussion
Reaction Pathway
Our QM/MM reaction coordinate calculations at the B3LYP/6-31G*:AMBER level revealed that the acylation stage of CocE-catalyzed (+)-cocaine hydrolysis reaction consists of two reaction steps. The first reaction step is the nucleophilic attack on the carbonyl carbon (Cζ) of (+)-cocaine benzoyl ester by Oγ atom in Ser117 site chain. The second reaction step is the dissociation between the benzoyl ester and ecgonine methyl ester of (+)- cocaine. The optimized geometries of the reactant, intermediates, and transition states are shown in Figures 2 and 3. Below we discuss each of these reaction steps in detail.
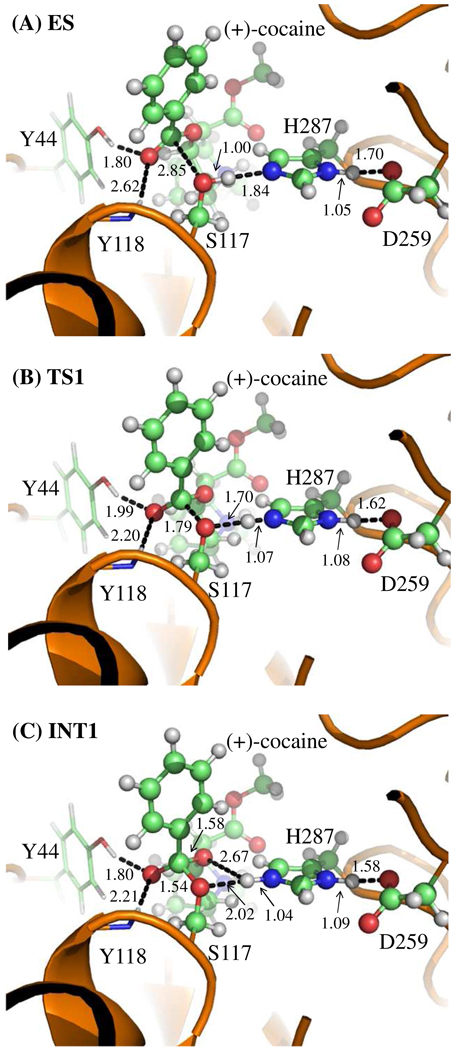
Figure 2.
Key configurations for reaction step 1, the nucleophilic attack by Oγ atom of Ser117. The geometries were optimized at QM/MM(B3LYP/6-31G*:AMBER) level. The key distances in the figure are in angstroms. Carbon, oxygen, nitrogen, and hydrogen atoms are colored in green, red, blue, and white, respectively. The backbone of the protein is rendered as a cartoon and colored in orange. The QM atoms are represented as ball and stick, and the surrounding residues rendered as stick. The figures below are represented using the same method.
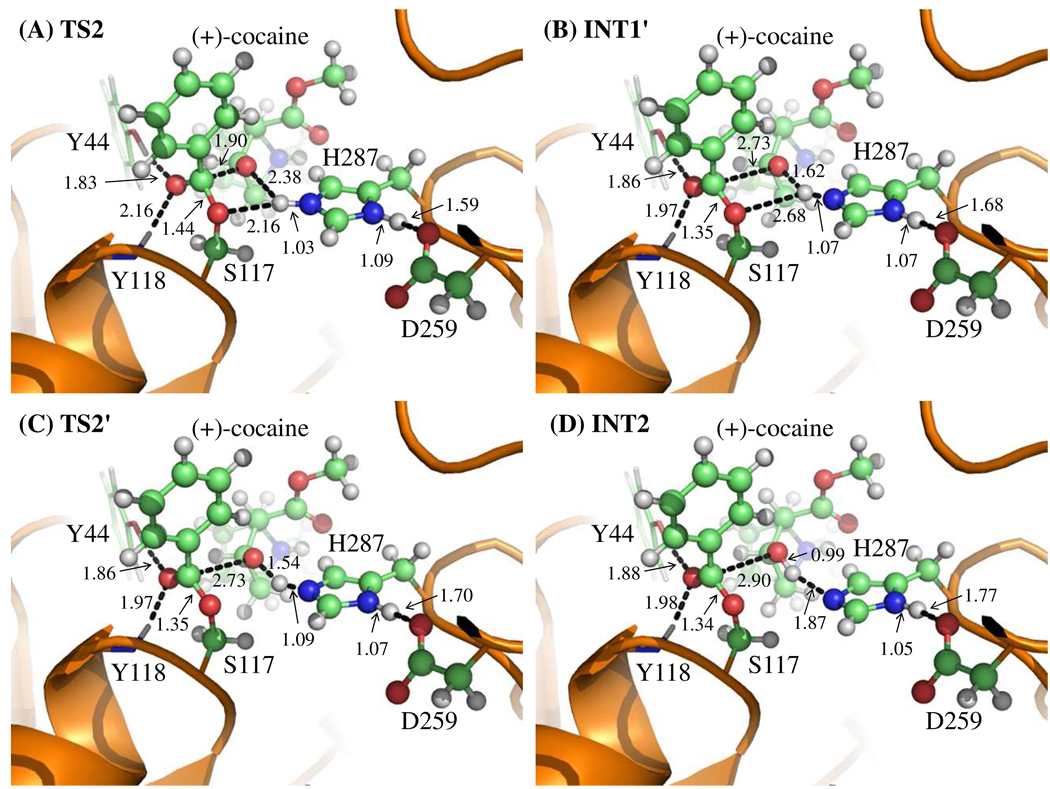
Figure 3.
Key configurations except INT1 for reaction step 2, the dissociation of (+)-cocaine benzoyl ester. The geometries were optimized at QM/MM(B3LYP/6-31G*:AMBER) level. The structure of INT1 is given in Figure 2C.
During the first step of the catalytic reaction, the nucleophilic attack process proceeds as the serine hydroxyl oxygen, i.e. Oγ atom of Ser117, gradually approaches the Cζ atom of (+)-cocaine benzoyl ester. Meanwhile, the serine hydroxyl hydrogen, i.e. Hγ atom of Ser117, gradually moves towards the nitrogen (Nε) atom of His287 side chain. Since this reaction step involves the breaking of Oγ–Hγ bond and formation of both Cζ–Oγ and Nε–Hγ bonds as shown in Scheme 1, the distances between Oγ and Hγ (ROγ–Hγ), between Cζ and Oγ (RCζ–Oγ), and between Nε and Hγ (RNε–Hγ) reflect the nature of the first chemical reaction step. Therefore, the reaction coordinate for the first reaction step was set as ROγ–Hγ − RCζ–Oγ − RNε–Hγ. As shown in the QM/MM-optimized geometries (Figure 2), as the Oγ atom of Ser117 gradually approaches the Cζ atom, the geometry of the reactant (ES), in which the Cζ atom is sp2 hybridized and is in a planar geometry with its three bonding atoms, gradually changes into a tetrahedral geometry centered on the sp3 hybridized Cζ atom in an intermediate (INT1) through a transition state (TS1).
During the dissociation of (+)-cocaine benzoyl ester, the ecgonine group of (+)-cocaine gradually departs from the (+)-cocaine benzoyl ester group in which the benzoyl ester bond Cζ–Oζ is broken. Meanwhile, the proton (Hγ) attached to Nε atom of His287 side chain transfers to the benzoyl ester oxygen atom (Oζ) of (+)-cocaine. The changes of the distances RCζ–Oζ, ROζ–Hγ, and RNε–Hγ reflect the nature of a dissociation process. Thus the reaction coordinate for the second reaction step was chosen as RCζ–Oζ + RNε–Hγ − ROζ–Hγ.
Contrary to what we purposed in Scheme 1 where only one transition state is hypothesized for reaction step 2, two transition states in current reaction process were found. This observation is similar to that in CocE-catalyzed (−)-cocaine hydrolysis where two transition states were characterized in the dissociation of (−)-cocaine benzoyl ester.32 The two transition states here are denoted by TS2 and TS2'. The intermediate between the two transition states is denoted by INT1'. The QM/MM-optimized geometries of the intermediates and transition states of current reaction process are given in Figure 3.
In the geometry of INT1 where the serine hydroxyl proton (Hγ) has been transferred to Nε atom of His287 in the reaction step 1, the distance (ROγ–Hγ) between Oγ atom of Ser117 side chain and Hγ atom of His287 side chain is 2.02 Å, indicating a strong hydrogen bond of Nε–Hγ…Oγ between Ser117 and His287 side chains. However, the distance (ROζ–Hγ) between Hγ and the leaving ester oxygen (Oζ) to which Hγ is about to be transferred is 2.67 Å, indicating a very weak hydrogen bond between the Hγ and Oζ atoms and an environment unsuitable for proton transfer from Nε atom of His287 to the leaving ester oxygen (Oζ) atom. In changing from INT1 to INT1', there are two major structural changes. One is the gradual breaking of the covalent bond Cζ–Oζ (RCζ–Oζ is 1.58 Å in INT1, 1.90 Å in TS2, and 2.73 Å in INT1'). The other is the formation of a hydrogen bond Nε–Hγ…Oζ indicated by the shorter and shorter distance ROζ–Hγ in going from INT1 to INT1' (2.67 Å in INT1, 2.38 Å in TS2, and 1.62 Å in INT1'). In the mean time, the hydrogen bond Nε–Hγ…Oγ formed between the transferring proton (Hγ) and the Oγ atom of Ser117 becomes progressively weaker (ROγ–Hγ is 2.02 Å in INT1, 2.16 Å in TS2, and 2.68 Å in INT1'), which is reasonable as the transferring proton (Hγ) is about to be transferred to the leaving ester oxygen (Oζ) in current reaction step.
The major difference between INT1' and TS2' is the position of transferring proton (Hγ) while the distance RCζ–Oζ remains unchanged, indicating that the reaction process associated with TS2' is primarily the proton (Hγ) transfer from Nε atom of His287 side chain to the leaving ester oxygen (Oζ) atom (ROζ–Hγ is 1.62 Å in INT1', 1.54 Å in TS2', and 0.99 Å in INT2; RNε–Hγ is 1.07 Å in INT1', 1.09 Å in TS2', and 1.87 Å in INT2). Therefore, the proton transfer in the current reaction process proceeds not simultaneously with but only after the breaking of C–O covalent bond.
Catalytic Role of Oxyanion Hole
It is interesting to know the catalytic role of the oxyanion hole consisting of the backbone amide of Tyr118 and the hydroxyl group of Tyr44 side chain. Based on the QM/MM reaction coordinate calculations, throughout the acylation stage of (+)-cocaine hydrolysis, the carbonyl oxygen (Oη) of (+)-cocaine forms two hydrogen bonds with the oxyanion hole. One is the hydrogen bond of O–Hη…Oη with hydroxyl hydrogen (Hη) atom of Tyr44 side chain and the other is the hydrogen bond of N–Hκ…Oη with the backbone NH group (Hκ atom) of Tyr118. As one can see from Figures 2 and 3, the hydrogen bond O–Hη…Oη between Oη atom and Tyr44 hydroxyl is very strong throughout the acylation stage with a distance of ~1.8 Å. The other hydrogen bond N–Hκ…Oη between Oη atom and Tyr118 backbone NH group is relatively weaker than the one with Tyr44 hydroxyl during the reaction. It is weak in ES with the distance of ~2.6 Å and then becomes stronger with the distance of ~2.1 Å in the subsequent states of the reaction. Therefore both hydrogen bonds stabilize the negative charge of carbonyl oxygen (Oη) developing during the hydrolysis, where the primary contribution to the stabilization comes from Tyr44.
Energetics and Kinetic Parameters
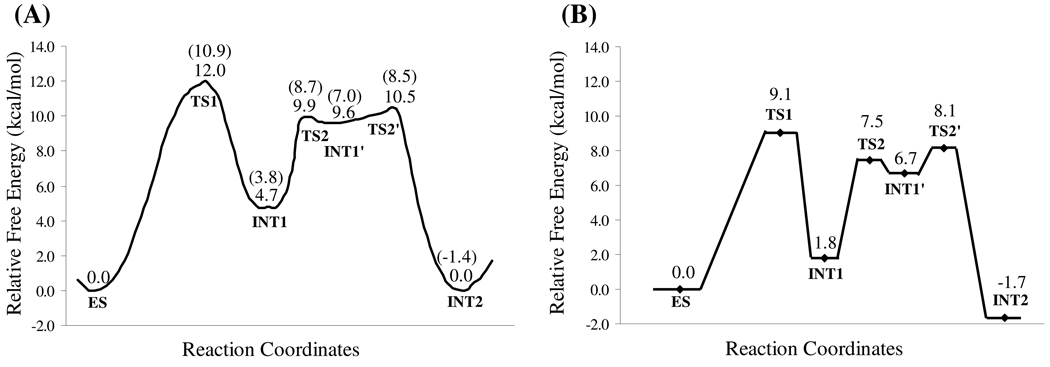
Using the QM/MM-optimized geometries at the QM/MM(B3LYP/6-31G*:AMBER) level, we carried out QM/MM single-point energy calculations at the QM/MM(MP2/6-31+G*:AMBER) level for each geometry along the minimum-energy path. For each geometry along the minimum-energy path, the ESP charges determined in the QM subsystem of the QM/MM single-point energy calculation were used in subsequent FEP simulations for estimating the free energy changes along the reaction path. Depicted in Figure 4A is the energy profile determined by the QM/MM-FE calculations excluding the zero-point and thermal corrections for the QM subsystem. The values given in the parentheses are the corresponding relative free energies with the zero-point and thermal corrections for the QM subsystem. It has been pointed out in our previous study32 that, although the counterions in CocE system are not directly involved in the reaction mechanism, the interaction (particularly the electrostatic interaction) between the QM subsystem and the large number of counterions in the CocE system is significant in determining free energy barrier of the reaction. Therefore, in the present study, we also estimated the electrostatic interaction between QM subsystem and counterions, which can be considered as the correction with the counterions to the free energies, by following the same computational strategy as in our previous study on the CocE-catalyzed (−)-cocaine hydrolysis where the coordinates of counterions in 100 snapshots were taken out (one snapshot in each 10 ps) from the MD trajectory.32 The relative free energies with both zero-point and thermal corrections for the QM subsystem and electrostatic corrections with the counterions are shown in Figure 4B. The calculated final free energy barriers are summarized in Table 1 in comparison with those calculated for CocE-catalyzed hydrolysis of (−)-cocaine.
Figure 4.
(A) Free energy profile determined by the MP2/6-31+G*:AMBER QM/MM-FE calculations excluding the zero-point and thermal corrections for the QM subsystem. The values in parentheses are relative free energies including zero-point and thermal corrections for the QM subsystem. (B) The relative free energies with both zero-point and thermal corrections for the QM subsystem and electrostatic corrections from counterions.
Table 1.
The final relative free energies for CocE-catalyzed hydrolyses of (+)- and (−)-cocaine.
| Reaction stage |
Geometry | Relative free energy (kcal/mol) | |
|---|---|---|---|
| (−)-cocainea | (+)-cocaine | ||
| ES | 0.0 | 0.0 | |
| TS1 | 2.3 | 9.1 | |
| Acylation | INT1 | −5.3 | 1.8 |
| TS2 | −2.6 | 7.5 | |
| INT1' | N/Ab | 6.7 | |
| TS2' | N/Ab | 8.1 | |
| INT2 | −13.1 | −1.7 | |
| INT2' | 0.0 | ||
| TS3 | 17.9 | ||
| Deacylation | INT3 | 14.8 | |
| TS4 | 17.5 | ||
| PD | −3.8 | ||
Data for (−)-cocaine are all from ref.32.
N/A, not applicable. The energy barrier associate with TS2' on the potential energy surface was too small (~0.1 kcal/mol) for the (−)-cocaine hydrolysis and the barrier disappeared after the FEP simulation was applied.
As shown in Figure 4B, the relative free energy (9.1 kcal/mol) associated with TS1 is slightly higher than those associated with the remaining two transition states in the acylation stage, namely TS2 (7.5 kcal/mol) and TS2' (8.1 kcal/mol). Therefore, the rate-determining step of the acylation stage of CocE-catalyzed (+)-cocaine hydrolysis is the first reaction step, i.e., the nucleophilic attack on the Cζ atom by Oγ atom of Ser117. As mentioned in the Introduction section, CocE-catalyzed hydrolyses of (+)- and (−)-cocaine share the same deacylation stage. Thus, the free energy profiles for the deacylation of both (+)- and (−)-cocaine by CocE are identical. Our previous study32 has shown that the calculated free energy barrier of deacylation in the CocE-catalyzed (−)-cocaine hydrolysis, which was found to be rate-determining, is ~17.9 kcal/mol. Therefore, the free energy barrier for the deacylation of (+)-cocaine should be also ~17.9 kcal/mol, and the deacylation is also rate-determining for CocE-catalyzed (+)-cocaine hydrolysis because its energy barrier is much higher than that of acylation stage (9.1 kcal/mol).
Now that the common deacylation step is rate-determining for both (+)- and (−)-cocaine hydrolyses catalyzed by CocE, these computational results predict that the catalytic rate constant (kcat) of CocE against (+)-cocaine should be the same as that of CocE against (−)-cocaine. The predicted reaction mechanisms and relative catalytic rate constants of CocE against (+)- and (−)- cocaine are remarkably different from those of BChE against (+)- and (−)-cocaine.22–23 Human BChE-catalyzed hydrolyses of (+)- and (−)-cocaine have different rate-determining steps and considerably different catalytic rate constants, with a difference in three-orders-of-magnitude 23
Kinetic parameters for CocE-catalyzed hydrolysis of (+)-cocaine are not available in literature, although kinetic parameters for CocE-catalyzed hydrolysis of (−)-cocaine were determined previously.14,18 To examine whether the computational prediction is correct, we also carried out experimental kinetic analysis on CocE-catalyzed hydrolyses of (+)- and (−)-cocaine at the same time under the same experimental conditions. The determined kinetic parameters are summarized in Table 2. As shown in Table 2, the kcat values for CocE-catalyzed hydrolyses of (+)- and (−)- cocaine were determined to be 1078±250 and 1082±181 min−1, respectively. The two kcat values are identical within the experimental fluctuations, which strongly supports the computational prediction that CocE-catalyzed hydrolyses of (+)- and (−)-cocaine have a common rate-determining reaction step. In addition, the determined two KM (Michaelis-Menten constant) values are also identical within the experimental fluctuations.
Table 2.
Kinetic parameters determined for CocE-catalyzed hydrolyses of (+)- and (−)-cocaine
| (+)-cocaine | (−)-cocaine | ||
|---|---|---|---|
| KM (µM) | kcat (min−1) | KM (µM) | kcat (min−1) |
| 15±4 | 1078±250 | 13±3 | 1082±181 |
Implication from the Mechanistic Insights for Design of an Improved Cocaine Esterase
The mechanistic differences between (+)- and (−)-cocaine hydrolyses catalyzed by CocE and their rate-determining steps are remarkably different from those catalyzed by human BChE. In the (+)- and (−)-cocaine hydrolyses catalyzed by human BChE, the rate-determining step for the (+)-cocaine hydrolysis is different from that for the (−)-cocaine hydrolysis; the rate-determining step for the (−)-cocaine hydrolysis is a reaction step before the deacylation. As a result, (+)-cocaine hydrolysis in human BChE is about three-orders-of-magnitude faster than the corresponding (−)-cocaine hydrolysis in the same enzyme. Thus, computational design of high-activity mutants of human BChE against (−)-cocaine has been focused on the reaction steps before the deacylation. Unlike BChE, the rate-determining steps for CocE-catalyzed hydrolyses of (+)- and (−)-cocaine are a common reaction step (the first step) in the deacylation stage such that the catalytic rate constants for both (+)- and (−)-cocaine hydrolyses are the same. Therefore, computational design of high-activity mutants of CocE against cocaine should be focused on the first reaction step of the deacylation.
The mechanistic differences between CocE-catalyzed (+)- and (−)-cocaine hydrolyses reside in the acylation stage which is not rate-determining and, thus, do not seem to provide direct clues to design a CocE mutant with an improved catalytic activity against (−)-cocaine. Nevertheless, the mechanistic differences between CocE-catalyzed (+)- and (−)-cocaine hydrolyses may indirectly provide beneficial clues to design high-activity mutants of CocE against (−)-cocaine. As reported in our previous study,32 the free energy barriers for the first and second reaction steps in the acylation of (−)-cocaine are ~2.3 and ~2.7 kcal/mol, respectively. As shown in Figure 4B and Table 1, the free energy barrier for the first reaction step of CocE-catalyzed hydrolysis of (+)-cocaine is ~9.1 kcal/mol, much higher than that for the first or second reaction step of the (−)-cocaine hydrolysis. A detailed analysis of the QM/MM-optimized geometries suggests that the significant difference in the free energy barrier for the first reaction step between the (+)- and (−)-cocaine hydrolyses may be attributed to the difference in the hydrogen bonding with the oxyanion hole in the transition state TS1. As discussed above, there are two hydrogen bonds between the carbonyl oxygen (Oη) of the substrate and the oxyanion hole consisting of the hydroxyl group (O–Hη) of Tyr44 side chain and the backbone NH group (Hκ atom) of Tyr118. In the TS1 geometry with (+)-cocaine, as depicted in Figure 2B, the optimized Hη…Oη and Hκ…Oη distances were 1.99 and 2.20 Å, respectively. In the corresponding TS1 geometry with (−)-cocaine, the optimized Hη…Oη and Hκ…Oη distances were 1.75 and 2.25 Å, respectively.32 The overall hydrogen bonding of the oxyanion hole with (−)-cocaine should be significantly stronger than that with (+)-cocaine. The possible effect of the hydrogen bonding on the TS1 stabilization may provide some indirect clues in rational design of high-activity mutants of CocE against cocaine, because the two similar hydrogen bonds also exist in the transition state (denoted by TS3) for the rate-determining step the deacylation.32 Apparently, certain amino acid mutations capable of enhancing the overall hydrogen bonding with the oxyanion hole in the TS3 structure could decrease the energy barrier for the rate-determining step and thus improve the catalytic activity of the enzyme against cocaine.
Conclusion
Results from the first-principle QM/MM-FE calculations demonstrate that the acylation stage of CocE-catalyzed hydrolysis of (+)-cocaine is initiated by the attack of the hydroxyl oxygen (Oγ) of Ser117 on carbonyl carbon (Cζ) of (+)-cocaine benzoyl ester. This process is facilitated by His287 through proton (Hγ) transfer from Ser117 hydroxyl to Nε atom of His287 side chain which increases the nucleophilicity of the Ser117 hydroxyl. His287 is in turn stabilized by the formation of another hydrogen bond between His287 and Asp259 side chains. The Ser117 nucleophile attacks the electron-deficient Cζ atom of (+)-cocaine benzoyl ester, forming a tetrahedral intermediate in which the carbonyl oxygen (Oη) of (+)-cocaine with developing negative charge is stabilized by two tyrosine residues (Tyr44 and Tyr118) in the oxyanion hole. Then His287 transfers a proton (Hγ) to the ester oxygen (Oζ) of the leaving ecgonine group, completing the acylation stage.
The QM/MM-optimized geometries indicate that the oxyanion hole stabilizes the negative charge of (+)-cocaine carbonyl oxygen (Oη) developing during the hydrolysis by providing two hydrogen bonds with Tyr44 and Tyr118. The hydrogen bond with Tyr44 is particularly strong and is the primary factor stabilizing the carbonyl oxygen (Oη) of (+)-cocaine benzoyl ester.
The highest energy barrier calculated for the acylation of (+)-cocaine is ~9.1 kcal/mol associated with the first reaction step of acylation. The calculated energy barrier of ~9.1 kcal/mol is much lower than the highest energy barrier for the deacylation (~17.9 kcal/mol associated with the first reaction step of deacylation). Therefore, the deacylation of (+)-cocaine, which is identical to that of (−)-cocaine, is rate-determining, revealing that CocE-catalyzed hydrolyses of (+)- and (−)-cocaine have a common rate-determining step. All of these results predict that the catalytic rate constant (kcat) of CocE against (+)-cocaine should be the same as that of CocE against (−)-cocaine, in contrast with the remarkable difference between human BChE-catalyzed hydrolyses of (+)- and (−)-cocaine. The computational prediction has been confirmed by performing experimental kinetic analysis on CocE-catalyzed hydrolysis of (+)-cocaine, for the first time, in comparison with CocE-catalyzed hydrolysis of (−)-cocaine.
The determined common rate-determining reaction step and detailed mechanistic differences in the acylation between CocE-catalyzed hydrolyses of (+)- and (−)-cocaine provide a valuable mechanistic base for future rational design of CocE mutants with an improved catalytic activity against cocaine. In particular, the common rate-determining reaction step indicates that rational design of a high-activity mutant of CocE should be focused on stabilization of the transition state structure (TS3) for the first reaction step of the deacylation. The mutation-caused stabilization of the rate-determining reaction step could lead to the decrease in the overall energy barrier and, thus, the increase in the catalytic rate constant.
Acknowledgments
This work was supported in part by NIH (grants R01 DA025100, R01 DA021416, and R01 DA013930). The entire work was performed at the University of Kentucky. Zhao worked in Zhan’s laboratory for this project at the University of Kentucky as exchange graduate students from Central China Normal University. The authors thank Drs. Roger K. Sunahara and Diwahar L Narasimhan at University of Michigan Medical School for providing the cDNA samples. The authors also acknowledge the Center for Computational Sciences (CCS) at University of Kentucky for supercomputing time on IBM X-series Cluster with 340 nodes or 1,360 processors.
References
- 1.Mendelson JH, Mello NK. N. Engl. J. Med. 1996;334(15):965–972. doi: 10.1056/NEJM199604113341507. [DOI] [PubMed] [Google Scholar]
- 2.Paula S, Tabet MR, Farr CD, Norman AB, Ball WJ. J. Med. Chem. 2003;47(1):133–142. doi: 10.1021/jm030351z. [DOI] [PubMed] [Google Scholar]
- 3.Singh S. Chem. Rev. 2000;100(3):925–1024. doi: 10.1021/cr9700538. [DOI] [PubMed] [Google Scholar]
- 4.Substance Abuse and Mental Health Services Administration. Drug Abuse Warning Network, 2004: National Estimates of Drug-Related Emergency Department Visits. Rockville, MD: Office of Applied Studies, Department of Health & Human Services; 2006. DAWN Series D-28; DHHS Publication No. SMA 06-4143. [Google Scholar]
- 5.Sparenborg S, Vocci F, Zukin S. Drug Alcohol. Depend. 1997;48(3):149–151. doi: 10.1016/s0376-8716(97)00072-0. [DOI] [PubMed] [Google Scholar]
- 6.Gorelick DA. Drug Alcohol. Depend. 1997;48(3):159–165. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 7.Gorelick DA, Gardner EL, Xi ZX. Drugs. 2004;64(14):1547–1573. doi: 10.2165/00003495-200464140-00004. [DOI] [PubMed] [Google Scholar]
- 8.Dickerson TJ, Janda KD. Recent Advances for the Treatment of Cocaine Abuse: Central Nervous System Immunopharmacotherapy. Drug Addiction. 2008:217–229. doi: 10.1208/aapsj070359. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Gao D, Cho H, Yang W, Pan Y, Yang G, Tai H-H, Zhan C-G. Angew. Chem. Int. Ed. 2006;45(4):653–657. doi: 10.1002/anie.200503025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vocci FJ, Acri J, Elkashef A. Am. J. Psychiat. 2005;162(8):1432–1440. doi: 10.1176/appi.ajp.162.8.1432. [DOI] [PubMed] [Google Scholar]
- 11.Zheng F, Yang W, Ko M-C, Liu J, Cho H, Gao D, Tong M, Tai H-H, Woods JH, Zhan C-G. J. Am. Chem. Soc. 2008;130(36):12148–12155. doi: 10.1021/ja803646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Y, Gao D, Yang W, Cho H, Yang G, Tai H-H, Zhan C-G. Proc. Natl. Acad. Sci. U.S.A. 2005;102(46):16656–16661. doi: 10.1073/pnas.0507332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bresler MM, Rosser SJ, Basran A, Bruce NC. Appl. Environ. Microbiol. 2000;66(3):904–908. doi: 10.1128/aem.66.3.904-908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen NA, Turner JM, Stevens J, Rosser SJ, Basran A, Lerner RA, Bruce NC, Wilson IA. Nat. Struct. Mol. Biol. 2002;9(1):17–21. doi: 10.1038/nsb742. [DOI] [PubMed] [Google Scholar]
- 15.Cooper ZD, Narasimhan D, Sunahara RK, Mierzejewski P, Jutkiewicz EM, Larsen NA, Wilson IA, Landry DW, Woods JH. Mol. Pharmacol. 2006;70(6):1885–1891. doi: 10.1124/mol.106.025999. [DOI] [PubMed] [Google Scholar]
- 16.Ko M-C, Bowen LD, Narasimhan D, Berlin AA, Lukacs NW, Sunahara RK, Cooper ZD, Woods JH. J. Pharmacol. Exp. Ther. 2007;320(2):926–933. doi: 10.1124/jpet.106.114223. [DOI] [PubMed] [Google Scholar]
- 17.Emily MJ, Michelle GB, Ziva DC, Diwahar N, Roger KS, James HW. Ann. Emerg. Med. 2008;54:409–420. [Google Scholar]
- 18.Gao D, Narasimhan DL, Macdonald J, Brim R, Ko M-C, Landry DW, Woods JH, Sunahara RK, Zhan C-G. Mol. Pharmacol. 2009;75(2):318–323. doi: 10.1124/mol.108.049486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brim RL, Nance MR, Youngstrom DW, Narasimhan D, Zhan C-G, Tesmer JJG, Sunahara RK, Woods JH. Mol. Pharmacol. 2010;77(4):593–600. doi: 10.1124/mol.109.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins GT, Brim RL, Narasimhan D, Ko MC, Sunahara RK, Zhan C-G, Woods JH. J. Pharmacol. Exp. Ther. 2009;331(2):445–455. doi: 10.1124/jpet.108.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narasimhan D, Nance MR, Gao D, Ko M-C, Macdonald J, Tamburi P, Yoon D, Landry DM, Woods JH, Zhan C-G, Tesmer JJG, Sunahara RK. Protein Eng. Des. Sel. 2010;23:537–547. doi: 10.1093/protein/gzq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan C-G, Gao DQ. Biophys. J. 2005;89(6):3863–3872. doi: 10.1529/biophysj.105.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhan C-G, Zheng F, Landry DW. J. Am. Chem. Soc. 2003;125(9):2462–2474. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H, Pang Y-P, Lockridge O, Brimijoin S. Mol. Pharmacol. 2002;62(2):220–224. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y, Atanasova E, Sui N, Pancook JD, Watkins JD, Brimijoin S. Mol. Pharmacol. 2005;67(1):204–211. doi: 10.1124/mol.104.006924. [DOI] [PubMed] [Google Scholar]
- 26.Gao DQ, Zhan C-G. J. Phys. Chem. B. 2005;109(48):23070–23076. doi: 10.1021/jp053736x. [DOI] [PubMed] [Google Scholar]
- 27.Hamza A, Cho H, Tai H-H, Zhan C-G. J. Phys. Chem. B. 2005;109(10):4776–4782. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H, El Yazal J, Lockridge O, Schopfer LM, Brimijoin S, Pang YP. J. Biol. Chem. 2001;276(12):9330–9336. doi: 10.1074/jbc.M006676200. [DOI] [PubMed] [Google Scholar]
- 29.Sun H, Shen ML, Pang YP, Lockridge O, Brimijoin S. J. Pharmacol. Exp. Ther. 2002;302(2):710–716. doi: 10.1124/jpet.302.2.710. [DOI] [PubMed] [Google Scholar]
- 30.Yang WC, Xue L, Fang L, Chen X, Zhan C-G. Chem. Biol. Interact. 2010;187(1–3):148–152. doi: 10.1016/j.cbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng F, Yang WC, Xue L, Hou SR, Liu JJ, Zhan C-G. Biochemistry. 2010;49(42):9113–9119. doi: 10.1021/bi1011628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Hamza A, Zhan C-G. J. Am. Chem. Soc. 2009;131(33):11964–11975. doi: 10.1021/ja903990p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner JM, Larsen NA, Basran A, Barbas CF, Bruce NC, Wilson IA, Lerner RA. Biochemistry. 2002;41(41):12297–12307. doi: 10.1021/bi026131p. [DOI] [PubMed] [Google Scholar]
- 34.Zheng F, Zhan C-G. Org. Biomol. Chem. 2008;6(5):836–843. doi: 10.1039/b716268e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan YM, Gao DQ, Yang WC, Cho H, Zhan C-G. J. Am. Chem. Soc. 2007;129(44):13537–13543. doi: 10.1021/ja073724k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao DQ, Zhan C-G. Proteins. 2006;62(1):99–110. doi: 10.1002/prot.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang YK. J. Chem. Phys. 2005;122(2):024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 38.Zhang YK, Lee TS, Yang WT. J. Chem. Phys. 1999;110(1):46–54. [Google Scholar]
- 39.Zhang YK, Liu HY, Yang WT. J. Chem. Phys. 2000;112(8):3483–3492. [Google Scholar]
- 40.Zhang YK. Theor. Chem. Acc. 2006;116(1–3):43–50. [Google Scholar]
- 41.Hu P, Zhang YK. J. Am. Chem. Soc. 2006;128(4):1272–1278. doi: 10.1021/ja056153+. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, Zhang Y, Yang W. J. Am. Chem. Soc. 2000;122(28):6560–6570. [Google Scholar]
- 43.Zhang Y, Kua J, McCammon JA. J. Am. Chem. Soc. 2002;124(35):10572–10577. doi: 10.1021/ja020243m. [DOI] [PubMed] [Google Scholar]
- 44.Cisneros GA, Liu H, Zhang Y, Yang W. J. Am. Chem. Soc. 2003;125(34):10384–10393. doi: 10.1021/ja029672a. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Kua J, McCammon JA. J. Phys. Chem. B. 2003;107(18):4459–4463. [Google Scholar]
- 46.Cheng Y, Zhang Y, McCammon JA. J. Am. Chem. Soc. 2005;127(5):1553–1562. doi: 10.1021/ja0464084. [DOI] [PubMed] [Google Scholar]
- 47.Corminboeuf C, Hu P, Tuckerman ME, Zhang Y. J. Am. Chem. Soc. 2006;128(14):4530–4531. doi: 10.1021/ja0600882. [DOI] [PubMed] [Google Scholar]
- 48.Wang L, Yu X, Hu P, Broyde S, Zhang Y. J. Am. Chem. Soc. 2007;129(15):4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S, Hu P, Zhang Y. J. Phys. Chem. B. 2007;111(14):3758–3764. doi: 10.1021/jp067147i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao C, Zhang Y. J. Phys. Chem. B. 2007;111(22):6229–6235. doi: 10.1021/jp068657f. [DOI] [PubMed] [Google Scholar]
- 51.Hu P, Wang S, Zhang Y. J. Am. Chem. Soc. 2008;130(12):3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng Y, Zhang Y, McCammon JA. Protein Sci. 2006;15(4):672–683. doi: 10.1110/ps.051852306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Case DA, Darden TA, TE Cheatham I, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER 8. San Francisco: University of California; 2004. [Google Scholar]
- 54.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery J JA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03. Wallingford, CT: Gaussian, Inc.; 2004. Revision C.02. [Google Scholar]