Abstract
Activation of the complement cascade induces inflammatory responses and marks cells for immune clearance. In the central complement-amplification step, a complex consisting of surface-bound C3b and factor B is cleaved by factor D to generate active convertases on targeted surfaces. We present crystal structures of the pro-convertase C3bB at 4 angstrom resolution and its complex with factor D at 3.5 angstrom resolution. Our data show how factor B binding to C3b forms an open “activation” state of C3bB. Factor D specifically binds the open conformation of factor B through a site distant from the catalytic center and is activated by the substrate, which displaces factor D’s self-inhibitory loop. This concerted proteolytic mechanism, which is cofactor-dependent and substrate-induced, restricts complement amplification to C3b-tagged target cells.
The human complement system contributes to a variety of processes, ranging from immunosurveillance to maintenance of cell homeostasis, which depend on a balance of complement activation and regulation (1). Besides a low level of steady activation through hydrolysis of the central complement component, C3, the cascade can be specifically triggered by various structures on the surfaces of foreign and apoptotic cells (1, 2). These events initiate distinct pathways that converge into a central amplification loop, which relies on an efficient enzyme complex that allows rapid turnover of C3 (3). This C3 convertase complex is generated when factor B (FB) interacts with surface-bound complement fragment C3b and forms the pro-convertase C3bB, which is cleaved by factor D (FD) to yield the active yet unstable C3 convertase C3bBb (4, 5). The C3 convertase cleaves C3 into anaphylatoxin C3a and opsonin C3b (2, 6-8). Newly generated C3b molecules become covalently attached to cell surfaces proximate to sites of activation and form additional convertases, thereby rapidly amplifying complement response on targeted cells (9-11). In order to prevent excessive activation and attack of host tissue, convertase activity has to be tightly controlled (12). Although recent studies provided insight into structure, function, stability, and regulation of the assembled C3 convertase (13, 14), molecular details about the convertase formation and activation step remained elusive.
The published crystal structure of free FB showed a “locked” conformation in which the scissile bond (Arg234–Lys235) is protected from being cleaved by FD (15). Moreover, a recent structure of FB in complex with the C3b homolog cobra-venom factor (CVF) indicated how the propeptide segment of FB, Ba (residues 1 to 234), mediates the Mg2+-dependent binding of the protease segment Bb (residues 235 to 739) to the C terminus of CVF through the metal-ion-dependent adhesion site (MIDAS) of the Von Willebrand factor type-A (VWA) domain in Bb (16). Unexpectedly, the structure of CVFB did not show conformational changes in FB that lead to exposure of its scissile bond. Single-particle reconstruction of negatively stained electron microscopy (EM) data for the C3bB complex, however, indicated the coexistence of two conformations: one corresponding to a closed form as observed in CVFB and one to an open form that putatively exposes the scissile bond in FB (17). Allosteric roles have been proposed for the regulatory MIDAS and C-terminal helix α7 of the VWA domain of Bb [structural elements that play a key role in signaling through the homologous I domains in integrins (18, 19)] and the three complement-control protein (CCP) domains and C-terminal helical linker αL of the Ba segment (15, 20, 21). Whereas it is likely that binding of FB to C3b induces conformational changes that expose the scissile bond (Arg234–Lys235) for cleavage by FD, the mechanisms involved in this process have not yet been elucidated.
FD is a single-domain serine protease, which circulates in plasma in a mature but self-inhibited form (22, 23). Even when the inhibition was prevented by mutagenesis, the activity of FD against peptides remained poor when compared with other serine proteases like trypsin or thrombin (24-26). Therefore, it is likely that proteolytic activity and specificity are controlled during the assembly and activation of the central enzyme complex.
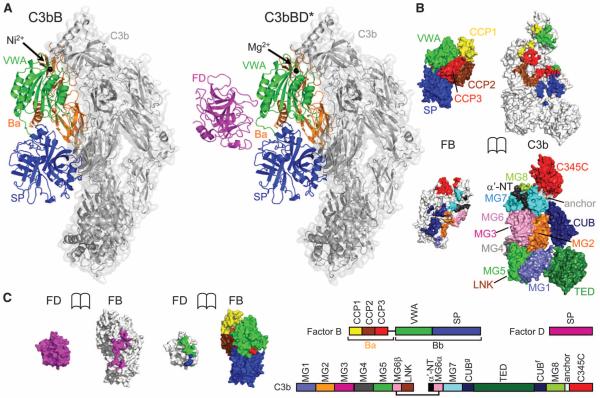
We determined crystal structures of the pro-convertase C3bB stabilized by Ni2+ (17, 27) at 4.0-Å resolution and of the pro-convertase C3bB in complex with a catalytically inactive FD mutant Ser183→Ala183 [abbreviated as S183A (28)] (S195A, chymotrypsin numbering is given in parentheses) in the presence of Mg2+ at 3.5-Å resolution, which we refer to as C3bBD* (Fig. 1A) (29). In addition, we solved the structures of two FD mutants, S183A (S195A) and R202A (R218A) in their unbound form. Refinement statistics and the quality of the final models are described in table S1 and fig. S1. C3b in both C3bB and C3bBD* resembles free C3b (fig. S2) (30, 31). Two polypeptide chains, termed β (residues 1 to 645) and α’ (residues 727 to 1641) chains, together form a core of eight macroglobulin (MG1 to MG8) domains with a linker domain (LNK) inserted into domain MG6, a “complement C1r/C1s, Uegf, Bmp1” (CUB) and a thioester-containing domain (TED) in between MG7 and MG8, and a C-terminal C345C domain attached to MG8 (Fig. 1). Most of the C3b domains (MG1 to MG8, LNK, and CUB) show only minor rearrangements (with rotations up to 7°) compared to free C3b; however, we observed larger rotations for the peripheral domains TED and C345C (up to 32° and 15° rotations for C3bB and C3bBD*, respectively) (fig. S2). FB consists of five domains grouped into two segments: three N-terminal CCP domains that together with the helical linker αL form the Ba segment and a central VWA domain and a C-terminal trypsinlike serine protease (SP) domain that form the protease segment Bb (Fig. 1). The CCP1-3 and VWA domains of FB bind C3b in an identical orientation as in CVFB (16) (Fig. 1B and fig. S2D). In contrast, the SP domain of FB in C3bB is rotated by 84°, which effectively shifts the catalytic serine residue by 65 Å and creates additional contacts between FB and C3b (Fig. 1, A and B). Furthermore, the structure of C3bBD* shows that FD binds this new conformation of FB without making any contacts to C3b (Fig. 1C), which is confirmed by surface plasmon resonance (SPR) measurements showing that C3b interacts with FB but not with FD (fig. S3).
Fig. 1.
Structures of C3bB and C3bBD*. (A) Overall structure of C3bB (left) and C3bBD* (right). C3b is shown as gray transparent surface; FB is shown as orange (Ba), green (VWA), and blue (SP) cartoons; and FD is represented as a magenta cartoon. The black spheres highlight the metal ions (Ni2+ for C3bB, Mg2+ for C3bBD*) at the MIDAS site. (B) Opened view of the footprint of the C3b-FB interaction, highlighting the domains of FB on the C3b surface (top) and the domains of C3b on FB (bottom). (C) Opened view of the footprint of the FB-FD interaction, highlighting the domains of FB on the FD surface (left) and the single domain of FD on FB (right). The scheme indicates the domain compositions and color codes of C3b, FB, and FD used in (B) and (C).
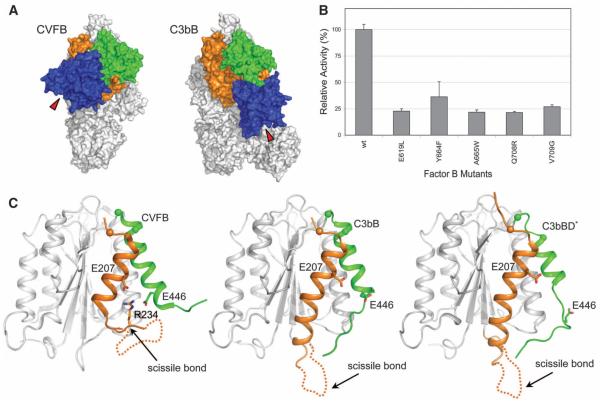
In the presence of the physiologically relevant ion Mg2+, two conformations of C3bB have been observed in EM experiments at a ratio of 35%:65%, which were referred to as closed and open forms, respectively (17). When using Ni2+, however, the equilibrium was shifted to a 2%:98% ratio (17). We used Ni2+ to crystallize the open form of C3bB. The crystal structure of C3bB (Ni2+) (Fig. 2A) fits well, with a correlation coefficient of 0.87 (fig. S4), to the shape of the open form of the pro-convertase seen in the EM experiments (17, 32). In contrast, the crystal structure of CVFB, which was confirmed by small-angle x-ray scattering solution studies (16), correlates with the closed form observed in EM. The closed conformation of CVFB possibly explains why it is processed ~100-fold slower than C3bB. However, despite the good agreement between the C3bB crystal structure and the EM reconstruction of the open form, our findings do not agree with the proposed structural model of domain rearrangements that was derived from the EM data (32). In particular, the x-ray data show a significant rotation of the SP domain rather than the CCP1-3 domains, as suggested by the EM model. The highly similar conformation of FB in the structure of C3bB in complex with FD (that is, C3bBD* crystallized in the presence of Mg2+) confirms that C3bB (Ni2+) adopts a physiological open conformation that is cleavable by FD. In all three structures (i.e., CVFB, C3bB, and C3bBD*), the C terminus of CVF or C3b is chelated to the metal ion in the regulatory MIDAS of the VWA domain in FB. The electron densities at 4 Å and 3.5 Å resolution for C3bB and C3bBD*, respectively, are consistent with high-affinity configurations for the MIDAS in both structures, as was observed for the structure of CVFB determined at 2.2 Å resolution (16). However, the limited resolution of the data precludes a detailed interpretation of the conformational changes that may be induced when Mg2+ is substituted by Ni2+ (fig. S5). The open form of C3bB is characterized by new interactions between the SP domain of FB and the MG2 and CUB domains of C3b, which result from the large rotation of the SP domain in FB. Mutations in FB at this new contact site reduced the C3bB cleavage rate (Fig. 2B and fig. S6), which shows that the interface observed in the crystal structure of C3bB in the open conformation is important for activity.
Fig. 2.
Comparison between the closed and open states in the pro-convertase C3bB. (A) Surface representation of the closed (CVFB, left) (16) and open (C3bB, right) conformations of the pro-convertase. Colors are the same as in Fig. 1A. Red triangles indicate the position of the catalytic site of FB during the conformational changes. (B) FD-mediated cleavage of C3bB performed by using SDS–polyacrylamide gel electrophoresis (PAGE) shows that mutations (28) in FB located at the interface between the SP and CUB domains in the open C3bB pro-convertase lower convertase formation rates. The histogram shows the relative pro-convertase activation rates compared with those of wild-type FB (see also fig. S6). Error bars represent deviations from the mean observed in multiple experiments (n>3). (C) Cartoon diagram of the VWA domain of FB, highlighting conformational changes in the transition from the closed (left) to the open state of the pro-convertase in absence (center) or in presence of FD (right). The αL helix is colored in orange and the α7 helix in green. The putative orientation of the loop containing the scissile bond of FB is shown with a dashed line. The positions of the C-α atoms located at the N-termini of αL and α7 helix are shown as spheres.
In both free FB and the closed CVFB complex, the P1 residue of the scissile bond (Arg234) is buried inside the VWA domain and forms salt bridges with Glu207 of helix αL and Glu446 of helix α7 (15, 16) (Fig. 2C). However, in the open C3bB structure concerted changes at the VWA-SP interface result in exposure of FB’s scissile loop (Fig. 2C). Rotation of the SP domain repositions the VWA-SP linker (residues 447 to 453) and elongates helix αL by two helical turns. This elongation is stabilized by residues Ile217 and Val220, which are docked into hydrophobic pockets of the VWA domain that were formerly occupied by residues Ile236 and Leu238 of the scissile loop (fig. S7A). A new interface is formed between four glutamates (residues 215 and 218 of αL and 422 and 424) of VWA and a cluster of four arginines (residues 580, 583, 683, and 685) in the SP domain (fig. S7B). In combination, these changes release Arg234 from its stabilizing interactions and expose the scissile loop (residues 224 to 239), which appears disordered in the structure of C3bB (fig. S7C). This mechanism differs markedly from that observed for the I domains of integrins, where a coupling of the MIDAS to helix α7 is implicated in the allosteric activation (18, 33). In the case of FB, however, the conformational changes involve predominantly the C-terminal end of helix αL, and to a lesser extent helix α7 and the interactions with the succeeding SP domain.
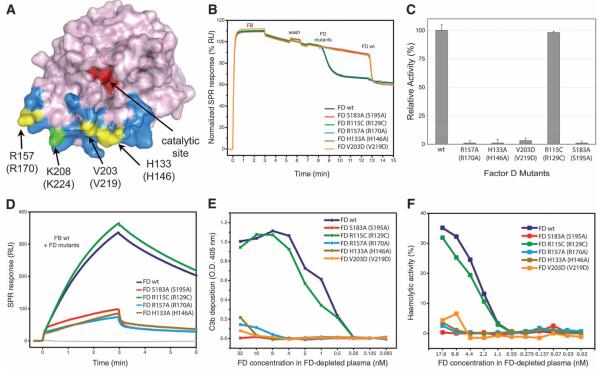
FD binds FB in the open C3bB configuration at the VWA-SP interface through a binding site 25 Å away from the catalytic center (Fig. 3A). This interaction of FD induces a downward and sideways shift (of ~4 Å) of the VWA helix α7 in FB and unwinds the C-terminal part of the helix (residues 442 to 446) (Fig. 2C). FD binds the unwound helix α7 and its surroundings, yielding a total interface area of ~700 Å2 dominated by 10 hydrogen bonds and nine salt bridges. In agreement with the structural findings, mutations at this site of FD [i.e., His133 (His146), Val203 (Val219), and Arg157 (Arg170)] abolish cleavage of FB (Fig. 3, B and C, and fig. S8) and render FD unable to amplify complement response (Fig. 3, D to F). Similarly, biotinylation of FD at residue Lys208 (Lys223) causes loss of activity (26), likely because biotin blocks the binding of FD to FB. In addition, Glu446 of FB, which plays an important role in binding the P1-Arg234 in the closed form, contacts His133 (His146) of FD in the C3bBD* complex, and mutation of this residue (i.e., FB E446A) strongly reduces convertase formation (fig. S9). This contact site in FD is formed by four loops: residues 132 to 135 (145 to 149), 155 to 159 (169 to 171), 173 to 176 (185 to 188), and 203 to 209 (220 to 224). In coagulation factor VIIa, loop 155 to 159 (169 to 171) is part of the zymogen activation domain (i.e., tissue-factor binding site). Loops 132 to 135 (145 to 149), 173 to 176 (185 to 188), and 203 to 209 (220 to 224) and the self-inhibitory loop of FD overlap with the Na+ ion–binding region in thrombin (34, 35). In FD, this exosite is critical for binding of the open form of FB in C3bB.
Fig. 3.
Analysis of FD exosite. (A) Surface diagram of FD highlighting its exosite in blue and its catalytic site in red (28). Exosite mutations are shown in yellow, whereas the biotinylation site associated with FD inactivation (26) is shown in green. (B) FD-mediated cleavage of C3bB pro-convertases monitored by SPR; FD mutants were injected onto surface-bound C3bB, and the cleavage activity was compared with that of wild type (wt). A drop in SPR response upon FD injection corresponds to the removal of the Ba fragment. Wild-type FD was injected at the end of each experiment to ensure cleavage sensitivity. (C) FD-mediated cleavage of C3bB performed by using SDS-PAGE with FD exosite mutants. The histogram shows the relative pro-convertase activation rates compared with those of wild-type FD. Error bars represent deviations from the mean observed in multiple experiments (n>3). (D) Effect of FD exosite mutations on the formation and decay of the C3bBb convertase monitored by SPR; wild-type FB was premixed with various FD mutants and injected over immobilized C3b. RU, resonance units. (E) Reconstitution of complement activity in FD-depleted plasma using FD mutants determined as C3b deposition on the enzyme-linked immunosorbent assay (ELISA) plate. O.D., optical density. (F) Hemolytic activity assays using FD-depleted plasma reconstituted with different mutants of FD: Lysis of rabbit erythrocytes was monitored by colorimetry and compared with 100% haemolysis in water.
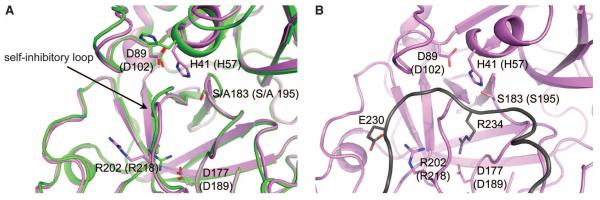
In the C3bBD* structure, the self-inhibitory loop of FD is rearranged, resulting in an active configuration of the catalytic triad in FD (Fig. 4A and fig. S10, A and B). In contrast, the structure of the unbound form of the corresponding FD mutant S183A (S195A), determined at 1.2 Å resolution (29), shows a self-inhibited state identical to the wild-type enzyme (22, 36) (fig. S10, A and B). We observe no induced structural changes at the exosite (fig. S10C). The C3bBD* structure and all eight available structures of free FD and mutants thereof (which occur in three different crystal forms) show well-defined and virtually identical conformations of the exosite loops. Thus, an induced activation, either structurally or dynamically, of FD through binding of FB at the exosite is not supported by the present data. Instead, the rearrangements in FD are restricted to the self-inhibitory loop [residues 196 to 202 (212 to 218)]. In the self-inhibited state, Arg202 (Arg218) of FD forms a salt bridge with Asp177 (Asp189) at the bottom of the S1 pocket, whereas the same residue points outward in C3bBD* (Fig. 4A). Mutation R202A (R218A) in FD increased the activity against peptides threefold (fig. S11A) because of rearrangements in the self-inhibitory loop that induce the active conformation of the catalytic triad (fig. S10A) (25). In contrast, the same R202A (R218A) mutation reduced cleavage of FB to ~20% and thus reduced the convertase formation rate (fig. S11, B to D), indicating a role for Arg202 (Arg218) in substrate specificity, as predicted earlier (23, 26, 36-38). However, published data showed complete inactivity for FD mutant R202Δ-V203G (R218Δ-V219G) toward both peptides and FB (37). We mimicked this by using a R202G (R218G) mutant of FD, which yielded almost no activity (fig. S11B).
Fig. 4.
Analysis of FD catalytic site. (A) Conformational changes observed in FD catalytic site in C3bBD* structure. Superposition of the structure of FD S183A (S195A) from C3bBD* (magenta) with wild-type free FD (green, PDB ID 1DSU) (22, 28) showing the displacement of the self-inhibitory loop and flipping of the side chain of His41 (His57) to the catalytic conformation. (B) Zoomed view of FD catalytic site with modeled FB scissile bond loop bound (dark gray). The model highlights the putative interaction between Glu230 of FB and Arg202 (Arg218) of FD and the P1 residue Arg234 making a salt bridge with Asp177 (Asp189).
Unexpectedly, we observed no electron density for the scissile loop of FB in C3bBD*. The apparent poor binding of this loop to FD is consistent with the observed low activity of FD toward short peptides even when the self-inhibitory loop is displaced (25). Starting from residues 223 and 240 that are visible in the electron density map, we could model the scissile loop of FB (224 to 239) in an extended fashion, placing residues 233 to 235 into the S2, S1, and S1’ binding pockets of FD. This modeling places the P5-residue Glu230 in the vicinity of the exposed Arg202 (Arg218) of FD (Fig.4B). Mutating Glu230 in FB (E230A) reduced the cleavage rate, indicating a putative interaction between Arg202 (Arg218) in FD and Glu230 in FB. (fig. S11B). Overall, we interpreted the specificity and activity of FD for cleaving FB in C3bB as primarily determined by its exosite-mediated binding to the open form of FB (with nanomolar affinity, fig. S3C), which allows the comparatively weaker interacting scissile loop of FB to induce activation of FD.
Lastly, cleavage of FB releases the propeptide segment Ba from the complex, inducing additional structural changes in the residual Bb fragment that remains bound to C3b (13). In this step, the helix αL of the Ba fragment is removed from its groove in the VWA domain, and helix α7 takes its place (fig. S7A). This results in a canonical arrangement of the VWA domain with the MIDAS-α7 presumably arranged in a high-affinity ligand binding configuration. Similar to the SP rearrangement in the pro-convertase, rearrangement of the α7 helix affects the VWA-SP linker, resulting in a rotation of ~140° for the SP domain (fig. S12). As derived from the crystal structure of the SCIN-inhibited convertase C3bBb (13), this arrangement allows cleavage of the substrate C3 thereby amplifying the complement response.
Our data reveal a highly concerted and specific activation mechanism based on cofactor-dependent and substrate-induced proteolysis, which provides an important “double-safety” catch to restrict complement amplification to C3b-tagged target cells. These detailed mechanistic insights into the central and transient, and therefore low-abundance, C3bBD complex of complement activation offer opportunities for the development of new therapeutics to fight complement-mediated diseases (39) such as age-related macular degeneration, atypical hemolytic uremic syndrome, membranoproliferative glomerulonephritis, and chronic inflammations that are associated with excessive or poorly controlled activation of complement.
Supplementary Material
Acknowledgments
Coordinates and structure factors of C3bB, C3bBD*, FD (S183A), and FD (R202A) have been deposited in the Protein Data Bank (PDB) with accession numbers 2XWJ, 2XWB, 2XW9, and 2XWA, respectively. We gratefully thank the European Synchrotron Radiation Facility (ESRF) and the Swiss Light source (SLS) for the provision of synchrotron radiation facilities and beamline scientists of the SLS, ESRF, and the European Molecular Biology Laboratory for assistance. This work was supported by a “Top” grant (700.54.304 to P.G.) by the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW) and NIH grants (AI030040, AI068730, AI072106, and GM062134 to J.D.L.).
Footnotes
Competing financial interests: P.G., F.F., D.R., and J.D.L. are co-inventors of a patent application titled “Structure of C3bB-factor D complex and use for rational drug design.”
References and Notes
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Nat. Immunol. 2010;11:785. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walport MJ. N. Engl. J. Med. 2001;344:1058. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 3.Pangburn MK, Müller-Eberhard HJ. Biochem. J. 1986;235:723. doi: 10.1042/bj2350723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gros P, Milder FJ, Janssen BJ. Nat. Rev. Immunol. 2008;8:48. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, Narayana SV, Volanakis JE. Immunol. Rev. 2001;180:123. doi: 10.1034/j.1600-065x.2001.1800111.x. [DOI] [PubMed] [Google Scholar]
- 6.Dunkelberger JR, Song WC. Cell Res. 2010;20:34. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 7.Law SK, Dodds AW. Protein Sci. 1997;6:263. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas PJ, van Strijp J. Immunol. Res. 2007;37:161. doi: 10.1007/BF02697367. [DOI] [PubMed] [Google Scholar]
- 9.Carroll MC. Mol. Immunol. 2004;41:141. doi: 10.1016/j.molimm.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Müller-Eberhard HJ. Annu. Rev. Immunol. 1986;4:503. doi: 10.1146/annurev.iy.04.040186.002443. [DOI] [PubMed] [Google Scholar]
- 11.van Lookeren Campagne M, Wiesmann C, Brown EJ. Cell. Microbiol. 2007;9:2095. doi: 10.1111/j.1462-5822.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- 12.Sjöberg AP, Trouw LA, Blom AM. Trends Immunol. 2009;30:83. doi: 10.1016/j.it.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Rooijakkers SH, et al. Nat. Immunol. 2009;10:721. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, et al. Nat. Immunol. 2009;10:728. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milder FJ, et al. Nat. Struct. Mol. Biol. 2007;14:224. doi: 10.1038/nsmb1210. [DOI] [PubMed] [Google Scholar]
- 16.Janssen BJ, et al. EMBO J. 2009;28:2469. doi: 10.1038/emboj.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torreira E, Tortajada A, Montes T, Rodríguez de Córdoba S, Llorca O. J. Immunol. 2009;183:7347. doi: 10.4049/jimmunol.0902310. [DOI] [PubMed] [Google Scholar]
- 18.Springer TA. Structure. 2006;14:1611. doi: 10.1016/j.str.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Nature. 2004;432:59. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya AA, Lupher ML, Jr., Staunton DE, Liddington RC. Structure. 2004;12:371. doi: 10.1016/j.str.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Hinshelwood J, Perkins SJ. J. Mol. Biol. 2000;298:135. doi: 10.1006/jmbi.2000.3632. [DOI] [PubMed] [Google Scholar]
- 22.Narayana SV, et al. J. Mol. Biol. 1994;235:695. doi: 10.1006/jmbi.1994.1021. [DOI] [PubMed] [Google Scholar]
- 23.Jing H, et al. J. Mol. Biol. 1998;282:1061. doi: 10.1006/jmbi.1998.2089. [DOI] [PubMed] [Google Scholar]
- 24.Kam CM, et al. J. Biol. Chem. 1987;262:3444. [PubMed] [Google Scholar]
- 25.Kim S, Narayana SV, Volanakis JE. J. Biol. Chem. 1995;270:24399. doi: 10.1074/jbc.270.41.24399. [DOI] [PubMed] [Google Scholar]
- 26.Taylor FR, et al. Biochemistry. 1999;38:2849. doi: 10.1021/bi982140f. [DOI] [PubMed] [Google Scholar]
- 27.Fishelson Z, Müller-Eberhard HJ. J. Immunol. 1982;129:2603. [PubMed] [Google Scholar]
- 28.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 29.Materials and methods are available as supporting material on Science Online.
- 30.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Nature. 2006;444:213. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 31.Wiesmann C, et al. Nature. 2006;444:217. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 32.Torreira E, Tortajada A, Montes T, Rodríguez de Córdoba S, Llorca O. Proc. Natl. Acad. Sci. U.S.A. 2009;106:882. doi: 10.1073/pnas.0810860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimaoka M, et al. Cell. 2003;112:99. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bock PE, Panizzi P, Verhamme IM. J. Thromb. Haemost. 2007;5(suppl. 1):81. doi: 10.1111/j.1538-7836.2007.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lechtenberg BC, Johnson DJ, Freund SM, Huntington JA. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14087. doi: 10.1073/pnas.1005255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S, Narayana SV, Volanakis JE. J. Immunol. 1995;154:6073. [PubMed] [Google Scholar]
- 37.Kim S, Narayana SV, Volanakis JE. Biochemistry. 1994;33:14393. doi: 10.1021/bi00252a004. [DOI] [PubMed] [Google Scholar]
- 38.Jing H, et al. EMBO J. 1999;18:804. doi: 10.1093/emboj/18.4.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ricklin D, Lambris JD. Nat. Biotechnol. 2007;25:1265. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.