Abstract
Prostate-Specific Antigen (PSA) is a serine protease biomarker that may play a role in prostate cancer development and progression. The inhibition of PSA’s enzymatic activity with small molecule inhibitors is an attractive and, as of yet, unexploited target. Previously, we reported a series of peptidyl aldehyde and boronic acid based inhibitors of PSA. In this study, the structural requirements in the P2 and P3 positions of peptide based PSA inhibitors are explored through the substitution of a series of natural and unnatural amino acids in these positions. This analysis demonstrated a preference for hydrophobic residues in the P2 position and amino acids with the potential to hydrogen bond in the P3 position. Using this information, a peptide boronic acid inhibitor with the sequence Cbz-Ser-Ser-Gln-Nle-(boro)-Leu was identified with a Ki for PSA of 25 nM. The attachment of a bulky metal chelating group to the amino terminal of this peptide did not adversely affect PSA inhibition. This result suggests that a platform of PSA-inhibitor chelates could be developed as SPECT or PET-based imaging agents for prostate cancer.
INTRODUCTION
Prostate cancer is uniformly lethal once it has escaped the confines of the prostate gland, resulting in the death of ~25,000 American men each year (1). There are two major clinical problems associated with the treatment of metastatic prostate cancer. First, all men undergoing androgen ablation therapy eventually relapse and no longer respond to androgen ablation (2, 3). At this point there is an urgent need for more effective therapies for patients with metastatic disease. Second, the clinical development of novel therapies is limited by the inability to adequately image the response of prostate cancer metastases. Unlike most solid tumor types, a hallmark of prostate cancer is the almost universal development of osteoblastic bone metastases in men failing androgen ablative therapy (4, 5). On this basis, 99mTc based bone imaging (i.e. bone scan) is the primary imaging modality used to evaluate the extent of prostatic disease and the response to therapy. Although the bone scan can measure the number of metastatic sites, it is relatively insensitive, detecting metastases at a relatively late stage (6) and does not accurately quantify response to therapy in “real-time”.
A unique attribute of prostate cancer cells that is shared with normal prostate epithelial cells is the ability to produce high levels of the differentiation marker Prostate-Specific Antigen (PSA) (7, 8). PSA is aptly named, in that it is exclusively produced by normal and malignant prostate epithelial cells and is not produced in any significant amounts by any other normal tissue in the human male. PSA is used extensively as a biomarker to screen for prostate cancer, to detect recurrence following local therapies and to follow response to systemic therapies for metastatic disease (7–9). Unlike other tissue differentiation markers, PSA continues to be expressed in high levels by prostate cancer cells even as they lose the morphologic characteristics of the normal prostate gland and become increasingly less differentiated (10). PSA expression is under androgen regulation in the normal prostate and androgen dependent prostate cancer (11). However, prostate cancer cells continue to express high levels of PSA even in patients with castration resistant prostate cancer and poorly differentiated disease.
PSA is a chymotrypsin-like serine protease with unique substrate specificity (12–14). Previous studies have documented that PSA present in the extracellular fluid surrounding prostate cancer cells is enzymatically active (15). In contrast, upon entering the circulation, PSA is rapidly inactivated by forming covalent complexes with the abundant serum protease inhibitors alpha-1-antichymotrypsin and alpha-2-macroglobulin with the end result being that there is no PSA activity in the blood (16,17). Thus, the exclusive presence of high levels of enzymatically active PSA only within the extracellular fluid of prostate cancers suggests that a PSA inhibitor based platform could be used to image metastatic prostate cancer sites.
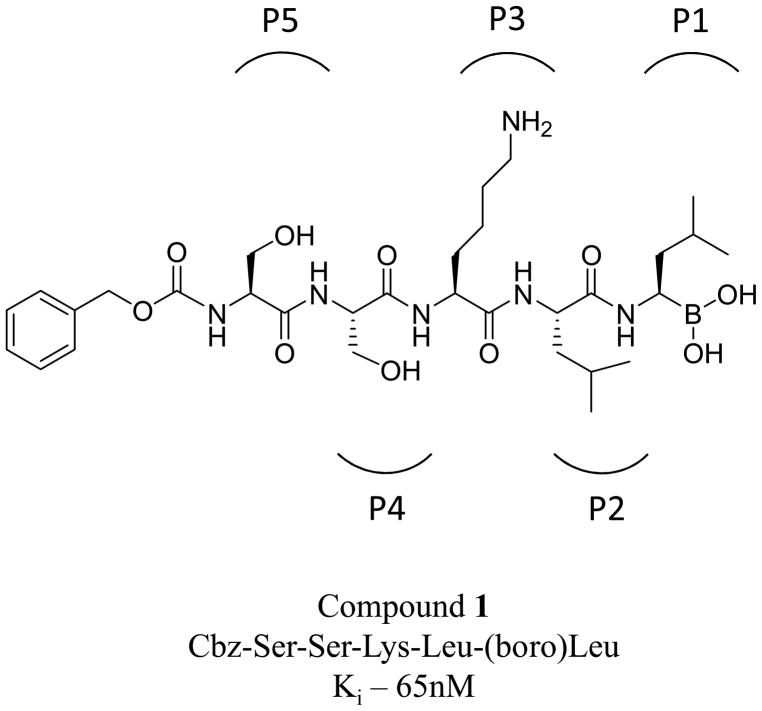
With this rationale in mind, in a previous report, we described the synthesis and characterization of potent and selective peptidyl boronic acid based inhibitors of PSA (18). Using an iterative screening approach based on a previously described PSA substrate and a homology model of the PSA catalytic site (19–20), we performed studies designed to evaluate the inhibitor length, amino acid sequence and D-isomer amino acid substitution on peptide aldehyde and peptidyl boronic acid based inhibitors of PSA (18). From these studies we identified a peptidyl boronic acid 1 with the sequence be Cbz-Ser-Ser-Lys-Leu- (boro)Leu (where Cbz, also abbreviated Z, is the carboxybenzoyl protecting group) as a potent and specific PSA inhibitor with a Ki of 65nM (Fig. 1).
Figure 1.
Chemical structure of Compound 1 used as the basis for the synthesis of the P2 and P3 inhibitor libraries. P5-P1 positions are indicated by brackets.
From these studies we learned that a peptide sequence length of at least 4 amino acids (i.e. P1–P4) was required for PSA inhibition and that D-isomer amino acid substitution was only permissible in the P5 position. In the modeling studies, it was determined that amino acids in the P5 position and beyond in the inhibitor were outside of the catalytic site for PSA and solvent exposed. This suggested that bulky adducts such as chelating groups, could be added to the amino terminus of the peptide without affecting the ability of the compound to act as a PSA inhibitor. In our current study, we expand upon the initial studies and perform a more exhaustive analysis to determine the preferred amino acid residues in the P2 and P3 positions of the inhibitor. Also, preliminary studies were performed exploring the effect the addition of a bulky transitional metal chelate group at the N-terminus of the sequence has on the ability of the peptidyl boronic acid to function as a PSA inhibitor. This acts as proof of concept that this peptidyl boronic acid platform could be used to generate PET or SPECT based PSA imaging agents for the detection of metastatic prostate cancer.
RESULTS
Previously, we generated a series of peptides containing a C-terminal aldehyde that were based on a known PSA-selective peptide substrate (18,19). In this study, we identified an aldehyde derivative, Z-SSKLL-H, that had a Ki of 6.5 μM for PSA inhibition. We subsequently generated the boronic acid derivative of this sequence Z-SSKL(boro)L and determined that the substitution of the boronic acid for the aldehyde group decreased the Ki ~ 100-fold to 65 nM. In order to further refine this sequence and identify PSA inhibitors with lower Ki values, we pursued a similar approach and constructed mini-libraries in which the P2 and P3 positions of this inhibitor were systematically modified. As before, a backbone-amide-linker (BAL) approach to create peptide aldehydes on solid phase resin was employed (21). The P2 position amino acid preference was investigated for several reasons: first, we wanted to find a residue that would increase the solubility of the compound in buffer and second, we were interested in trying to incorporate novel unnatural amino acids that would help us to further define the PSA pharmacophore and increase the specificity of the inhibitor for PSA.
For the P2 position investigation, twenty-seven natural and unnatural amino acids, spanning the entire range of size and hydrophobicity, were incorporated into the leucine aldehyde inhibitor sequence (i.e. Z-SSK-X-L-H where X = the substituted amino acid) (Table 1). From this analysis, the preferred amino acid for the P2 position was found to be norleucine 2 with a Ki of 3.5μM for PSA. The second best was norvaline 3 (4.4μM) and the third was the original leucine containing inhibitor 4 (6.5μM). Subsequently, we modified the aldehyde to produce the boronic acid derivative 43 of the norleucine aldehyde [i.e. Z-SSK-n-(boro)L] and determined that this inhibitor had a Ki of 48nM for PSA inhibition, which is slightly lower than the original inhibitor Ki for 1 of 65nM.
Table 1.
Results of the P2 – P3 peptide aldehdye library screen
| Compound | Ki (μM) | |
|---|---|---|
| P2-Peptide Aldehyde | ||
| 2 | Nle, Norleucine | 3.6 ± 0.3 |
| 3 | Nva, Norvaline | 4.4 ± 0.4 |
| 4 | Leu, Leucine | 6.5 ± 0.3 |
| 5 | Phe, Phenylalanine | 11.9 ± 0.8 |
| 6 | Met(o), Methionine sulfoxide | 12.8 ± 1.1 |
| 7 | Tyr, Tyrosine | 13.1 ± 0.9 |
| 8 | Met, Methionine | 13.7 ± 1.2 |
| 9 | His, Histidine | 18.6 ± 1.1 |
| 10 | Gln, Glutamine | 21.8 ± 1.3 |
| 11 | Hse, Homoserine | 29.4 ± 2.2 |
| 12 | Ile, Isoleucine | 37.4 ± 4.1 |
| 13 | Asn, Asparagine | 41.9 ± 3.8 |
| 14 | Phg, Phenylglycine | > 50 |
| 15 | Cys(tBu), S-t-butylcysteine | > 50 |
| 16 | Lys, Lysine | > 50 |
| 17 | Cha, Cyclohexylalanine | > 50 |
| 18 | Ser, Serine | > 50 |
| 19 | Val, Valine | > 50 |
| 20 | Thr, Threonine | >100 |
| 21 | Ala, Alanine | >100 |
| 22 | Nal, 2-Napthylalanine | >100 |
| 23 | 4-Br-Phe, 4-bromophenylalanine | >500 |
| 24 | Pro, Proline | >500 |
| 25 | Hyp, Hydroxyproline | >500 |
| 26 | Gly, Glycine | >1000 |
| 27 | Asp, Aspartic Acid | >1000 |
| 28 | Glu, Glutamic Acid | >1000 |
| P3-Peptide Aldehyde | ||
| 29 | Lys, Lysine | 3.6 ± 0.3 |
| 30 | Gln, Glutamine | 3.9 ± 0.2 |
| 31 | Met, Methionine | 7.5 ± 0.7 |
| 32 | Hse, Homoserine | 8.8 ± 0.7 |
| 33 | Cha, Cyclohexylalanine | 13.1 ± 1.2 |
| 34 | Asn, Asparagine | 18.2 ± 1.4 |
| 35 | Ser, Serine | 19.9 ± 1.2 |
| 36 | Met(o), Methionine sulfoxide | 25.9 ± 1.8 |
| 37 | Thr, Threonine | 43.8 ± 3.9 |
| 38 | Ala, Alanine | >500 |
| 39 | Pro, Proline | >1000 |
| 40 | Gly, Glycine | >1000 |
| 41 | Glu, Glutamic acid | >1000 |
| 42 | Asp, Aspartic Acid | >1000 |
In the next series of experiments, the P2 position was fixed as norleucine and peptide aldehydes were made incorporating different amino acids at the P3 position of the PSA inhibitor sequence (i.e. Z-SS-X-n-L-H) (Table 1). These studies were, in part, designed to identify amino acids that could replace the P3 lysine in the starting inhibitor in an attempt to make inhibitors that would be less susceptible to degradation by trypsin-like proteases in the circulation. Therefore, this library was more focused, with only thirteen amino acids tested. Based on our stability goal, basic residues were omitted altogether. The results of earlier modeling studies led us to also omit most hydrophobic residues from the library. The substitution of acidic amino acids in the P3 position was found to be highly deleterious producing Ki values above 1mM. In contrast, glutamine 30 and homoserine 32 were well tolerated at the P3 position with glutamine the most potent surveyed with a Ki of 3.9μM. Asparagine 34 and serine 35, both shorter by one methylene group, were modest inhibitors with Ki values around 19μM.
On the basis of these two libraries, a new PSA inhibitor sequence with glutamine in the P3 position and norleucine in the P2 was made into a boronic acid 44 (i.e. Z-SS-Q-n-(boro)L and tested for PSA inhibition, (Table 2). This new inhibitor was found to possess an improved Ki for PSA compared to the original Z-SSKL(boro)L inhibitor with a Ki of 27 nM. The inhibitor was less specific against chymotrypsin than the original inhibitor with a Ki of 211 nM (data not shown), making it roughly 8 fold more specific for PSA vs. chymotrypsin. Replacing the benzyloxycarbonyl N-terminal capping group with a more water soluble morpholinocarbonyl cap (45) did not affect the activity of the inhibitor and slightly improved the Ki to 25nM, (Table 2).
Table 2.
Ki values of the P2–P3 screen boronic acids and PSA imaging agents
| Compound | Ki (nM) | |
|---|---|---|
| P2 – P3 Screen Inihibitors | ||
| 43 | Z-SSKn(boro)L | 48.4 ± 3.6 |
| 44 | Z-SSQn(boro)L | 27.5 ± 2.4 |
| 45 | Mu-SSQn(boro)L | 25.3 ± 1.1 |
| Imaging Agents | ||
| 46 | SAAC-SSQn(boro)L | 15.6 ± 1.3 |
| 47 | (Re) SAAC-SSQn(boro)L | 28.4 ± 2.1 |
Addition of a Single Amino Acid Chelating (SAAC) Group
While a number of different radionuclides have been used for radiolabeling antibodies, proteins and peptides, Technetium-99m (99mTc) is the preferred choice. There are many advantages to using 99mTc that include easy availability, low cost, ease of handling, reasonable half-life of 6 hrs, 140 keV γ-emission, excellent imaging characteristics and favorable dosimetry. In addition, for this particular application, 99mTc can be easily attached to peptides using a number of different chelating groups (22).
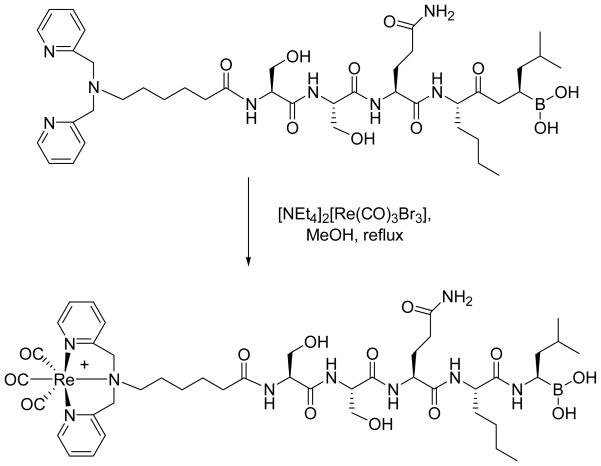
Previously, the syntheses of single amino acid chelates (SAAC) were reported that could be used for labeling peptides and other biomolecules with [Tc(CO)(3)]+ and [Re(CO)(3)]+ (23–25). These compounds are tridentate ligands derived initially from N-α-Fmoc Lysine (23,24). This tridentate structure is superior to other bidentate metal chelating agents, which display unfavorable pharmacokinetics due to the coordination of such complexes to plasma proteins after exchange of the substitution labile water/halide molecule. Based on this rationale, we generated the SAAC functionalized boronic acid based peptide 46 by coupling the SAAC group to the N-terminus of the SSQ-n-(boro)L inhibitor (Fig. 2). Without the presence of a chelated transition metal the SAAC inhibitor had a Ki for PSA of 15 nM (Table 2). To mimic the size and polarity of 99mTc, Re was substituted to determine if metal binding to the ligand would change the ability of the compound to act as an inhibitor of PSA. With the chelated Re, the SAAC inhibitor 47 continued to function well as an inhibitor of PSA with a Ki value of 28 nM. Thus, as predicted by our model of PSA’s catalytic site, the addition of a bulky adduct to the amino terminus of this 5 amino acid peptidyl boronic acid had no significant affect on PSA inhibition. In addition, the chelation of Re to the SAAC had no effect on the activity of the inhibitor. Thus, it can be assumed that 99mTc, when chelated with the SAAC, would have a similar Ki and act as an effective inhibitor of PSA in vivo that could potentially be used to image sites of metastatic prostate cancer.
Figure 2.
Chemical structure of the PSA inhibitor SSQn(boro)L coupled to a single amino acid chelating group at the amino terminus and method used to load with rhenium.
Effect of Backbone N-Methylation on PSA inhibition
The overarching goal of this study is to define a PSA inhibitor that could serve as a platform for both the imaging and targeted therapy of prostate cancer. Therefore, the serum stability of the molecule needs to be considered in the overall design. Previously, using a D-isomer amino acid scanning approach, we documented that the substitution of D-isomer amino acids was only permitted at the P5 position in a series of peptidyl boronic acid inhibitors (18). As an additional means to increase the in vivo stability of these inhibitors, we explored N-methylating the peptide bonds in the body of the inhibitor. Many studies have documented that N-methylation can increase the stability and bioavailability of peptide based molecules in vivo (26). Based on these results, we used a resin approach developed by Kesseler et al. to N-methylate the peptide backbone at every feasible position of the inhibitor sequence Z-SSKL(boro)L (Table 3) (27). The first available site to be methylated was the peptide bond between the P2 leucine and P3 lysine. This bond represents a potential scissile bond for trypsin-like protease mediated degradation of the inhibitor. N-methylation at this position increased the Ki of compound 48 to 2.5 μM, which was 38-fold higher than the parent inhibitor 1 with a Ki of 0.065 μM. An even more drastic change in inhibition was documented when the amido proton residing between residues P4 and P3 was replaced with a methyl group. The Ki of 49 with this substitution was found to be over 19 μM for PSA. Concurring with the D-isomer incorporation data, N-methylation of the P5-P4 peptide bond in 50 had little effect on the molecule since the replaced amido proton has little or no hydrogen bonding capacity. Out of all the N-methylated inhibitors, this inhibitor was the only one with a sub-micromolar Ki and was only ~4-fold less potent than 1. N-methylating all of the three available sites resulted in an inhibitor that almost completely lacked activity with a Ki over 50μM.
Table 3.
Ki values for PSA inhibition by N-methylated boronic acids
| Compound | Ki (μM) | |
|---|---|---|
| 48 | Z-SSK* L(boro)L | 2.5 ± 0.1 |
| 49 | Z-SS* KL(boro)L | 19.9 ± 1.4 |
| 50 | Z-S* SKL(boro)L | 0.2 ± 0.01 |
| 51 | Z-S* SK* L(boro)L | 5.6 ± 0.1 |
| 52 | Z-S* S* K* L(boro)L | > 50 |
indicates the position of the N-methylated peptide backbone
DISCUSSION
In this study we evaluated the effect substituting different amino acids in the P2 and P3 positions of a previously described peptide-based PSA inhibitor would have on PSA inhibition. The data from the P2 focused library provided overwhelming evidence that PSA prefers hydrophobic residues in the P2 position, with seven of the top ten inhibitors having hydrophobic residues in the P2 position. Both negatively and positively charged residues were not tolerated with Ki values > 50 μM. It should also be noted that residues containing beta branched side chains, i.e. isoleucine, valine and threonine, were not effective inhibitors and it appears that much as we observed previously with the P1 position, beta-branching is not tolerated. These results are in agreement with our previous modeling studies which demonstrated that P2 residue side chains can dock against the face of the imidazole ring of the catalytic His41 residue while on the opposite side of the P2 pocket, hydrophobic side chains can be in close contact with the hydrophobic Trp205 residue (18).
In our model of the PSA catalytic site interaction with the peptide inhibitors, the P3 residue serves as a lid to the specificity pocket and docks at a location just above the P1 binding site in the specificity pocket (18, 20). In this model, we observed that the P3 lysine residue in the Z-SSKLL-H inhibitor 3 interacts with the carboxyl group of the Glu208 side chain in the protease active site (18). The distance between the two groups (3.5Å) is close enough for the formation of a salt bridge, thus leading to favorable binding interactions between the protease and the inhibitor. In our previous study, omission of the lysine residue or replacing it with its mirror image, abrogated this important interaction and yielded poor inhibitors. The results of the P3 substitution in this current study supports these insights as amino acids with the potential to hydrogen bond (e.g. glutamine, methionine, homoserine) ranked as the best of the group.
Much like D-isomer amino acid substitution, N-methylation of the peptide backbone yielded inhibitors that were more resistant to proteolysis but not effective PSA inhibitors. N-methylating the peptide bond between the P2 and P3 residues, the bond representing the scissile bond of trypsin-like mediated proteolysis, resulted in an inhibitor with a 38-fold increase in Ki for PSA compared to the non-methylated peptide. One explanation for this change in Ki comes from our homology model which predicts that the backbone atoms of the P3-P1 residues within the inhibitor are in the proper orientation to form a short anti-parallel beta sheet with residues 204–206 of the PSA catalytic site. Replacement of the amido proton with a methyl group disrupts the interconnecting hydrogen bonding network resulting in the observed lack of inhibitory ability. A large increase in the inhibition constant was observed when the peptide bond between the P3 and P4 residues was methylated. As predicted by our PSA homology model, the amido proton in this position forms the strongest hydrogen bond with the active site of the protease and N-methylation has a deleterious effect on inhibitor binding. These data further document the important enthalpic contributions that are provided by the hydrogen bonding between the inhibitor amido protons and the protease active site.
In this study we also evaluated whether the addition of a bulky adduct to the amino terminus of the peptide would have an effect on PSA inhibition. Previously, our model of PSA’s catalytic site demonstrated that the P5 side chain is docked in the lower groove area of the catalytic site but it is exposed to the solvent and has no specific interactions with the protease residue. This observation suggested that the P5 residue and additional N-terminal moieties should not provide a significant contribution towards the overall binding affinity of inhibitor molecule. On this basis, we evaluated the SAAC group as a prototype chelating group to assess the effect on PSA inhibition. The addition of this group did not have a deleterious effect, but instead produced a slight improvement in the Ki. Chelation of the metal Re, as a model system representative of chelated radionuclide metals, also produced no significant effect on the Ki. These results suggest that a variety of chelating groups, such as the DOTA group, currently used in a variety of imaging applications could be incorporated into the PSA inhibitor structure to generate both SPECT and PET based imaging agents for the detection of PSA-producing prostate cancers. These concepts are currently undergoing further development and testing in our laboratory.
EXPERIMENTAL
General Information
PSA was purchased from Calbiochem (San Diego, CA). The enzymatic activity of this PSA was determined in previously reported studies. The fluorescent PSA substrate, Morpholinocarbonyl-Ser-Arg-Lys-Gln-Gln-Tyr-aminomethyl coumarin (Mu-SRKSQQY-AMC), was selected as the PSA substrate based on a previous characterization study (13) and was custom synthesized by California Peptides (Napa Valley, CA). All Fmoc and Boc protected amino acids used in the synthesis of PSA inhibitors were purchased from AnaSpec (San Jose, CA). Cbz protected amino acids were purchased from Novabiochem (San Diego, CA). Unless noted, all other reagents were from Sigma-Aldrich (St. Louis, MO).
PSA Inhibitor and Imaging Agent Synthesis
The leucine aldehyde and leucine boronic acid based peptide inhibitors were synthesized as previously described (18). For the boronic acid inhibitors, precursor protected peptides were synthesized using standard Fmoc solid-phase synthesis conditions on an AAPPTEC Apex 396 40 well peptide synthesizer using HOBt/DIC double coupling conditions The single amino acid chelate (SAAC) group was made from 6-aminohexanoic acid and pyridine-2-carboxaldehyde according to the method described by Levadala et al (28). The SAAC group was then added as the final residue during the solid phase synthesis of the inhibitor precursor peptide and this SAAC peptide was subsequently coupled to the leucine boronic acid derivative. The SAAC inhibitor was labeled with Re as previously described (23). High resolution mass spectrometery (HRMS) was performed on the final products using a VG-70S Magnetic Sector Mass Spectrometer in FAB ionization mode (Table 4). Peptide aldehyde and boronic acid final products were HPLC purified using a Waters Delta 600 semi-prep system with a Phenomenex Luna 10u C18 250 × 10 mm semi-prep column. The HPLC gradient profile was linear starting at 100% solvent A (0.1% TFA in H2O) and changing to 100% solvent B (0.1% TFA in acetonitrile) over 25 min with a flow rate of 8 ml/min. The Re labeled SAAC inhibitor was purified from the unlabeled SAAC inhibitor using an isocratic solvent mixture of 50/50 solvent A and B with a flow rate of 6 ml/min (Table 4).
Table 4.
Analytical properties of the PSA inhibitors
| No. | tRa | HRMS [M+] | No. | tRa | HRMS [M+] |
|---|---|---|---|---|---|
| 1 | 11.2 | 680.3937 | 27 | 12.8 | 666.3227 |
| 2 | 14.8 | 664.3801 | 28 | 12.4 | 680.3402 |
| 3 | 14.3 | 650.3698 | 29 | 14.8 | 664.3801 |
| 4 | 15.4 | 664.3798 | 30 | 12.8 | 664.3481 |
| 5 | 13.8 | 698.3672 | 31 | 13.7 | 667.3301 |
| 6 | 13.1 | 698.3324 | 32 | 12.8 | 637.3452 |
| 7 | 13.2 | 714.3598 | 33 | 14.2 | 689.4021 |
| 8 | 14.5 | 682.3371 | 34 | 13.1 | 650.3307 |
| 9 | 12.8 | 688.3574 | 35 | 13.7 | 623.3178 |
| 10 | 13.4 | 679.3556 | 36 | 12.4 | 683.3221 |
| 11 | 14.3 | 652.3446 | 37 | 12.9 | 637.3354 |
| 12 | 15.6 | 664.3814 | 38 | 13.8 | 607.3304 |
| 13 | 14 | 665.3393 | 39 | 13.4 | 648.3525 |
| 14 | 15.4 | 684.3488 | 40 | 12.4 | 593.3094 |
| 15 | 16.1 | 710.3678 | 41 | 13.1 | 665.3278 |
| 16 | 13.4 | 679.392 | 42 | 12.6 | 651.3189 |
| 17 | 15.7 | 704.4131 | 43 | 10.5 | 680.3941 |
| 18 | 14.1 | 638.3281 | 44 | 11.9 | 680.3607 |
| 19 | 14.9 | 650.3641 | 45 | 9.2 | 659.2854 |
| 20 | 14.5 | 652.3489 | 46 | 12.1, 4.2b | 841.5104 |
| 21 | 15.3 | 622.3338 | 47 | 12.8b | 1112.4084 |
| 22 | 15.8 | 748.3982 | 48 | 11.5 | 694.4081 |
| 23 | 13.7 | 776.2782 | 49 | 11.1 | 694.4081 |
| 24 | 15 | 634.3381 | 50 | 11.6 | 694.4078 |
| 25 | 15.1 | 664.3439 | 51 | 11.8 | 708.4248 |
| 26 | 13.8 | 608.3174 | 52 | 13.1 | 722.4402 |
tR is retention time determined by semi-preparative HPLC.
tR was determined using an isocratic flow of 50/50 solvent A and B.
Enzymatic Assays and Inhibition Kinetics
The assay for PSA activity was performed as previously described (20). The PSA concentration per assay was 2.5 μg/ml (2 nM active PSA) with a PSA substrate concentration of 300 μM. Substrate hydrolysis ± inhibitor over a range of concentrations was monitored by measuring fluorescence change secondary to AMC release Complete hydrolysis of the substrate was maintained below 5% to ensure that the substrate concentration was essentially constant. The inhibition constant values were determined using the method of progress curves of Nagase and Salvesen (29) as previously described (18). To assess inhibitor specificity, chymotrypsin (2 nM) was assayed with 25 μM of suc-AAF-AMC in the presence or absence of inhibitor over a range of concentrations.
Acknowledgments
This work was supported by a Prostate SPORE grant (P50CA58236) to SRD, a grant from the One-in-Six Foundation, Akron, OH to SRD and NIH grants CA92871 and CA134675 to MGP.
References
- 1.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- 2.Yu JX, Chao L, Chao J. J Biol Chem. 1994;269:18843–18848. [PubMed] [Google Scholar]
- 3.Wu Q, Parry G. Front Biosci. 2007;12:5052–5059. doi: 10.2741/2447. [DOI] [PubMed] [Google Scholar]
- 4.Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Hampton GM. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 5.Ernst T, Hergenhahn M, Kenzelmann M, Cohen CD, Bonrouhi M, Weninger A, Klären R, Gröne EF, Wiesel M, Güdemann C, Küster J, Schott W, Staehler G, Kretzler M, Hollstein M, Gröne HJ. Am J Pathol. 2002;160:2169–2180. doi: 10.1016/S0002-9440(10)61165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takayama TK, McMullen BA, Nelson PS, Matsumura M, Fujikawa K. Biochemistry. 2001;40:15341–15348. doi: 10.1021/bi015775e. [DOI] [PubMed] [Google Scholar]
- 7.Nelson PS, Gan L, Ferguson C, Moss P, Gelinas R, Hood L, Wang K. Proc Natl Acad Sci USA. 1999;96:3114–3119. doi: 10.1073/pnas.96.6.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 9.Yousef GM, Diamandis EP. Clin Biochem. 2003;36:443–452. doi: 10.1016/s0009-9120(03)00055-9. [DOI] [PubMed] [Google Scholar]
- 10.Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, Kim R, Rubin MA, Pienta KJ. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 11.Williams SA, Singh P, Isaacs JT, Denmeade SR. Prostate. 2006;67:312–329. doi: 10.1002/pros.20531. [DOI] [PubMed] [Google Scholar]
- 12.Watt KWK, Lee P-J, M’Timkulu T, Chan W-P, Loor R. Human prostate-specific antigen: Structural and functional similarity with serine proteases. Proc Natl Acad Sci USA. 1986;83:3166–3170. doi: 10.1073/pnas.83.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lilja H. J Clin Invest. 1985;76:1899–1903. doi: 10.1172/JCI112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBeau AM, Singh P, Isaacs JT, Denmeade SR. Biochemistry. 2009 Mar 26; doi: 10.1021/bi9001858. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denmeade SR, Sokoll LJ, Chan DW, Khan SR, Isaacs JT. Prostate. 2001;48:1–6. doi: 10.1002/pros.1075. [DOI] [PubMed] [Google Scholar]
- 16.Lilja H, Christensson A, Dahlen U. Clin Chem. 1991;37:1618–1625. [PubMed] [Google Scholar]
- 17.Stenman UH, Leinonen J, Alfthan H. Cancer Res. 1991;51:222–226. [PubMed] [Google Scholar]
- 18.LeBeau AM, Singh P, Isaacs JT, Denmeade SR. Chem Biol. 2008;15:665–674. doi: 10.1016/j.chembiol.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denmeade SR, Lou W, Malm J, Lovgren J, Lilja H, Isaacs JT. Cancer Res. 1997;57:4924–4930. [PMC free article] [PubMed] [Google Scholar]
- 20.Singh P, Williams SA, Shah MH, Lectka T, Pritchard GJ, Isaacs JT, Denmeade SR. Proteins. 2007;70:1416–1428. doi: 10.1002/prot.21676. [DOI] [PubMed] [Google Scholar]
- 21.Kappel JC, Barany G. J Peptide Sci. 2005;11:525–535. doi: 10.1002/psc.614. [DOI] [PubMed] [Google Scholar]
- 22.Okarvi SM. Med Res Rev. 2004;24:357–397. doi: 10.1002/med.20002. [DOI] [PubMed] [Google Scholar]
- 23.Banerjee SR, Wei L, Levadala MK, Lazarova N, Golub VO, Oconnor CJ, Stephenson KA, Valliant JF, Babich JW, Zubieta J. Inorg Chem. 2002;41:5795–5802. doi: 10.1021/ic020391d. [DOI] [PubMed] [Google Scholar]
- 24.Banerjee SR, Levadala MK, Lazarova N, Wei L, Stephenson KA, Valliant JF, Babich JW, Maresca KP, Zubieta J. Inorg Chem. 2002;41:6417–6425. doi: 10.1021/ic020476e. [DOI] [PubMed] [Google Scholar]
- 25.Stephenson KA, Banerjee SR, Sogbein OO, Levadala MK, McFarlane N, Boreham DR, Maresca KP, Babich JW, Zubieta J, Valliant JF. Bioconjugate Chem. 2005;16:1189–1195. doi: 10.1021/bc0500591. [DOI] [PubMed] [Google Scholar]
- 26.Biron E, Chatterjee J, Ovadia O, Langenegger D, Brueggen J, Hoyer D, Schmid HA, Jelinek R, Gilon C, Hoffman A, Kessler H. Angew Chem Int Ed Engl. 2008;47:2595–2599. doi: 10.1002/anie.200705797. [DOI] [PubMed] [Google Scholar]
- 27.Biron E, Chatterjee J, Kessler H. J Pept Sci. 2006;3:213–219. doi: 10.1002/psc.711. [DOI] [PubMed] [Google Scholar]
- 28.Levadala MK, Banerjee SR, Maresca KP, Babich JW, Zubieta J. Synthesis. 2004;11:1759–1766. [Google Scholar]
- 29.Salvesen GS, Nagase N. In: Proteolytic Enzymes. 2. Benyon R, Bond JS, editors. Oxford: Oxford University Press; 2001. pp. 131–147. [Google Scholar]