Abstract
During genomic revolution, algorithmic and analytical methods for organizing, integrating, analyzing, and querying biological sequence data proved invaluable. Today, increasing availability of high-throughput data pertaining functional states of biomolecules, as well as their interactions, enables genome-scale studies of the cell from a systems perspective. The past decade witnessed significant efforts on the development of computational infrastructure for large-scale modeling and analysis of biological systems, commonly using network models. Such efforts lead to novel insights into the complexity of living systems, through development of sophisticated abstractions, algorithms, and analytical techniques that address a broad range of problems, including the following: (1) inference and reconstruction of complex cellular networks; (2) identification of common and coherent patterns in cellular networks, with a view to understanding the organizing principles and building blocks of cellular signaling, regulation, and metabolism; and (3) characterization of cellular mechanisms that underlie the differences between living systems, in terms of evolutionary diversity, development and differentiation, and complex phenotypes, including human disease. These problems pose significant algorithmic and analytical challenges because of the inherent complexity of the systems being studied; limitations of data in terms of availability, scope, and scale; intractability of resulting computational problems; and limitations of reference models for reliable statistical inference. This article provides a broad overview of existing algorithmic and analytical approaches to these problems, highlights key biological insights provided by these approaches, and outlines emerging opportunities and challenges in computational systems biology.
In post-genomic biology, the nature and scale of data that pertain to the structure, function, and organization of biomolecules present novel opportunities for exploratory research. Along with these opportunities, the large volume and high dimensionality of data pose significant challenges in terms of management, annotation, and integration of data, as well as transformation of data into biological knowledge through large-scale mining and analysis. As demonstrated by the large-scale application of sequence alignment tools, such as BLAST1 and CLUSTAL,2 computational models and algorithms prove extremely useful in the development of tools for exploring, manipulating, and interpreting large data sets. Furthermore, algorithmic and analytical approaches render the study of complex biological systems tractable, through development of sophisticated abstractions. 3This article presents a broad overview of computational algorithms and analytical techniques that provide useful insights into the complexity of biological systems.
GENOME-SCALE DATA ON BIOMOLECULES AND THEIR INTERACTIONS
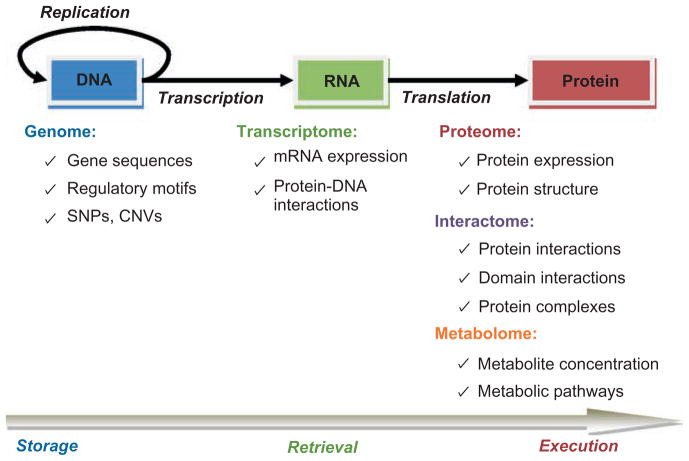
Recent technological advances in biological data collection and acquisition enable interrogation of biological systems at multiple levels, generating genome-scale data on the structure, abundance, activity, and interactions of biomolecules. These diverse data sources, often referred to as omic data, are illustrated in the context of the central dogma of molecular biology in Figure 1.
FIGURE 1.
Description of omic data sets in the context of central dogma.
Genome
At the level of DNA, genomic data uncover the information that is stored in the genomes of organisms and passed across generations. These include sequences of genes coding for functional proteins, regulatory motifs that serve as markers for the regulation of the expression of specific genes, as well and individual differences in the genetic composition of populations, such as single nucleotide polymorphisms (SNPs—common individual differences at a single nucleotide base4) and copy number variations (CNVs—multiplicity or lack of certain DNA segments in genomic sequences5). Genome sequencing is traditionally achieved through exploitation of the natural process of DNA replication. On the other hand, identification of SNPs6 and CNVs,7 as well as precise sequences of small genomes (e.g., human immunodeficiency virus) are often carried out using DNA microarrays, which exploit the natural process of hybridization. However, sequencing technology and associated computational techniques are being transformed by the emergence of short-read sequence data, also known as next-generation sequencing.130
Transcriptome
DNA microarrays are also commonly used to monitor the retrieval of genomic information under various conditions.8 More specifically, the relative amount of mRNA molecules that are present in a sample can be measured simultaneously for thousands of mRNA sequences (transcriptome), enabling comparison of the expression of thousands of genes in a given sample or across samples.9 Although the expression of a gene at the transcriptomic level serves as a proxy to the abundance of the corresponding protein in the sample, it does not necessarily capture the functional activity of the protein10 because protein expression is also regulated after transcription, through several mechanisms including mRNA degradation, alternative splicing, and post-translational modification.11
Proteome
Proteomic screening, on the other hand, captures molecular activity at the functional level.12 A common method, 2-D polyacrylamide gel electrophoresis enables separation of proteins in a given sample based on their electrochemical properties (e.g., isoelectric point or mass). Separated proteins can then be identified using mass spectrometry (MS).13 Although proteomic screening techniques are useful in quantifying the expression, as well as modification of proteins at the functional level, established proteomic screening techniques can only monitor the expression of a limited subset of proteins in the cell at a time. Furthermore, techniques such as flow cytometry allow screening of protein activity at the resolution of thousands of individual cells; however, this comes at the price of a very limited coverage of the proteome.14
Interactome
In addition to abundance at the level of single molecules, current high-throughput screening techniques enable identification of physical interactions between proteins. A common method, yeast two-hybrid (Y2H) screening identifies interactions between pairs of proteins by exploiting the modularity of the activating and binding domains of eukaryotic transcription factors.15 Namely, in Y2H, the activating and binding domains of a specific transcription factor are separated, and each domain is fused to one of the two (prey and bait) proteins. Subsequently, the interaction between the two proteins is captured by the expression of a reporter gene that is the target of the transcription factor. Tandem affinity purification (TAP), on the other hand, identifies interactions between a single bait protein and multiple other proteins.16 This is achieved by tagging the protein of interest and introducing it to the host. Once the bait protein is retrieved along with other proteins attached to it, these interacting partners are identified using MS.
Experimentally identified protein–protein interactions (PPIs) are organized into PPI networks, which provide a high-level and static description of cellular organization, commonly referred to as the interactome. Currently, established PPI network models assume binary interactions between pairs of proteins, which is naturally descriptive of the outcome of Y2H screening. On the other hand, multiple interactions identified by TAP are represented by either a star network around the bait protein (spoke model) or a clique of all proteins retrieved by the bait protein, including itself (matrix model).17 An important limitation of high-throughput PPIs, however, is their incomplete and noisy nature.15,18 Furthermore, these interactions only represent a snapshot of the dynamical organization of proteins in the cell.19 Currently available PPI data sets are also highly prone to ascertainment bias.
Metabolome
Metabolism, i.e., chains of chemical reactions that transform various forms of matter and energy into one another, is one of the fundamental processes in living systems. The organization of metabolic reactions is generally abstracted using metabolic network models, which represent the complex web of relationships between metabolites (compounds consumed and/or produced by reactions) and enzymes (gene products that catalyze reactions). Today, several well-characterized metabolic pathways for diverse species are available in public databases. However, large-scale analyses of the kinetics of metabolic networks are bound by data availability and computational complexity. Nevertheless, flux balance analyses that rely on steady-state assumption provide significant insights into the dynamics of metabolism.20 The logistic support for such analyses comes from monitoring of the abundance of metabolites via nuclear magnetic resonance and MS, as well as monitoring of the abundance and functional activity of enzymes through transcriptomic and proteomic screening.
NETWORK INFERENCE: GENETIC REGULATION AND CELLULAR SIGNALING
The cell adapts to its environment by recognition and transduction of a broad range of environmental signals, which in turn activate response mechanisms by regulating the expression of proteins that take part in the corresponding processes.21 Mechanisms of cellular signaling and genetic regulation also play key roles in cellular communication in multicellular organisms, including developmental processes. A fundamental challenge in systems biology is therefore to reconstruct networks that describe cellular signaling and regulation, with a view to deriving maps of interconnectivity and functional relationships between molecules.22 These maps are then used to derive chemically accurate representations of biochemical events within signaling networks, through detailed mathematical models that capture the dynamics of cellular systems.
Inference of protein interactions
Transduction of cellular signals is generally carried out through a complex web of interactions between proteins. Therefore, an important step toward reconstructing cellular networks involves identification of PPIs. As discussed in the previous section, although high-throughput screening techniques such as Y2H and TAP can identify physical interactions between many proteins, the interactions identified by these techniques are often incomplete and highly noisy.18 For this reason, many bioinformatics approaches also utilize other sources of molecular data for in silico identification of PPIs.23
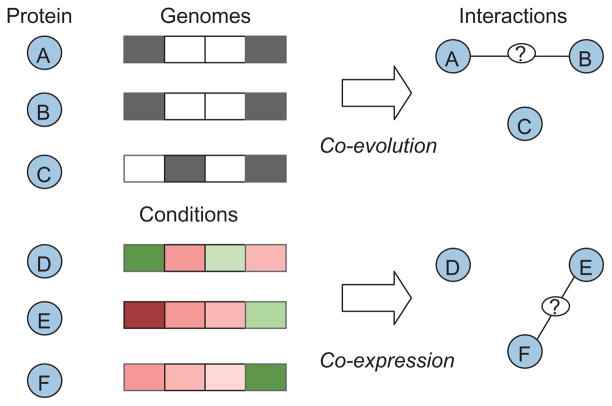
Common approaches to computational prediction of PPIs are illustrated in Figure 2. As seen in the figure, the recurring idea in computational prediction of PPIs is the premise that functionally associated proteins are likely to consistently appear together in different contexts. At the evolutionary level, such correlations are detected through assessment of co-evolution between proteins—that is, the degree of correlation between the conservation of two or more proteins in diverse species.24 For this purpose, a phylogenetic profile is constructed for each protein of interest, which is a vector of values indicating the presence of a homolog of the protein in a range of species, for which comprehensive genomic data are available. Subsequently, the correlation between phylogenetic profiles, often quantified in terms of their mutual information,25 is used to assess the likelihood of functional association between the corresponding proteins. This approach is further enhanced by embedding the phylogenetic vector in phylogenetic trees that represent evolutionary histories thereby capturing the underlying evolutionary relationships more accurately.26,27 Similarly, detection of evolutionary events such as gene fusion (i.e., two independent proteins in one organism are part of a single polypeptide chain in another organism) or conserved gene neighborhoods also provides a handle to the identification of interacting proteins.23,68
FIGURE 2.
Illustration of the general principles of common computational methods for predicting protein–protein interactions. In the upper panel, black and white boxes, respectively, indicate existence and absence of a homolog in the corresponding genome. In the lower panel, the red and green shades of boxes, respectively, indicate the degree of up- and down-regulation of the coding gene with respect to the corresponding condition.
At a finer timescale, correlation of gene expression profiles is often utilized to identify interacting proteins, based on the premise that interacting proteins are likely to be co-expressed under different conditions.29 It should be noted, however, that some interactions are permanent (e.g., protein complexes that are maintained through most conditions), whereas some are transient; transient interactions exhibit weaker relationship with correlation of gene expression.19 This observation suggests that, for accurate identification of functional relationships between proteins, it is important to consider local correlations in expression, i.e., those that manifest themselves only in a subset of various conditions.30 In general, the problem of identifying multiple genes with correlated expression in a subspace of the sample space is known as biclustering or co-clustering, and is studied extensively in the bioinformatics literature.31
Availability of a wide variety of experimental and computational methods for predicting interactions naturally calls for the integration of interactions identified by various methods. This can be achieved using statistical models that characterize the likelihood of predicted interactions based on gold standard interactions.32 Similarly, classification based approaches treat pairs of proteins as data items, predictions of various inference schemes as features, interactions as labels to be assigned to pairs of proteins, and known interactions as training data. They then learn models that specify the relationship between the outcome of various predictions and the existence of interactions between pairs of proteins.33 Large-scale integration of PPIs for model organisms demonstrates that the integrated network represents functional association between pairs of proteins better than any method alone,32 suggesting that different methods capture different aspects of functional association between proteins. Once interactions between proteins are characterized at genome scale, these interactions are mined in conjunction with other data sources to identify signaling pathways.34
Inference of domain interactions
Proteins are composed of multiple domains, which are often regarded as their primary structural and functional units. As similar domains can be utilized by different proteins that are involved in diverse processes, domains are often classified into domain families, based on their evolutionary, structural, and functional relationships.35 Consequently, identification of domain–domain interactions (DDIs) that mediate PPIs is likely to provide structural insights on the nature of interactions, leading to insights that can be transferred across different processes.36 Indeed, it is shown that, if phylogenetic profiles are constructed by taking into account the conservation of domains (as opposed to whole proteins), the performance of phylogenetic profiling in predicting interactions can be improved significantly.37 However, information on the domain decomposition of many proteins may not be readily available. Consequently, computational approaches incorporate domain information by constructing and clustering phylogenetic profiles at the residue level123 or identifying correlated mutations between residues through alignment of multiple protein sequences.39 A key advantage of these methods is that, they can identify novel domains and DDIs concurrently.
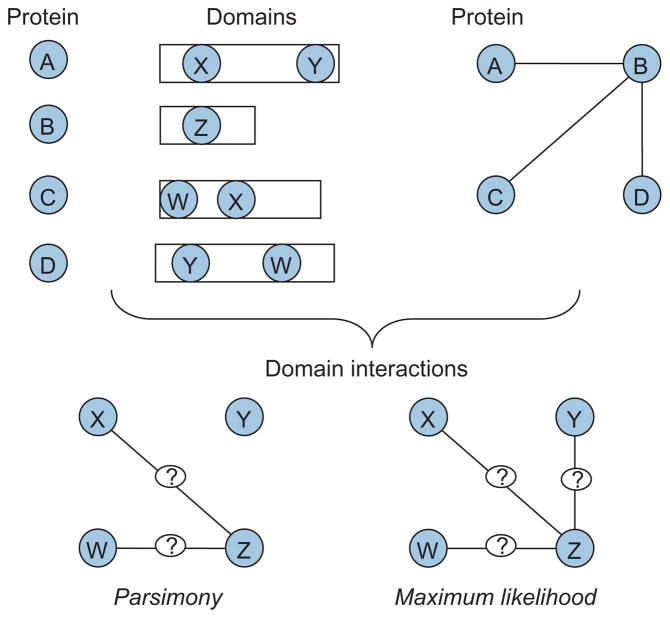
If the domain decomposition of a large number of proteins is available, then PPIs identified via high-throughput screening can also be used to infer DDIs.40 As illustrated in Figure 3, this is achieved by assuming that the observed PPIs are generated by a hidden model that specifies DDIs, and reconstructing the hidden model by optimizing an objective function derived from a particular assumption on the generating principles of the model (e.g., maximum likelihood41 or parsimony42). Furthermore, through consideration of experimental PPIs and phylogenetic profiles together, DDIs can be inferred more precisely.43 Recent studies show that organization of DDIs into networks is likely to provide useful information on the functional relationships between biomolecules.129
FIGURE 3.
Inferring domain–domain interactions (DDIs) from protein–protein interactions (PPIs). Given the domain decomposition of proteins and a set of PPIs, DDI inference methods target identification of DDIs that mediate these interactions. Different formulations of the problem optimize different criteria, leading to different solutions for DDI inference problem.
Inference of regulatory networks
At the transcriptional level, gene expression is regulated through interaction of transcription factor proteins with the DNA at specific locations. The combinatorial relationship between transcription factors and their target genes are organized into transcriptional regulatory networks, providing qualitative models of genetic regulation at the level of transcription.45 Although transcriptional networks can be reconstructed through identification of specific protein–DNA interactions,46 correlations between expression levels of genes also provide valuable information for the inference of regulatory interactions that extend beyond transcriptional regulation.28 The simplest model for gene regulatory networks is based on Boolean networks, where the expression of each gene in the network is represented by a binary variable and the regulatory effect of the genes that regulate a particular gene is represented as a Boolean function.47 Assuming that regulation is synchronized across well-defined time steps and quantizing gene expression properly into binary values,48 one can construct truth tables for each gene. By subsequent mining of these truth tables, the minimum set of regulators that can explain the variation in the expression of each gene can be identified, along with their effects.49
Boolean networks provide simple, yet useful models of causal relationships in the cell, and they can be surprisingly powerful in predicting cellular behavior in various contexts.50 However, they do not account for many important factors, including the quantitative and asynchronous nature of cellular signaling and regulation, as well as variables that are not measured. Bayesian networks utilize stochasticity to account for such factors that are otherwise intractable.51 They represent the expression of a gene as a random variable and characterize the relationship between a gene and its regulators in terms of the conditional probability distribution of this random variable with respect to the expression of regulators. Consequently, inference of the structure of regulatory networks is reduced to the problem of identifying Markov blankets for all genes in the network. Here, the Markov blanket of a random variable is defined as the minimum set of random variables that satisfy the following property: distribution of the variable of interest is independent of all other variables in the network, given the variables in this set, its Markov blanket.52 Identification of Markov blankets is a computationally difficult problem that is studied extensively, and there exists a wealth of publicly available software tools that can be used for this purpose. However, in general, existing tools do not scale to thousands of genes in terms of the computational resources they require. Once the structure of the network is inferred, the parameters that characterize the dependency between genes and their regulators can be identified using various model fitting algorithms.
Bayesian networks are also used successfully to identify signaling pathways for a small number of signaling proteins based on high-resolution proteomic data.14 An important limitation of Bayesian network models, however, is that they represent probabilistic dependencies rather than causal relationships. Dynamic Bayesian networks overcome these difficulties by laying out the network across time steps and conditioning a gene’s expression on the expression of its regulators in the previous time step.53 In a dual manner, probabilistic Boolean networks introduce stochasticity to Boolean networks by modeling the Boolean function that characterizes the expression of a gene as a random variable.54
UNDERSTANDING COMMONALITIES: CANONICALIZATION OF NETWORKS
One of the defining characteristics of complex systems is their modularity, which has important implications in their robustness and adaptability.55,56 Indeed, biological systems are observed to exhibit modularity at multiple levels, and evolutionary mechanisms such as gene duplication facilitate recurrent use of similar principles in different processes.57,58 Consequently, identification of common and coherent patterns in large-scale molecular networks is likely to provide insights into the richness of the design principles of cellular circuitry, which, through canonicalization of common patterns, has the potential to uncover the periodic table of systems biology.3
Identification of functional modules
Functional modules are generally defined as groups of proteins that perform a distinct biological function together. In PPI networks, functional modules manifest themselves as subnetworks with high connectivity, while being somewhat isolated from the rest of the network.59 Consequently, graph clustering algorithms are commonly utilized to modularize networks. An important challenge in functional module identification is the establishment of formal criteria for assessing the likelihood of a subnetwork to be a functional module. Subnetwork density, i.e., the fraction of observed interactions among all possible interactions between a given set of proteins, is often used for this purpose. Similarly, edge connectivity, i.e., the minimum number of edges that must be removed to break the subnetwork apart, is a useful measure in assessing the modularity of a subnetwork.60 A major problem associated with these modularity measures is that they are rather arbitrary, i.e., they are not directly associated with a quantitative model of underlying biological processes. However, assessment of the significance of these quantities with respect to a reference statistical model may provide a statistical basis to establish the potential biological relevance of a module.61 For example, with respect to a particular generating model for the network, one can develop an analytical framework to characterize the distribution of the size of the largest subnetwork with given density.62 Indeed, assessment of the modularity of a subnetwork with respect to such a distribution is shown to significantly improve the quality of identified modules compared to those identified via ad hoc measures, in terms of the functional coherence of identified modules.62
The functional coherence of a group of proteins is often assessed with respect to established standardized libraries of molecular function (e.g., Gene Ontology63). Using functions assigned to individual molecules in these libraries, the coherence of each module is assessed by the significance of the observed enrichment of a particular function in the module, based on hypergeometric models.64,65 Identification of functions that are significantly enriched in network modules is useful in functionally annotating the modules, as well as calibrating and comparing module identification algorithms.
From an algorithmic perspective, the problem of finding dense subnetworks in a network is computationally intractable in its most general setting. Therefore, most of the existing algorithmic approaches utilize heuristics that either greedily grow subnetworks starting from a seed protein66,67 or recursively partition the subnetwork until the resulting subnetworks are sufficiently dense.60 Furthermore, algebraic models that are based on random walks128 or spectral network decomposition69 are also effective in modularizing networks, through continuous relaxation of the problems. Because PPI networks are incomplete, noisy, and provide only a static description of cellular organization, module identification algorithms are also enriched via incorporation of knowledge on gene expression, to capture dynamic organization of modularity in biological systems.70
Network alignment
Availability of interaction data for multiple species is commonly utilized to identify network structures that are conserved throughout evolution.71 Such conserved subnetworks are likely to underlie modular processes that are essential to the respective taxa. The problem of local network alignment, i.e., identification of subnetworks with approximate matches in multiple networks (in terms of conservation of proteins, as well as interactions between them), leads to computationally challenging problems as the mapping of homolog proteins between different species (networks) is not one-to-one.72 Consequently, the resulting computational problem is a generalization of the intractable subgraph isomorphism problem.
While aligning pairs of networks, existing algorithms generally construct a Cartesian-product graph, in which each node represents a pair of homolog proteins, one from each species.73 Homology between two proteins is often quantified in terms of the sequence similarity between proteins. Because an important objective of network alignment is the use of network information to enhance identification of orthologs, these algorithms treat sequence homology in a flexible way. Subsequently, by assigning weights to the edges of the product graph based on the conservation of respective interactions, these algorithms reduce the problem into one of finding heavy subgraphs in the product graph. Here, assignment of conservation scores to interactions requires sound modeling of the constraints on the conservation and divergence of interactions. Existing approaches to tackling this challenge include assignment of likelihood scores to subgraphs based on Bayesian modeling of the existence and observation of interactions in modular and conserved subnetworks, which can be decomposed into edge weights.74 On the other hand, assignment of match (conserved interaction), mismatch (missing homologous interaction), and duplication (homologous proteins in the same network) scores based on network evolution models57,58 enables tuning and adjustment of alignment parameters based on empirically derived statistics on conservation of interactions75,76—a generalization of the framework employed by popular sequence alignment algorithms (e.g., BLAST) and aminoacid similarity matrices (e.g., PAM, BLOSUM). Existing network evolution models are quite powerful in capturing the basic structural properties of extant networks, including degree distribution, clustering coefficient distribution, and subgraph distribution.77–81
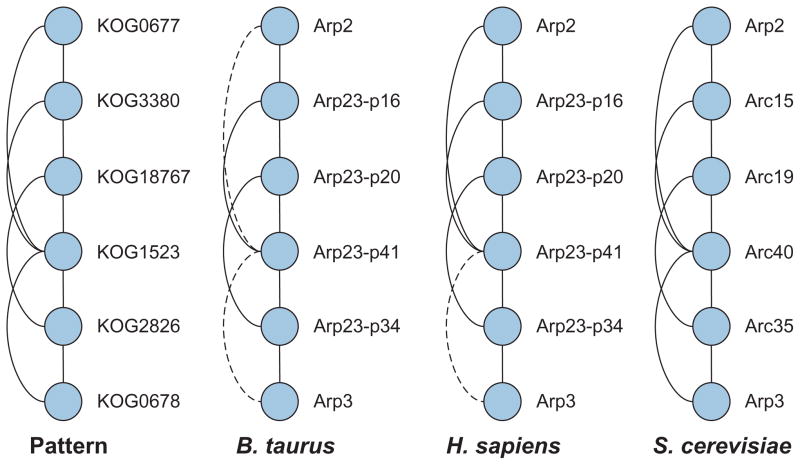
As the size of a product graph grows exponentially with the number of networks, pairwise network alignment algorithms do not scale well to large numbers of networks. This problem is alleviated through construction of a layered representation of multiple networks that represents groups of potentially orthologous proteins as subgraphs (as opposed to vertices).82 Furthermore, by summarizing PPI networks through contraction of nodes that correspond to ortholog proteins, and by using dedicated frequent subgraph mining algorithms on the resulting uniquely labeled graphs, the problem of finding exact subgraph matches in multiple networks is rendered tractable.83 A conserved subgraph identified by frequent subgraph mining on the PPI networks of nine eukaryotic species is shown in Figure 4. Finally, by formulating multiple graph alignment problem as one of assigning nodes into equivalence classes, the complexity of the problem is rendered linear in the number of networks.84 Local network alignment algorithms are also extended to the application of small subgraph match queries, and efficient algorithms are developed for searching for paths,85 trees and graphs with bounded tree width,86 and general subnetworks.87
FIGURE 4.
Arp 2/3 complex, which plays a significant role in the regulation of actin cytoskeleton, is identified as a conserved subnetwork through mining of protein–protein interaction networks of multiple organisms, using a fast algorithm that relies on contraction of ortholog proteins.126 The conserved subnetwork is shown on the left with nodes annotated by cluisters of ortholog groups (COG) identifiers.127 The occurrence of the subnetwork in three eukaryotic organisms is shown on the right. Dashed links indicate indirect interactions. Such knowledge discovery based analyses are likely to lead to the construction of canonical module libraries.
The idea of network alignment is also applied to global alignment of networks that belong to different species, with a view to enhancing identification of orthologous proteins in multiple species.88 Global network alignment algorithms aim to assign pairwise similarity scores to pairs of proteins from different networks to reflect the topological similarity of the proteins in the corresponding networks.89 The principle here is that two proteins can be considered topologically similar if their interacting partners are topologically similar; therefore, the problem can be formulated as a mutually reinforcing relation, which lends itself to iterative solution of an algebraic system.
Network motifs
An important feature of molecular networks is that certain subnetworks with coherent topological properties are significantly over-represented in these networks.90 Many of these common motifs are shared with other natural, social, and built networks, including electronic circuits, World Wide Web, and food webs, indicating that such topological structures may have essential functionality in the system. Indeed, algorithmic studies at multiple levels of complexity, from evolutionary timescale to cellular dynamics, demonstrate the key role of network motifs in biological systems: (1) proteins that are clustered into coherent motifs are likely to be conserved together;91 (2) network motifs provide signatures that are useful in comparison and classification of networks,92 also highlighting the basic mechanisms of network evolution;93 and (3) abundance of network motifs correlates significantly with their dynamic properties, in terms of stability and robustness.94
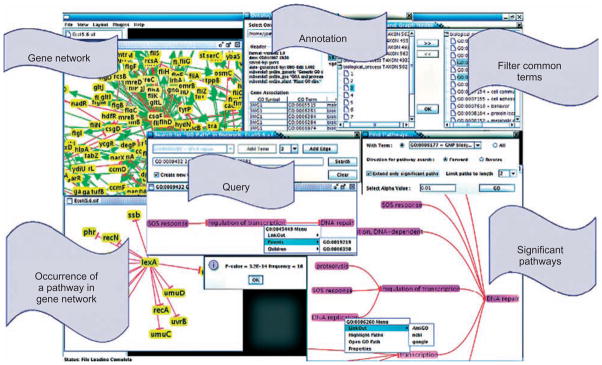
Functional annotation of the nodes of network motifs gives rise to functional network motifs, providing high-level descriptions of the crosstalk between different processes.44,95 One such software tool that enables exploration of canonical regulatory pathways in various organisms is demonstrated in Figure 5. These canonical representations of the wiring of cellular networks are useful in projecting information across different processes or species in many ways, including inference of molecular function96 and identification of novel pathways.97 It should be noted, however, that the identification of statistically significant network motifs poses significant computational challenges.44,98,99
FIGURE 5.
Screenshots from a sample computational tool, NARADA,44 that enables identification and browsing of canonical network patterns in regulatory networks. NARADA takes gene regulatory networks and functional annotation of individual genes as input and processes queries on regulatory pathways that involve specific biological processes (e.g., what are the processes that regulate ciliary or flagellar motility in E. coli? Are these regulatory pathways mediated by other processes?). NARADA is available as an open source at http://www.cs.purdue.edu/~jpandey/narada/. With the availability of such sophisticated tools, browsing basic biological information becomes a visually rich and interactive activity, moving beyond basic text and database searches.
Network based functional inference
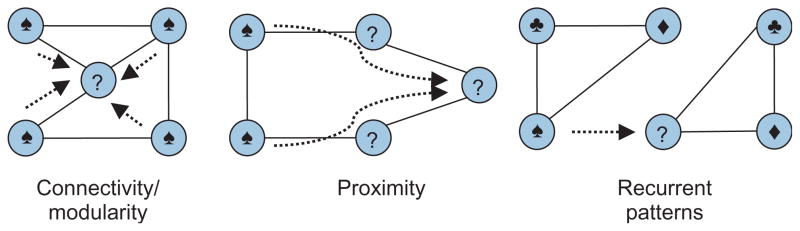
A common application of canonical network analysis is the prediction of molecular function based on partial knowledge of the functions of some of the proteins in the network.100 As illustrated in Figure 6, current computational approaches to this problem exploit three distinct observations on the functional coherence of molecular networks:
FIGURE 6.
Overview of common approaches to network based functional annotation. In each hypothetical example, the proteins with known function are annotated by a symbol that represents their function. Proteins with unknown function are labeled with question marks. As seen on the left, connectivity/modularity based schemes transfer function based on direct interactions. As seen in the middle, proximity based schemes diffuse function through the network. Finally, as seen on the right, pattern based schemes derive templates of functional interactions and interpolate these patterns accordingly to infer novel functions for proteins.
Connectivity: As functional modularity is closely related to network connectivity, proteins that are highly connected to each other in PPI networks are likely to be functionally associated.101
Proximity: Generalizing this observation further, multiple lines of evidence suggest that functional similarity of proteins correlates with their proximity in PPI networks.44,100
Recurrent patterns: As similar design principles of cellular signaling are used recurrently in various contexts, partial occurrences of recurrent patterns in networks can be interpolated to assign new functions to proteins involved in these patterns.38
A basic approach to connectivity based functional inference is to assign a function to a protein if the function is significantly enriched in its neighborhood.102 This approach is generalized to incorporate network topology by identifying network modules, annotating the modules based on functional enrichment of proteins in the module, and projecting this annotation to other proteins in the module.100 Further improvement on this approach is achieved through incorporation of proximity, by letting functions diffuse across the network.103,104
Pattern based functional inference algorithms consider the problem from a different angle. Rather than relying primarily on the interactions between functionally similar proteins, these approaches exploit the recurrence of interactions between different functional classes. Although this approach leads to challenging computational problems in that it requires identification of over-represented subnetworks, it captures information that cannot be captured by traditional functional annotation schemes and it enables propagation of knowledge across different organisms.38
UNDERSTANDING DIFFERENCES: NETWORKS AND PHENOTYPE
Recent advances in high-throughput molecular screening enable studies of phenotypic differences in terms of their signatures in cellular mechanisms. While genetic studies (gene association, haplotype mapping, etc.) are useful in discovering genetic differences that relate to certain phenotypes, differential analysis of molecular expression (gene expression, protein expression, metabolomics) helps to elucidate the variation in the activity of cellular systems. However, cellular systems are orchestrated through combinatorial organization of thousands of biomolecules.22 This complexity is reflected in the diversity of phenotypic effects, which generally present themselves as weak signals in terms of the changes in the expression of single molecules. For this reason, studies often focus on identification of multiple markers that together differentiate various phenotypes.
Integrating genomic data with network information
Interpretation of the findings of gene association studies in the context of molecular networks may highlight the cellular mechanisms that underlie various phenotypes, including human disease.105 Indeed, preliminary studies on the relationship between genes that are implicated in similar phenotypes indicate that these genes tend to interact with each other; they are likely to be expressed in similar tissues, and their mRNA expression profiles are often correlated.106 Capitalizing on these findings, several algorithmic approaches utilize network information to identify novel genetic markers.107 For example, if the linkage interval for a particular phenotype spans a large number of genes and some genes that are associated with similar phenotypes are known, then these two sources of information can be integrated within network context to rank the genes that are potentially associated with the phenotype.108
Molecular networks also provide a basis for interpreting genetic interactions.109 In the context of synthetic lethal interactions in yeast (i.e., pairs of genes such that the cell survives without any one of these genes, but dies if both are knocked out), two hypotheses are tested systematically to explain the network mechanisms behind genetic interactions: (1) within-pathway model assumes that many of the pairs of genes that are involved in a single pathway are synthetic lethal, indicating cooperation and (2) between-pathway model assumes that several pairs of genes, each from one of two ‘parallel’ pathways are synthetic lethal, indicating complementation. Through generalization of module identification algorithms, many clusters of genetic interactions that correspond to one of the two categories are identified, suggesting that both models can explain a certain fraction of observed genetic interactions.109
Integrating molecular expression data with network information
Molecular networks provide static and qualitative descriptions of the wiring of cellular systems. Molecular expression data, on the other hand, provides quantitative information on the functional states of constituent molecules under different conditions/samples, or over time. Consequently, it is natural to integrate these two sources of information to gain insights into alterations of the dynamic organization of cellular systems.70 Toward this end, preliminary studies particularly focus on the functional behavior of metabolic networks, as metabolism is one of the relatively well-characterized processes in biological systems.20 Systematic studies of the behavior of gene expression with respect to metabolic networks of model organisms indicate that divergent reactions (in which the product of an enzyme is consumed by two different enzymes) often act as switches, where the expression of the upstream enzyme is correlated with only one of the downstream enzymes.110 Furthermore, metabolic networks can be dissected into tissue-specific pathways based on coupled analysis of flux models and tissue-specific gene expression within an optimization framework that maximizes the inconsistency between the flux through reactions and the expression of the genes coding for enzymes that catalyze corresponding reactions.111 Similarly, in the context of transcriptional regulation, the transcriptional network is dissected into subnetworks based on the expression of transcription factors in various processes, revealing that the transcription networks that correspond to different processes exhibit different topological properties.112 Recently, tissue-specificity of protein interaction networks is also explored, demonstrating that tissue-specific proteins make only a few interactions, whereas proteins that are universally expressed actually have many tissue-specific interactions.113
Identification of implicated subnetworks
In the context of a particular phenotype or perturbation, identification of implicated (or, more specifically, differentially expressed or dysregulated) subnetworks enables discovery of multiple phenotype markers, i.e., multiple genes that are linked to each other in the network and are differentially expressed in samples that belong to different phenotypes, when considered together. In one of the early algorithmic studies, Ideker et al.114 propose a method for identifying differentially expressed subnetworks with respect to GAL80 deletion in yeast. This method is based on quantifying the differential expression of each gene individually and subsequently searching for connected subgraphs with high aggregate significance of differential expression. Variations of this method are shown to be effective in identifying multiple gene markers in various other diseases, including melanoma,115 diabetes,116 and others. A similar approach is to binarize differential expression of genes in the network and formulate dysregulated pathway identification as a problem of finding maximal subnetworks with a limited number of genes that are not differentially expressed.117
Although such network based approaches are useful in retrieving weak signals by considering interactions between multiple genes, they do not capture the sample-specific variation in the expression of different genes.118 Recent approaches assess the coordinate differential expression of multiple genes by aggregating the expression of a group of genes for each sample and then quantifying the mutual information between this aggregate expression profile and phenotype.119 It has been shown that classifiers that are trained by subnetwork markers identified via such integrative approaches are more successful in predicting breast cancer metastasis, as compared to those that incorporate single-gene markers.119 Anastassiou118 further elaborates these information-theoretic measures to capture the synergy of the dysregulation of a group of proteins, i.e., the overall differential expression of the subnetwork that is not explained by the differential expression of individual genes in the subnetwork. This approach leads to construction of phenotype-specific synergy networks that provide global insights into the coordination of multiple genes in the manifestation of a particular phenotype.120 It should be noted, however, that the task of identifying multiple genes with synergistic differential expression leads to intractable computational problems.118 As synergy by definition does not exhibit monotonicity properties, these problems may not be adequately addressed by greedy approximations.
Incorporation of proteomic data
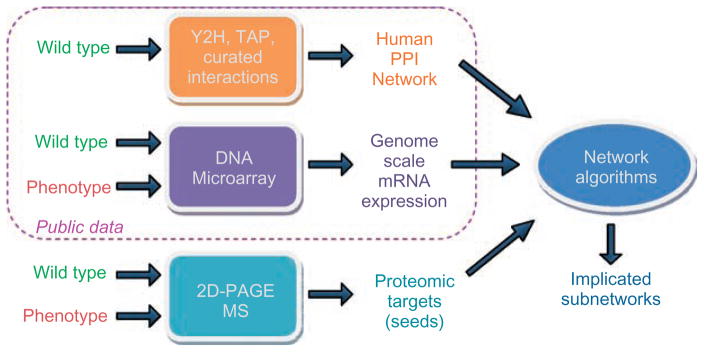
mRNA expression provides genome-scale information on gene expression, whereas proteomic screening provides information with relatively less coverage at the functional level. Knowledge of molecular interactions is useful in integrating these two complementary data sources to identify multiple phenotype markers. In a recent study on human colorectal cancer (CRC), Nibbe et al.122 propose a general framework for the integration of omic data sets, which is illustrated in Figure 7. In this framework, they first identify proteomic targets with significant fold change in late stages of CRC and map these proteins on a network of PPIs to extract candidate subnetworks that are significantly associated with these proteomic seeds. Subsequently, using genome-scale mRNA expression data, they quantify the synergistic differential expression of these candidate subnetworks, extracting known as well as novel markers for CRC. The basic premise here is that small changes in the expression of multiple gene products in the neighborhood of a protein may be synergistically associated with significant changes in the functional activity of the corresponding protein. Indeed, systematic analyses in the context of CRC show that differential mRNA expression is not necessarily correlated with proximity to proteomic targets on a single gene basis, but it is significantly correlated with the synergistic differential expression when multiple genes are considered.
FIGURE 7.
Framework for the integration of omic data for the discovery of subnetworks implicated in complex phenotypes. Proteomic screening provides functional data for a limited set of proteins, transcriptomic screening provides genome-scale data on mRNA expression, and curated or high-throughput protein–protein interactions provide a framework for the integration of these two complementary, valuable sources of data. This framework also illustrates how researchers can couple specific data sets generated in their labs with public data to broaden the scope of their analyses.
CONCLUSIONS AND OUTLOOK
Scientific and technological developments in the genomic era establish systems biology as a fundamental interdisciplinary science that marks the transformation of scientific research into a synergistic effort across multiple disciplines. To this end, developments in biotechnology are coupled with parallel developments in information technology. Besides their contribution to the development of tools for handling data, computational approaches play a key role in systems biology through development of sophisticated abstractions, computational models, and algorithms. In the past decade, such approaches utilized novel data sources in innovative ways to characterize the basic working principles of cellular systems, identify common patterns in cellular organization, effectively transfer knowledge across platforms, and identify and interpret the markers, signatures, and mechanisms that underlie differences among living systems. A summary of publicly available tools for the problems discussed in this article is given in Table 1. As more data become available and algorithmic approaches mature, high-level, large-scale analyses will generate a knowledge base, which, in turn, will enrich low-level, detailed models of biological systems, making it possible to precisely characterize the dynamics of cellular processes at larger scales. To this end, models and algorithms that facilitate effective integration of multiple data sources are likely to dominate the next generation of algorithms for systems biology.
TABLE 1.
A Summary of Common Problems in Network Biology and Corresponding Computational Tools Available in the Public Domain
| Problem | Tools |
|---|---|
| Prediction of protein/domain interactions | In silico two-hybrid,39 MORPH,26 Coevolutionary-Matrix,123 GPE,42 MLE,124 DPEA41 |
| Inference of gene regulatory networks | REVEAL,47 BN/PBN,49 DBN53 |
| Identification of functional modules and signaling pathways | SiDeS,62 MCODE8, HCS,60 SEEDY,67 PathFinder,34 MATISSE125 |
| Identification of functional subnetworks, annotation of network modules, network based inference of protein function | Narada,44 NetGrep,10 Ontologizer,64 VAMPIRE,65 GAIN,104 PROTAN37 |
| Network alignment | MaWISh,76 PathBLAST,73 NetworkBLAST,74 Graemlin,75 MULE,68 IsoRank88 |
| Querying protein–protein interaction networks for subgraph matches | QPath,56 QNet,86 TORQUE87 |
| Identification of subnetworks implicated in a particular phenotype | Chuang et al.,99 GNEA,116 DEGAS117 |
Although integrative approaches to network analysis have already been commonly applied, many data classes are not yet fully utilized. In particular, mRNA expression data are still used as the principal indicator of molecular expression and activity, although techniques that enable monitoring of protein expression, protein states and modification, and metabolic activity are available. Incorporation of such sources of information will enable modeling of cellular activity more accurately; however, such data sets often do not provide information at the genome scale. Network data provide the ideal substrate for the use of such data sets to extract information beyond their scale. In the near future, with the availability of more comprehensive and reliable interaction data, proteomic and metabolomic data are likely to be more commonly utilized in large-scale analysis of cellular systems. Furthermore, availability of information at multiple levels of cellular organization will also facilitate annotation of high-throughput interactions, providing more detailed models of cellular networks.
Acknowledgments
This work is supported in part by an endowment from Theodore and Dana Schroeder Assistant Professorship in Computer Science and Engineering. This work was also supported, in part, by the National Institutes of Health Grant, UL1-RR024989 Supplement, from the National Center for Research Resources (Clinical and Translational Science Awards). The author would also like to thank Rod Nibbe (CWRU), Mark Chance (CWRU), Shankar Subramaniam (UCSD), and Ananth Grama (Purdue) for many useful discussions.
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 3.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 4.Wang DG, Fan JB, Siao CJ, Berno A, Young P, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 5.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 6.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 7.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;(suppl 7):39. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulze A, Downward J. Navigating gene expression using microarrays-a technology review. Nat Cell Biol. 2001;3:E190–E195. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- 9.Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet. 2002;32(suppl):502–508. doi: 10.1038/ng1033. [DOI] [PubMed] [Google Scholar]
- 10.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4(9):117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mata J, Marguerat S, Bähler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem Sci. 2005;30:506–514. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson LP, Smith RD. Proteome analysis by mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:399–424. doi: 10.1146/annurev.biophys.32.110601.141854. [DOI] [PubMed] [Google Scholar]
- 14.Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 15.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 17.Scholtens D, Vidal M, Gentleman R. Local modeling of global interactome networks. Bioinformatics. 2005;21:3548–3557. doi: 10.1093/bioinformatics/bti567. [DOI] [PubMed] [Google Scholar]
- 18.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 19.Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papin JA, Price ND, Wiback SJ, Fell DA, Palsson BO. Metabolic pathways in the post-genome era. Trends Biochem Sci. 2003;28:250–258. doi: 10.1016/S0968-0004(03)00064-1. [DOI] [PubMed] [Google Scholar]
- 21.Beckerman M. Molecular and Cellular Signaling. New York: Springer; 2005. [Google Scholar]
- 22.Papin JA, Hunter T, Palsson BO, Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 23.Valencia A, Pazos F. Computational methods for the prediction of protein interactions. Curr Opin Struct Biol. 2002;12:368–373. doi: 10.1016/s0959-440x(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 24.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci U S A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Kasif S, DeLisi C. Identification of functional links between genes using phylogenetic profiles. Bioinformatics. 2003;19:1524–1530. doi: 10.1093/bioinformatics/btg187. [DOI] [PubMed] [Google Scholar]
- 26.Jothi R, Kann MG, Przytycka TM. Predicting protein-protein interaction by searching evolutionary tree automorphism space. Bioinformatics. 2005;21(suppl 1):241–250. doi: 10.1093/bioinformatics/bti1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 2001;14:609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- 28.D’Haeseleer P, Liang S, Somogyi R. Genetic network inference: from co-expression clustering to reverse engineering. Bioinformatics. 2000;16:707–726. doi: 10.1093/bioinformatics/16.8.707. [DOI] [PubMed] [Google Scholar]
- 29.Stuart JM, Segal E, Koller D, Kim SK. A Gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y, Church GM. Biclustering of expression data. Proc Int Conf Intell Syst Mol Biol. 2000;8:93–103. [PubMed] [Google Scholar]
- 31.Madeira SC, Oliveira AL. Biclustering algorithms for biological data analysis: a survey. IEEE/ACM Trans Comput Biol Bioinform. 2004;1:24–45. doi: 10.1109/TCBB.2004.2. [DOI] [PubMed] [Google Scholar]
- 32.Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 33.Srinivasan BSS, Shah NHH, Flannick JAA, Abeliuk E, Novak AFF, et al. Current progress in network research: toward reference networks for key model organisms. Brief Bioinform. 2007;8(5):318–332. doi: 10.1093/bib/bbm038. [DOI] [PubMed] [Google Scholar]
- 34.Bebek G, Yang J. PathFinder: mining signal transduction pathway segments from protein-protein interaction networks. BMC Bioinform. 2007;8:335. doi: 10.1186/1471-2105-8-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aloy P, Russell RB. Structural systems biology: modelling protein interactions. Nat Rev Mol Cell Biol. 2006;7:188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- 37.Kim Y, Subramaniam S. Locally defined protein phylogenetic profiles reveal previously missed protein interactions and functional relationships. Proteins. 2006;62:1115–1124. doi: 10.1002/prot.20830. [DOI] [PubMed] [Google Scholar]
- 38.Kiraç M, Özsoyoğlu G. Protein Function Prediction Based on Patterns in Biological Networks. Res Comput Mol Biol. 2008:197–213. [Google Scholar]
- 39.Pazos F, Valencia A. In silico two-hybrid system for the selection of physically interacting protein pairs. Proteins Struct Function Genet. 2002;47:219–227. doi: 10.1002/prot.10074. [DOI] [PubMed] [Google Scholar]
- 40.Lee H, Deng M, Sun F, Chen T. An integrated approach to the prediction of domain-domain interactions. BMC Bioinform. 2006;12:7. doi: 10.1186/1471-2105-7-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riley R, Lee C, Sabatti C, Eisenberg D. Inferring protein domain interactions from databases of interacting proteins. Genome Biol. 2005;6:R89. doi: 10.1186/gb-2005-6-10-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guimaraes KS, Przytycka TM. Interrogating domain-domain interactions with parsimony based approaches. BMC Bioinform. 2008;9:171. doi: 10.1186/1471-2105-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jothi R, Cherukuri PF, Tasneem A, Przytycka TM. Co-evolutionary analysis of domains in interacting proteins reveals insights into domain-domain Interactions mediating protein-protein interactions. J Mol Biol. 2006;362:861–875. doi: 10.1016/j.jmb.2006.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandey J, Koyutürk M, Kim Y, Szpankowski W, Subramaniam S, et al. Functional annotation of regulatory pathways. Bioinformatics. 2007;23:i377–i386. doi: 10.1093/bioinformatics/btm203. [DOI] [PubMed] [Google Scholar]
- 45.Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman & Hall/CRC; 2006. [Google Scholar]
- 46.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 47.Akutsu T, Miyano S, Kuhara S. Identification of genetic networks from a small number of gene expression patterns under the Boolean network model. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing; Maui, HI. 1999. pp. 17–28. [DOI] [PubMed] [Google Scholar]
- 48.Shmulevich I, Zhang W. Binary analysis and optimization-based normalization of gene expression data. Bioinformatics. 2002;18:555–565. doi: 10.1093/bioinformatics/18.4.555. [DOI] [PubMed] [Google Scholar]
- 49.Lähdesmäki H, Shmulevich I, Yli-Harja O. On Learning Gene Regulatory Networks Under the Boolean Network Model. Machine Learning. 2003;52:147–167. [Google Scholar]
- 50.Genoud T, Métraux JP. Crosstalk in plant cell signaling: structure and function of the genetic network. Trends Plant Sci. 1999;4:503–507. doi: 10.1016/s1360-1385(99)01498-3. [DOI] [PubMed] [Google Scholar]
- 51.Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- 52.Friedman N, Linial M, Nachman I, Pe’er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- 53.Kim SY, Imoto S, Miyano S. Inferring gene networks from time series microarray data using dynamic Bayesian networks. Brief Bioinform. 2003;4:228–235. doi: 10.1093/bib/4.3.228. [DOI] [PubMed] [Google Scholar]
- 54.Shmulevich I, Dougherty ER, Kim S, Zhang W. Probabilistic Boolean Networks: a rule-based uncertainty model for gene regulatory networks. Bioinformatics. 2002;18:261–274. doi: 10.1093/bioinformatics/18.2.261. [DOI] [PubMed] [Google Scholar]
- 55.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(suppl 6761):C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 56.Przulj N, Wigle DA, Jurisica I. Functional topology in a network of protein interactions. Bioinformatics. 2004;20:340–348. doi: 10.1093/bioinformatics/btg415. [DOI] [PubMed] [Google Scholar]
- 57.Bebek G, Berenbrink P, Cooper C, Friedetzky T, Nadeau J, et al. The degree distribution of the generalized duplication model. Theor Computer Sci. 2006;369:239–249. [Google Scholar]
- 58.Pastor-Satorras R, Smith E, Solé RV. Evolving protein interaction networks through gene duplication. J Theor Biol. 2003;222:199–210. doi: 10.1016/s0022-5193(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 59.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hartuv E, Shamir R. A clustering algorithm based on graph connectivity. Inform Processing Lett. 2000;76:175–181. [Google Scholar]
- 61.Itzkovitz S, Milo R, Kashtan N, Ziv G, Alon U. Subgraphs in random networks. Phys Rev E. 2003;68:026127. doi: 10.1103/PhysRevE.68.026127. [DOI] [PubMed] [Google Scholar]
- 62.Koyutürk M, Szpankowski W, Grama A. Assessing significance of connectivity and conservation in protein interaction networks. J Comput Biol. 2007;14:747–764. doi: 10.1089/cmb.2007.R014. [DOI] [PubMed] [Google Scholar]
- 63.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grossmann S, Bauer S, Robinson PNN, Vingron M. Improved detection of overrepresentation of gene-ontology annotations with parent-child analysis. Bioinformatics. 2007;24:1650–1651. doi: 10.1093/bioinformatics/btm440. [DOI] [PubMed] [Google Scholar]
- 65.Hsiao A, Ideker T, Olefsky JM, Subramaniam S. VAMPIRE microarray suite: a web-based platform for the interpretation of gene expression data. Nucleic Acids Res. 2005;33:W627–W632. doi: 10.1093/nar/gki443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bader JS. Greedily building protein networks with confidence. Bioinformatics. 2003;19:1869–1874. doi: 10.1093/bioinformatics/btg358. [DOI] [PubMed] [Google Scholar]
- 68.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 69.Bu D, Zhao Y, Cai L, Xue H, Zhu X, et al. Topological structure analysis of the protein-protein interaction network in budding yeast. Nucl Acids Res. 2003;31:2443–2450. doi: 10.1093/nar/gkg340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanisch D, Zien A, Zimmer R, Lengauer T. Co-clustering of biological networks and gene expression data. Bioinformatics. 2002;18(suppl 1):S145–S154. doi: 10.1093/bioinformatics/18.suppl_1.s145. [DOI] [PubMed] [Google Scholar]
- 71.Sharan R, Ideker T. Modeling cellular machinery through biological network comparison. Nat Biotechnol. 2006;24:427–433. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- 72.Koyutürk M. Comparative Analysis of Biological Networks. New York: VDM Verlag Dr. Müller; 2009. [Google Scholar]
- 73.Kelley BP, Sharan R, Karp RM, Sittler T, Root DE, et al. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc Natl Acad Sci U S A. 2003;100:11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharan R, Ideker T, Kelley B, Shamir R, Karp RM. Identification of protein complexes by comparative analysis of yeast and bacterial protein interaction data. J Comput Biol. 2005;12:835–846. doi: 10.1089/cmb.2005.12.835. [DOI] [PubMed] [Google Scholar]
- 75.Flannick J, Novak A, Srinivasan BS, McAdams HH, Batzoglou S. Graemlin: general and robust alignment of multiple large interaction networks. Genome Res. 2006;16:1169–1181. doi: 10.1101/gr.5235706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koyutürk M, Kim Y, Topkara U, Subramaniam S, Szpankowski W, et al. Pairwise alignment of protein interaction networks. J Comput Biol. 2006;13:182–199. doi: 10.1089/cmb.2006.13.182. [DOI] [PubMed] [Google Scholar]
- 77.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 78.Chung F, Lu L, Dewey TG, Galas DJ. Duplication models for biological networks. J Comput Biol. 2003;10:677–687. doi: 10.1089/106652703322539024. [DOI] [PubMed] [Google Scholar]
- 79.Çolak R, Hormozdiari F, Moser F, Schonhuth A, Holman J, Ester M, Sahinalp C. Dense graphlet statistics of protein interaction networks and random networks. Pacific Symposium on Biocomputing. 2009 doi: 10.1142/9789812836939_0018. [DOI] [PubMed] [Google Scholar]
- 80.Hormozdiari F, Berenbrink P, Przulj N, Sahinalp SC. Not all scale-free networks are born equal: The role of the seed graph in PPI network evolution. PLoS Comput Biol. 2007;3(7):e118. doi: 10.1371/journal.pcbi.0030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Przulj N, Corneil DG, Jurisica I. Modeling interactome: scale-free or geometric? Bioinformatics. 2005;20:3508. doi: 10.1093/bioinformatics/bth436. [DOI] [PubMed] [Google Scholar]
- 82.Kalaev M, Bafna V, Sharan R. Fast and accurate alignment of multiple protein networks. Res Comput Mol Biol. 2008;12:246–256. doi: 10.1089/cmb.2009.0136. [DOI] [PubMed] [Google Scholar]
- 83.Koyutürk M, Grama A, Szpankowski W. An efficient algorithm for detecting frequent subgraphs in biological networks. Bioinformatics. 2004;20:200–207. doi: 10.1093/bioinformatics/bth919. [DOI] [PubMed] [Google Scholar]
- 84.Flannick J, Novak A, Do JB, Srinivasan BS, Batzoglou S. Automatic parameter learning for multiple network alignment. Res Comput Mol Biol. 2008;4955:214–231. doi: 10.1089/cmb.2009.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shlomi T, Segal D, Ruppin E, Sharan R. QPath: a method for querying pathways in a protein-protein interaction network. BMC Bioinform. 2006;7:199. doi: 10.1186/1471-2105-7-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dost B, Shlomi T, Gupta N, Ruppin E, Bafna V, et al. QNet: a tool for querying protein interaction networks. J Comput Biol. 2008;15:913–925. doi: 10.1089/cmb.2007.0172. [DOI] [PubMed] [Google Scholar]
- 87.Bruckner S, Hüffner F, Karp RM, Shamir R, Sharan R. Topology-free querying of protein interaction networks. Res Comput Mol Biol. 2009;13:74–89. doi: 10.1089/cmb.2009.0170. [DOI] [PubMed] [Google Scholar]
- 88.Singh R, Xu J, Berger B. Global alignment of multiple protein interaction networks with application to functional orthology detection. Proc Nat Acad Sci. 2008;105:12763–12768. doi: 10.1073/pnas.0806627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh R, Xu J, Berger B. Pairwise global alignment of protein interaction networks by matching neighborhood topology. Res Comput Mol Biol. 2007;11:16–31. [Google Scholar]
- 90.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 91.Wuchty S, Oltvai ZN, Barabási AL. Evolutionary conservation of motif constituents in the yeast protein interaction network. Nat Genet. 2003;35:176–179. doi: 10.1038/ng1242. [DOI] [PubMed] [Google Scholar]
- 92.Natasa Przulj. Biological network comparison using graphlet degree distribution. Bioinformatics. 2007;23:e177–e183. doi: 10.1093/bioinformatics/btl301. [DOI] [PubMed] [Google Scholar]
- 93.Middendorf M, Ziv E, Wiggins CH. Inferring network mechanisms: the Drosophila melanogaster protein interaction network. Proc Natl Acad Sci. 2005;102:3192–3197. doi: 10.1073/pnas.0409515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Prill RJJ, Iglesias PAA, Levchenko A. Dynamic properties of network motifs contribute to biological network organization. PLoS Biol. 2005;3(11):e343. doi: 10.1371/journal.pbio.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Banks E, Nabieva E, Peterson R, Singh M. Net-Grep: fast network schema searches in interactomes. Genome Biol. 2008:9. doi: 10.1186/gb-2008-9-9-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kiraç M, Özsoyoglu G, Yang J. Annotating proteins by mining protein interaction networks. Bioinformatics. 2006;22:e260–e270. doi: 10.1093/bioinformatics/btl221. [DOI] [PubMed] [Google Scholar]
- 97.Çakmak A, Özsoyodlu G. Mining biological networks for unknown pathways. Bioinformatics. 2007;23:2775–2783. doi: 10.1093/bioinformatics/btm409. [DOI] [PubMed] [Google Scholar]
- 98.Alon N, Dao P, Hajirasouliha I, Hormozdiari F, Sahinalp CS. Biomolecular network motif counting and discovery by color coding. Bioinformatics. 2008;24:i241–i249. doi: 10.1093/bioinformatics/btn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Çakmak A, Özsoyoglu G. Taxonomy-superimposed graph mining. ACM Int Conf Extend Database Technol. 2008;261:217–228. [Google Scholar]
- 100.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vázquez A, Flammini A, Maritan A, Vespignani A. Global protein function prediction from protein-protein interaction networks. Nat Biotechnol. 2003;21:697–700. doi: 10.1038/nbt825. [DOI] [PubMed] [Google Scholar]
- 102.Samanta MP, Liang S. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc Natl Acad Sci U S A. 2003;100:12579–12583. doi: 10.1073/pnas.2132527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deng M, Zhang K, Mehta S, Chen T, Sun F. Prediction of protein function using protein-protein interaction data. J Comput Biol. 2003;10:947–960. doi: 10.1089/106652703322756168. [DOI] [PubMed] [Google Scholar]
- 104.Karaoz U, Murali TM, Letovsky S, Zheng Y, Ding C, et al. Whole-genome annotation by using evidence integration in functional-linkage networks. Proc Natl Acad Sci U S A. 2004;101:2888–2893. doi: 10.1073/pnas.0307326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, et al. The human disease network. Proc Natl Acad Sci. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oti M, Snel B, Huynen MA, Brunner HG. Predicting disease genes using protein-protein interactions. J Med Genet. 2006;43:691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lage K, Karlberg OE, Størling ZM, Olason PI, Pedersen AG, Rigina O, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 109.Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ihmels J, Levy R, Barkai N. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat Biotechnol. 2004;22:86–92. doi: 10.1038/nbt918. [DOI] [PubMed] [Google Scholar]
- 111.Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 112.Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 113.Bossi A, Lehner B. Tissue specificity and the human protein interaction network. Mol Syst Biol. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;(supp 1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 115.Nacu S, Critchley-Thorne R, Lee P, et al. Gene expression network analysis and applications to immunology. Bioinformatics. 2007;23:850–858. doi: 10.1093/bioinformatics/btm019. [DOI] [PubMed] [Google Scholar]
- 116.Liu M, Liberzon A, Kong SW, Lai WR, Park PJ, et al. Network-based analysis of affected biological processes in type 2 diabetes models. PLoS Genet. 2007;3:e96. doi: 10.1371/journal.pgen.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ulitsky I, Karp RM, Shamir R. Detecting disease-specific dysregulated pathways via analysis of clinical expression profiles. Res Comput Mol Biol. 2008;12:347–359. [Google Scholar]
- 118.Anastassiou D. Computational analysis of the synergy among multiple interacting genes. Mol Syst Biol. 2007;3:83. doi: 10.1038/msb4100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Watkinson J, Wang X, Zheng T, Anastassiou D. Identification of gene interactions associated with disease from gene expression data using synergy networks. BMC Syst Biol. 2008;2:10. doi: 10.1186/1752-0509-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nibbe RK, Chance M, Koyutürk M. Integrating proteomic, transcriptomic, and interactomic data to identify subnetworks implicated in human colorectal cancer. PLoS Comput Biol. Submitted. [Google Scholar]
- 122.Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance M. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol Cell Prot. 2009 doi: 10.1074/mcp.M800428-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim Y, Koyutürk M, Topkara U, Grama A, Subramaniam S. Inferring functional information from domain co-evolution. Bioinformatics. 2006;22:40–49. doi: 10.1093/bioinformatics/bti723. [DOI] [PubMed] [Google Scholar]
- 124.Deng M, Mehta S, Sun F, Chen T. Inferring domain-domain interactions from protein-protein interactions. Genome Res. 2002;12:1540–1548. doi: 10.1101/gr.153002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst Biol. 2007;1:8. doi: 10.1186/1752-0509-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Koyutürk M, Kim Y, Subramaniam S, Szpankowski W, Grama A. Detecting conserved interaction patterns in biological networks. J Comput Biol. 2006;13:1299–1322. doi: 10.1089/cmb.2006.13.1299. [DOI] [PubMed] [Google Scholar]
- 127.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, et al. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pandey J, Koyutürk M, Subramaniam S, Grama A. Functional coherence in domain interaction networks. Bioinformatics. 2008;24:i28–i34. doi: 10.1093/bioinformatics/btn296. [DOI] [PubMed] [Google Scholar]
- 130.Schuster C. Next generation sequencing transforms today’s biology. Nature Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
FURTHER READING
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Joyce AR, Palsson BØ. The model organism as a system: integrating ‘omics’ data sets. Nat Rev Mol Cell Biol. 2006;7:198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- Bornholdt S. Less is more in modelling large genetic networks. Science. 2005;310:449–451. doi: 10.1126/science.1119959. [DOI] [PubMed] [Google Scholar]