Abstract
An important goal of metabolomics is to characterize the changes in metabolic networks in cells or various tissues of an organism in response to external perturbations or pathologies. The profiling of metabolites and their steady state concentrations does not directly provide information regarding the architecture and fluxes through metabolic networks. This requires tracer approaches. NMR is especially powerful as it can be used not only to identify and quantify metabolites in an unfractionated mixture such as biofluids or crude cell/tissue extracts, but also determine the positional isotopomer distributions of metabolites derived from a precursor enriched in stable isotopes such as 13C and 15N via metabolic transformations. In this article we demonstrate the application of a variety of 2-D NMR editing experiments to define the positional isotopomers of compounds present in polar and non-polar extracts of human lung cancer cells grown in either [U–13C]-glucose or [U–13C,15N]-glutamine as source tracers. The information provided by such experiments enabled unambiguous reconstruction of metabolic pathways, which is the foundation for further metabolic flux modeling.
Keywords: Stable isotope resolved metabolomics, Isotope editing
Introduction
Metabolomics technologies are rapidly being established as a valuable set of tools for understanding biochemical regulation in organisms under normal and stress conditions. The rapid improvements in instrumentation and chemometrics have made it possible to identify and quantify a large number of metabolites in unfractionated cell or tissue lysates or biofluids, using appropriate analytical platforms (Fan et al. 1986; Maaheimo et al. 2001; Emmerling et al. 2002; Lindon et al. 2004; Fan et al. 2008; Lane et al. 2008). For most practical purposes, the two major platforms are mass spectrometry and NMR.
Although moderate sample throughput is straightforward with these technologies, of the order 25–50 samples per day when running under automation, the extraction of information from the extremely rich data sets remains problematic. One solution has been largely to ignore the identities of the compounds present, and simply focus on the differences between sets of samples using unsupervised statistics (Lindon et al. 2004; Trygg et al. 2007). However, to derive biochemical information and mechanistic understanding, the identities of the compounds must be known.
Furthermore, because biochemical networks are highly interlinked, intracellular concentrations are maintained within fairly narrow limits (homeostasis), and many metabolites participate in numerous common pathways, determination of metabolite concentrations alone is not sufficient to establish the biosynthetic routes unambiguously, as illustrated in Fig. 1. To place a metabolite in a given segment of a metabolic network, it is essential to use tracer technology, for which MS and NMR are ideally suited when the tracers are stable isotopes of common biological elements, such as 2H, 13C, and 15N.
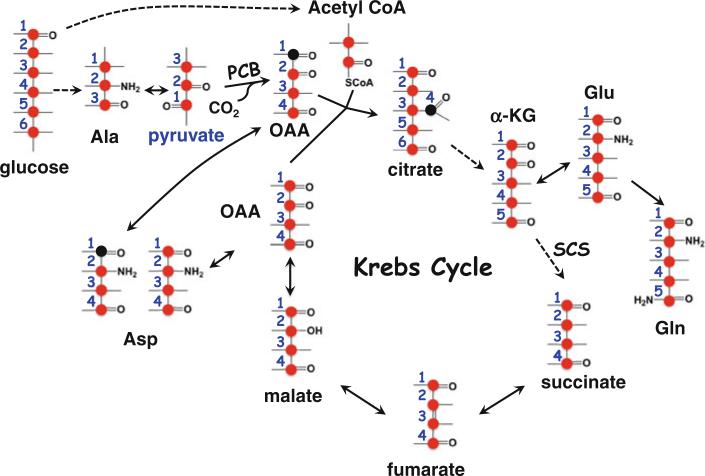
Fig. 1.
Tracers are necessary for delineating metabolic pathways. In the scheme depicted, pyruvate, lactate, α-ketoglutarate (α-KG), malate and Glu participate in multiple pathways including glycolysis, the Krebs cycle, serinolysis, and glutaminolysis. Without labeled tracers, it is impractical to resolve the specific pathway(s) involved in their production. With the use of 13C6-glucose, the synthesis of pyruvate, lactate, α-KG, malate, and Glu via glycolysis and the Krebs cycle can be distinguished from that via serinolysis or glutaminolysis by the 13C labeling pattern of these metabolites. Furthermore, the 13C positional isotopomers of Glu (i.e. 13C-2,3-Glu and 13C-4,5-Glu) can be used to delineate respectively the Krebs cycle with or without pyruvate carboxylation (PC) input. In essence, chemical identity of metabolites does not equate to their biochemical origin, which requires labeled isotopomers for their resolution
Mass spectrometry is a universal detector of ionized compounds, and some mass spectrometers have very high resolution (sub ppm resolution) and sensitivity (fmol or less detection limit). For this reason it is often the technique of choice for high throughput metabolomics. MS can easily distinguish molecules that differ by one atomic mass unit, and thus can be used to measure mass isotopologues of metabolites. The highest resolution spectrometers such as Fourier transform ion-cyclotron-resonance (FT-ICR) MS can resolve differences in mass equivalent to differences in nuclear binding energy. Despite these advantages, MS cannot readily discern stable isotopes such as 13C labeled at different atomic positions, e.g. Ala labeled at C-1, 2, or 3 positions. The exceptions are metabolites that can be extensively fragmented to provide information on labeled positions, but this is not a general case and can lead to a significant loss in sensitivity. In contrast, NMR readily determines each positional isotopomer as a distinct species without the need for fragmentation, i.e. at natural abundance appropriate NMR techniques produce distinct resonances corresponding to each single 13C position. The disadvantage of NMR is its relatively low sensitivity, such that at least nmol amounts of each compound is needed for reliable measurement. Nevertheless, the unique capabilities of NMR for positional isotopomer determination are complementary to MS for the analysis of mass isotopologues, both of which are essential to resolving the complexity of metabolic networks.
A major step forward in the application of NMR to structure analysis was the development of 2-D methods that enabled much more straightforward and rigorous structural elucidation while improving the resolution, thereby greatly expanding the size range and complexity of molecules that could be studied (Ernst et al. 1990; Cavanagh et al. 2007). The combination of multidimensional methods with substitution of magnetically active stable isotopes, especially 13C and 15N, along with indirect detection further extended the size and complexity of molecular systems that could be studied (Clore and Gronenborn 1998a, b; Gardner and Kay 1998; Cavanagh et al. 2007). In most instances, uniform labeling was used, usually via the metabolism of microorganisms in which the protein is over expressed (Gardner and Kay 1998), but later semi-selective labeling approaches became common (Lundstrom et al. 2007). The incorporation of the alternative isotopes makes it possible to design a wide variety of spectral editing experiments (Gardner and Kay 1998). In metabolic studies however, the goal is to delineate which atoms in individual metabolites have been substituted during the metabolic transformations, which in essence is an inverse problem. Nevertheless, the use of stable isotope tracing in defining metabolic pathways, has a long history, dating back to 1935 (Rittenberg and Schoenheimer 1937).
Another important difference between macromolecular NMR and metabolic NMR is that in the former usually a pure compound of known composition and stoichiometry is studied. In contrast, in metabolomics it is the identity of the components, and their isotopomer distributions in a mixture that are to be determined.
Nevertheless, stable isotope tracing in unfractionated mixtures of metabolites (loosely defined as biomolecules of molecular mass M < 1,000 Da that are subject to enzymatic transformation) has been enabled by the same developments in high field, multinuclear and multi-dimensional NMR techniques, which have proven utility in macromolecular structural elucidation. We have applied some of the common 2-D editing experiments to determine phosphorylated metabolites (Gradwell et al. 1998), and to profile composition of labeled isotopomers indirectly using proton detection in 13C and 15N tracer studies in microbes (Fan et al. 2009a, b), plants (Fan 1996), and human tumor metabolism (Fan et al. 2005, 2008, 2009a, b, 2010; Lane et al. 2009). Here we extend this approach using additional editing techniques to determine the structural topology of the stable isotopic enrichment in metabolites directly in a complex mixture.
Materials and methods
Materials
Human lung adenocarcinoma A549 cells were cultured in RPMI media containing 10.75 mM [U–13C]-glucose (Sigma–Aldrich, St. Louis) and 10% dialyzed fetal calf serum as previously described (Fan et al. 2005). Polar metabolites were extracted using 60% cold acetonitrile, freeze dried and then redissolved in 99.9% D2O for NMR analysis (Fan 2010). 30 nmol DSS-d6 (Sigma–Aldrich, St. Louis) were added to samples (771 μM methyl protons) as a shift reference and concentration standard. [U13C,15N]-Glutamine was purchased from Sigma–Aldrich (St. Louis) and used without further purification. All other reagents were of cell culture grade.
Polar lipids were extracted from cells using methanol containing 1 mM butylated hydroxytoluene (BHT), vacuum-dried, and redissolved in 99.98% methanol d4 for NMR analysis, as previously described (Lane et al. 2009). Chemical shifts were referenced to the methanol resonance at 3.31 ppm.
Methods
NMR spectra were recorded at 293 K on Varian Inova spectrometers at either 14.1 T or 18.8 T.
1D NMR spectra were recorded with weak presaturation of the solvent resonance with an acquisition time of 2 s and a recycle time of 3 s.
The following isotope edited 2D experiments were carried out to quantify enrichment at specific atoms and/or to detect specific isotopomers: 1H{13C}-HSQC, HSQC-TOCSY, HCCH-TOCSY, HACACO; 1H{31P} HSQC-TOCSY; 1H{15N}-HSQC-TOCSY and 1H-1H TOCSY.
High-resolution 1H{13C}-HSQC spectra were recorded with acquisition time of 0.15 s in t2 and 40 ms in t1. After linear prediction and zero filling the resolution in F1 was 6–8 Hz/point, which is adequate to resolve 1JCC of ca. 45 Hz. GARP decoupling was applied during the proton acquisition.
The HACACO experiment was performed with different 13C offsets to select either for the Cα or the Cβ/Cγ of Glu/Asp (Jeng and Dyson 1996; Qin et al. 1996), and recording the HA–CO and H–CA/CB planes.
Results and discussion
Analysis of polar metabolites
1H detection
The primary advantage of proton detection over direct X detection is sensitivity. We have applied a number of common proton-detected 2-D NMR experiments to identify and quantify positional enrichment of 13C and 15N in metabolites present in unfractionated extracts of only a few mg of cancer cells by dry weight. This approach circumvented the need for chromatographic separation.
As we have previously described, cross-peak satellite patterns in TOCSY spectra provide quantitative estimates of the enrichments at different atomic positions of metabolites, where the protons are directly bonded to 13C or 15N (Lane and Fan 2007; Fan and Lane 2008; Lane et al. 2008). Figure 2 shows an example TOCSY spectrum of a polar extract of A549 cells grown in the presence of [U–13C]-glucose. The red rectangles connect the cross-peaks of protons attached to 12C. The green dashed boxes trace the 13C satellite peaks arising from the attached 13C atoms for a pair of correlated protons. For example, lactate and the ribose moieties of the free nucleotides show isotopomers with either no 13C (central cross-peak) or both atoms are 13C (square array) (Lac-H2/3sat, 5′AXP-H1′/2′sat, UDPGH1′/2′sat, and NAD+ - H1′/2′sat in Fig. 2b). In contrast, the cross-peaks of glutamate and the glutamyl moiety of glutathione (GSH) displayed more complex patterns, that correspond to a mixture of four isotopomers comprising the fragments 12C312C4, 13C313C4 (Glu-H3/4sat), 12C313C4 (Glu-H4sat), and 13C312C4 (Glu-H3sat). As these cross-peaks can be volume integrated, the relative abundance of the positional labeling can be determined with reasonable accuracy, as shown in Table 1 (Lane and Fan 2007). The uracil rings of uridinediphosphoglucose (UDPG) also exhibited similar cross-peak patterns as those of Glu (see above), indicating 13C at either C5 (UDPG-H5sat) or C6 (UDPG-H6sat) and at both C5 and C6 (UDPG-H5/6sat, Fig. 2d). The ribose rings of NAD+ showed a fully labeled plus unlabeled cross-peak patterns (Fig. 2a), but the nicotinamide ring displayed no labeled pattern (Fig. 2d), as expected since it is derived from the vitamin niacin.
Fig. 2.
Positional 13C enrichment detected from satellite patterns in 1H–1H TOCSY spectrum of an A549 cell extract. The cells were grown in the presence of 10.75 mM [U–13C]-glucose for 24 h, and the metabolites were extracted as described in the “Methods”. NMR spectra were recorded at 800 MHz and 20°C. A,C 1-D 1H spectrum recorded with an acquisition time of 2 s, a relaxation delay of 3 s, and pre-saturation of the HOD signal. Data were processed by zerofilling once and apodizing with an unshifted Gaussian function and a 1 Hz line broadening exponential. B,D The TOCSY spectrum was recorded with acquisition times of 0.512 s in t2 and 0.038 s in t1, linear-predicted in F1, zero-filled to 8 k by 2 k points, and apodized with an unshifted Gaussian function and a 1 Hz line broadening exponential in both dimensions. Total experiment time was 11.2 h. Assignments were based on standards recorded under identical conditions (Fan and Lane 2008). Red rectangles trace the 1H–1H connectivities while dashed green boxes depict the 13C satellite cross-peak patterns (e.g. Lac-H3sat). Lac lactate, Mal malate, GSH reduced glutathione, GSSG oxidized glutathione, GAB γ-aminobutyrate. Metabolite numbering was according to (Fan and Lane 2008)
Table 1.
2-D 1H TOCSY quantification of different 13C positional isotopomers of glycolytic and Krebs cycle metabolites in an example A549 cell extract
| Isotopomersa | Fraction |
|---|---|
| 12C-2, 12C 4-Glu-GSH | 0.725 |
| 13C-2,12C-4-Glu-GSH | 0.024 |
| 13C -4,12C-2-Glu-GSH | 0.150 |
| 13C-2, 13C-4-Glu-GSH | 0.102 |
| 12C-2, 12C-3-Glu-GSH | 0.839 |
| 13C -2,12C-3-Glu-GSH | 0.067 |
| 13C -3,12C-2-Glu-GSH | 0.026 |
| 13C-2, 13C-3-Glu-GSH | 0.069 |
| 12C-3, 12C-4-Glu-GSH | 0.647 |
| 13C -3,12C-4-Glu-GSH | 0.008 |
| 13C -4,12C-3-Glu-GSH | 0.296 |
| 13C -3, 13C-4-Glu-GSH | 0.049 |
| 12C-3, 12C-4-Glu | 0.619 |
| 13C -4,12C-3-Glu | 0.381 |
| 12C-2, 12C-3-Lactate | 0.042 |
| 13C -2,12C-3-Lactate | 0.016 |
| 13C -3,12C-2-Lactate | 0.005 |
| 13C -2, 13C-3-Lactate | 0.937 |
| 12C-2, 12C-3-Ala | 0.137 |
| 13C -2, 13C-3-Ala | 0.863 |
| 12C-1′, 12C-2′-5′AXP | 0.117 |
| 13C-1′, 13C-2′-AXP | 0.883 |
| 12C-1′, 12C-2′-5′CXP | 0.222 |
| 13C-1′, 13C-2′-5′ CXP | 0.778 |
| 12C-1′, 12C-2′-UDPG | 0.073 |
| 13C-1′, 13C-2′-UDPG | 0.927 |
| 12C-U5, 12C-6-UDPG | 0.614 |
| 13C -U5,12C-U6-UDPG | 0.106 |
| 13C -U6,12C-U5-UDPG | 0.135 |
| 13C -U5, 13C-6-UDPG | 0.145 |
| 12C-A1′, 12C-A2′-NAD+ | 0.214 |
| 13C -A1′, 13C-A2′-NAD+ | 0.786 |
Isotopomer assignment and numbering are as described in (Fan et al. 2008)
GSH reduced glutathione, AXP adenine nucleotides, CXP cytosine nucleotides, UDPG uracildiphospho glucose, NAD+ nicotinamide adenine dinucleotide
13C editing
As 1H-1H TOCSY experiments do not detect 13C with no attached protons (e.g. quaternary or carbonyl carbons), the 1H{13C}-HSQC experiment is a useful, complementary experiment. When recorded at high resolution in F1, the HSQC experiment shows the 13C–13C couplings from which it is possible to infer the presence of label at quaternary or carbonyl carbons (Fig. 3). However, this experiment provides little information concerning the level of enrichment, unless careful peak shape analysis is carried out as is necessary for direct 13C detection experiments (Lu et al. 2002; Sherry et al. 2004). Nevertheless, the appearance of 13C doublets for Glu-C4 implies that this metabolite was present mainly as either 13C413C5 or 13C313C4, but as Glu-C3 showed no splitting (Fig. 3a, b), it is most likely that 13C413C5 was the more abundant species. This is consistent with the condensation of glucose-derived 13C2–acetyl CoA with unlabeled OAA, which subsequently leads to the production of 13C-4,5-Glu in the first turn of the Krebs cycle (Fan et al. 2010). It is also notable that the glutamyl moiety of reduced GSH showed the same splitting pattern as free glutamate, implying de novo biosynthesis of GSH from the free glutamate pool. Similarly, the splitting patterns of the 2′- to 4′carbons of the nucleotide ribose were triplets while that of the 1′ carbon was a doublet (Fig. 3c, d), which is consistent with uniform labeling of the nucleotide riboses via the pentose phosphate pathway (PPP).
Fig. 3.
High resolution 1H{13C}-HSQC shows 13C–13C couplings in labeled metabolites of A549 extract. The sample was the same as in Fig. 2. The HSQC spectrum was recorded with acquisition times of 0.15 s in t2 and 0.025 s in t1 with a recycle time of 1.5 s. GARP decoupling was applied during t2. The data were zerofilled once in t2, linear predicted and zerofilled in t1 to a final digital resolution of 5.9 Hz/pt. The data were apodized using an unshifted Gaussian and a line broadening exponential of 1 Hz in F2 and 4 Hz in F1. Total experiment time was 32 h. a, c are projections on the 13C axis. b aliphatic region, d sugar and pyrimidine C5 region. Cit citrate, all other abbreviations and numbering are as in Fig. 2. The 13C chemical shift was referenced indirectly to 1H, which was 0.5 ppm higher than those referenced directly to DSS (Fan 1996)
The splitting patterns of C3 and C2 of lactate were a doublet and triplet, respectively (Fig. 3a–d), which indicates that lactate was uniformly 13C labeled, and therefore derived from [U–13C]-glucose via glycolysis and to a lesser extent via the non-oxidative branch of the PPP, which is typically active in cancer cells (Boren et al. 2001; Lee et al. 1998). Moreover, the triplet pattern of C2 of Asp (Fig. 3a, b) suggests the presence of 13C1,13C2,13C3–Asp. These data complemented the isotopomer information obtained from the TOCSY analysis described above.
Additional 2-D 13C-edited experiments (including HACACO, HCCH-TOCSY, and HSQC-TOCSY) were performed for verifying resonance assignment and for discerning the exact 13C-labeling pattern in some metabolites, which was ambiguous from the TOCSY and HSQC analysis. For example, it is unclear whether the doublet of C2,5 of citrate (Fig. 3a, b) represents 13C1,13C2/13C5,13C6,13C2,13C3/13C3,13C5 or both (cf. Fig. 4 for numbering of citrate). It is also difficult to ascertain whether 13C3,13C4-Asp was present based on the TOCSY and HSQC data. This information is crucial to reconstructing the exact pathway(s) from which the labeled isotopomers are derived. For example, using [U–13C]-glucose as tracer, 13C3,13C4-Asp plus 13C1,13C2-Asp are products of the Krebs cycle after one turn while 13C1,13C2,13C3-Asp is derived from pyruvate carboxylation, an important anaplerotic reaction for the Krebs cycle (Fan et al. 2010).
Fig. 4.
HACACO experiments directly detect 13C-labeled carbonyls. The sample was similarly prepared as in Fig. 2. The spectrum was recorded to detect the CO–HA,HB,HG plane with a 13C offset set to 55 ppm. The acquisition times were 0.12 s in t2, and 0.042 s in t1 with a recycle time of 1.3 s. The data were zerofilled once in t2, and linear predicted and zerofilled in t1. The data were apodized using an unshifted Gaussian and a line broadening exponential of 1 Hz in F2 and 4 Hz in F1. Total experiment time was 5.9 h. The chemical shift displayed was 2.1 ppm higher than that referenced to DSS for Gly. The abbreviations and numbering were the same as in Fig. 2. Also shown are the structures and numbering of citric acid and UDPG
To address the labeling of the carbonyl groups in the amino acids, we recorded HACACO experiments that correlate the carbonyl carbon with either the HA, or the side chain carbonyl with HG; these two carbonyls have significantly different 13C chemical shifts, as shown in Fig. 4. The cross-peaks for C1 to H2 (Asp-2 → 1) and C4 to H3 of Asp (Asp-3 → 4) gave credence to the presence of 13C1,13C2-Asp and 13C3,13C4-Asp. These two products of one turn of the Krebs cycle arise because of the reversible reaction from the symmetric succinate and fumarate to the non-symmetric malate and oxalacetate, an example of metabolic scrambling of label. The cross-peak of C1 to H2 of citrate (Cit-2 → 1) indicates the presence of 13C1,13C2-citrate, which is also a product of the Krebs cycle after one turn. Further evidence for one turn of Krebs cycle activity came from the cross-peak of C5 to H4 of Glu (Glu-4 → 5), which arose from the isotopomer 13C4,13C5-Glu. Also detected were the cross-peaks of C1 to H2 (Glu-2 → 1) and C1 to H3 of Glu (Glu-3 → 1), which corresponded to 13C1,13C2-Glu and 13C1,13C2,13C3-Glu, respectively. These two isotopomers could be produced from the Krebs cycle after two and three turns (Fan et al. 2010). It should be noted that the intensity of the Glu-4 → 5 cross-peak was higher than those of Glu-2 → 1 and Glu-3 → 1, which suggests that more cells have undergone only one turn of the Krebs cycle.
1H{13C} HSQC-TOCSY and 1H-1H HCCH-TOCSY are complementary experiments that can help determine the enrichment positions and the number of contiguous 13C in the chain, as illustrated in Figs. 5 and 6, respectively. The HSQC-TOCSY provides the 13C-edited proton TOCSY connectivity, and where there are multiple 13C in the same compound, the specific cross-peak patterns substantiate the 1H and 13C assignments. The spectrum can be recorded in the same amount of time as the HSQC experiment, but more definitively shows the number of 13C sites in each metabolite, as aided by the 1H connectivities. The HSQC-TOCSY experiment can also provide clearer correlations for 13C labeled isotopomers than the TOCSY experiment since spectral contributions from unlabeled species are greatly reduced via the 13C selection in the HSQC part of the experiment. For example, the 13C-labeling pattern of Pro was not evident in the TOCSY spectrum (Fig. 2b), but was clearly observed in the HSQC-TOCSY spectrum (Fig. 5b), i.e. the presence of 13C at 2, 3, and 4 positions. Likewise, the presence of 13C3-malate and 13C1 to C6-glucose was clear in the HSQC-TOCSY (Fig. 5b, d) but not in the TOCSY spectrum (Fig. 2b).
Fig. 5.
1H{13C} HSQC-TOCSY verifies 13C isotopomer assignment and reduces spectral crowding. The sample was the same as in Fig. 4. The HSQC-TOCSY spectrum was recorded with GARP decoupling during t2 (0.14 s) centered at 60 ppm in the 13C dimension, and a recycle time of 1.5 s. The acquisition time in t1 was 0.0125 s. The spin lock strength was 8 kHz for 50 ms. The data were zerofilled in t2, and linear predicted and zerofilled in t1 to 8 k × 2 k points. The data were apodized using an unshifted Gaussian and a line broadening exponential of 1 Hz in F2 and 4 Hz in F1. Total experiment time was 6 h. a, b aliphatic region. a is the 13C projection of b. c, d sugar and pyrimidine C5 region. c is the 13C projection of b. The abbreviations and numbering were the same as in Fig. 2
Fig. 6.
1H-1H HCCH-TOCSY reveals numbers of contiguous 13C atoms in isotopomers. The sample was similarly prepared as in Fig. 2 except that the cells were extracted in 10% trichloroacetic acid. The HCCH-TOCSY spectrum was recorded at 14.1 T with GARP decoupling during t2 (0.12 s) centered at 60 ppm in the 13C dimension, and a recycle time of 1.5 s. The acquisition tine in t1 was 0.05 s. The spin lock strength was 8 kHz for 11 ms. The data were zerofilled in t2, and linear predicted and zerofilled in t1 to 8 k × 2 k points. The data were apodized using an unshifted Gaussian and a line broadening exponential of 1 Hz in F2 and 4 Hz in F1. Total experiment time was 11.4 h. a high resolution 1-D 1H spectrum. b Projection of the 2D spectrum onto the F2 axis showing spectral simplification via 13C editing. c 2-D spectrum of the same region in a, b. GlyOH3P glycerol-3-phosphate, all other abbreviations and numbering were the same as in Fig. 2
The HCCH-TOCSY experiment also reduces spectral crowding by selecting only those isotopomers that contains at least two contiguous 13C in the same molecule. This experiment confirmed that the ribose moieties of the free nucleotide pool were predominantly the 13C5 isotopomers (Fig. 6c), which indicates their production from the oxidative branch of the pentose phosphate pathway (PPP). However, if the non-oxidative branch of the PPP were significant with the use of non 13C sources, different labeling patterns in the ribose moieties would have been observed. Therefore the NMR data provided a simple means to discriminate between the two branches of the PPP compared with the GC–MS based approaches (Lee et al. 1998). In addition, the connectivity from H1 to H3 of the low-abundance glycerol-3-phosphate (GlyOH3P in Fig. 6c) was discernable, which indicates the presence of 13C3-GlyOH3P, a precursor to glycerolipid synthesis. Moreover, the connectivity from H2 to H4 of the glutamyl moiety of the glutathiones (GSH + GSSG) was observed, which corresponds to the isotopomer 13C2,13C3,13C4-Glu. This isotopomer cannot be generated from the Krebs cycle alone even after three turns (Fan et al. 2010). However, 13C5-Glu, which contains the 13C2,13C3,13C4 fragment, can be produced with the input of pyruvate carboxylation (PC) in the first turn of the cycle, according to the scheme in Fig. 7. Finally, the connectivity from H2 to H3 of Asp is consistent with the presence of 13C1,13C2,13C3-Asp, which was also deduced from the HSQC spectrum (Fig. 3a, b).
Fig. 7.
Labeling scheme for the Krebs cycle with pyruvate carboxylation using [U–13C]-glucose as tracer. Red circles are 13C atoms, black circles are 12C atoms. Double and single headed arrows represent reversible and irreversible reactions, respectively; dashed arrows depict multiple reaction steps. OAA oxalacetate, SCS succinyl CoA synthetase, PCB pyruvate carboxylase
The isotopomer pattern of Asp generated via mitochondrial Krebs cycle activity should be closely related to that of the uracil ring, which is synthesized from aspartate. The combination of 2-D TOCSY and 13C-edited experiments enabled such biosynthetic linkage to be made. Based on HSQC, HACACO, and HCCH-TOCSY data, the 13C isotopomers of Asp detected were 13C1,13C2-Asp, 13C3,13C4-Asp, and 13C1,13C2,13C3-Asp. Upon incorporation into the uracil ring, these Asp isotopomers should yield the following 13C isotopomers respectively (see Fig. 4 for uracil ring numbering): 12C5,13C6-uracil, 13C5,12C6-uracil, and 13C5,13C6-uracil. The TOCSY satellite cross-peak pattern of the uracil ring (cf. Fig. 2d) revealed the presence of all three uracil 13C isotopomers, thereby confirming the biosynthetic route from the tracer [U–13C]-glucose through glycolysis, the Krebs cycle with and without PC to pyrimidine ring biosynthesis. This route requires not only an active Krebs cycle in these cells, but also a functioning mitochondrial electron transport chain, as a critical uracil biosynthetic enzyme, dihydroorotate dehydrogenase, is a mitochondrial enzyme (Evans and Guy 2004) (Nagy et al. 1992). This is in contrast to the common notion that mitochondrial functions in cancer cells are compromised (King et al. 2006). It should also be noted that no labeled isotopomers were detected for any of the essential amino acids such as valine, leucine and isoleucine or other essential metabolites such as phosphocholine or nicotinamide, which confirmed the absence of contaminating microbial activity.
31P editing
In addition to 13C-edited experiments for determining positional isotopomer profiles in A549 cell extracts, 31P-edited 2-D experiments are useful for discerning phosphorylated metabolites without a priori knowledge (Gradwell et al. 1998). There are numerous such metabolites whose concentrations differ substantially among tissue or cell types (Fan and Lane 1992; Gradwell et al. 1998; Vizan et al. 2007). Cancer cells are often characterized by high concentrations of phosphocholine, which has been detected by NMR in vivo (Glunde et al. 2004; Eliyahu et al. 2007). They can also be abundant in nucleotides, which is a critical parameter for tumor cell proliferation. Here, we employed 2-D 1H{31P} HSQC-TOCSY experiments to determine the identities and relative amounts of phosphorylated compounds in the A549 cell extracts (Fig. 8). Because the phosphomonoesters typically have a pK near 7, the 31P chemical shifts are extremely sensitive to solution conditions (pH, ionic strength, and divalent cations) in cell or tissue lysates. Further complicating the assignment of the 31P signals, the resolution in the 31P dimension for common phosphomonoesters is relatively poor. We therefore used HSQC-TOCSY to obtain multiple 31P–1H correlations in each molecule, thereby making identification of the phosphorylated compounds more reliable. As Fig. 8 shows, the assignment of the highly abundant phosphocholine (P-choline) was substantiated by the connectivity between its two methylene protons and verified by comparison with the standard spectrum (Fan and Lane 2008) (and unpublished data). The same was true for the assignment of 5′ATP, glycerol-3-phosphate, and fructose-6-phosphate (F6P). We estimate that these experiments detect > 50 nmol of phosphorylated compound, which translates roughly to a concentration of 20 μM in the cell, in line with reported concentrations of common phosphometabolites in cells (Fersht 1999; Vizan et al. 2007). It should also be noted that glycerol-3-P and F6P could not be unambiguously assigned in 1H and 13C-edited experiments due to their low abundance and/or spectral crowding (cf. Fig. 2). However, their presence was clear in the 31P-edited experiment (Fig. 8).
Fig. 8.
Spectral editing via 1H{31P} HSQC-TOCSY for the assignment of phosphorylated metabolites. The sample was the same as in Fig. 4. The 1H-{31P} HSQC-TOCSY spectrum was recorded with acquisition time of 0.14 s in t2 and 0.014 s in t1, with a recycle time of 1.35 s and GARP decoupling during t2 The spin lock strength was 8 kHz for 50 ms. Total experiment time was 13.9 h. The 31P spectrum was referenced to external methylene diphosphonate (Gradwell et al. 1998). a 1-D high resolution 1H spectrum. b 2-D spectrum. c 1-D projection on the 31P axis
15N editing
Nitrogen metabolism is often dominated by transamination in liver and kidney, which ultimately leads to removal of potentially toxic ammonia via the urea cycle. On the other hand, in several cell types, including cancer cells, glutamine metabolism via glutaminase is up regulated and important for cell proliferation and survival, which makes glutamine a conditionally essentially amino acid. The glutaminase reaction generates ammonium ions and glutamate, which can be used in a wide variety of metabolic reactions, including anaplerotic incorporation of the carbon skeleton into the Krebs cycle (Portais et al. 1996; DeBerardinis et al. 2007; Yuneva 2008). The amido nitrogen of Gln is also the immediate donor for nucleobase biosynthesis and aminosugars such as glucosamine and galactosamine. Thus, 15N/13C-Gln is a useful precursor for tracing numerous metabolic pathways and the resulting products can be detected by 1H{15N}-HSQC experiments. Figure 9 shows an example spectrum, where the incorporation of the amido N from 13C5, 15N2-Gln into the purine bases in A549 cells was evident. The HSQC experiment utilized the substantial 2JNH between H8, N9 and H2, N1 for observing N–H connectivity in adenine rings of nucleotides (Ippel et al. 1996). As the detected protons do not exchange with solvent, this experiment was carried out in D2O. In addition, the 15N satellites of H8 and H2 were resolved in the 1-D 1H spectrum, allowing a simple determination of the degree of enrichment following 13C5,15N2-Gln tracer administration to A549 cells (data not shown). Thus, the HSQC analysis enabled the reconstruction of nitrogen pathway from Gln to the synthesis of purine bases. Other 15N products such as those resulting from transamination can also be detected but only in H2O, as previously described (Fan et al. 1997).
Fig. 9.
Spectral editing via 1H{15N}-HSQC detects 15N-isotopomers of nucleotides. A549 cells were grown for 24 h in the presence of 5 mM unlabeled glucose and 4 mM [U–13C,15N]-glutamine and extracted as described in the “Methods”. The spectrum was recorded at 14.1 T at 20°C with acquisition times of 0.15 s in t2 and 0.021 s in t1 and a recycle time of 1.6 s. The value of JNH was set to 10 ms. The data were zerofilled in t2, linear predicted and zerofilled in t1 to a final 8 k by 2 k points, apodized with an unshifted Gaussian and a 1 Hz line broadening exponential in both dimensions. Total experiment time was 0.91 h
Analysis of lipid metabolites
In addition to 2-D NMR analysis of water-soluble metabolites, the same set of experiments can be used to determine isotopomer patterns of lipids, including the membrane phospholipids and cholesterol. Figure 10 shows a TOCSY spectrum of a methanolic extract of A549 cells grown in [U–13C]-glucose. This solvent extracted primarily polar lipids and cholesterol (Lane et al. 2009). The 13C cross-peak pattern of the glycerol backbone (traced by dashed green boxes) indicates the presence of both all 12C and all 13C-labeled isotopomers. There was no evidence for metabolic scrambling of the isotope in the three carbons, which suggests that they were derived from glucose via glycolysis. This is consistent with the observation of the precursor 13C3-GlyOH3P by HCCH-TOCSY (cf. Fig. 6c). Also evident in Fig. 10a are the 13C cross-peaks of the fatty acyl chains, including those of ωCH3, 2,3,4,8–CH2, and unsaturated protons (CH=CH). Unlike those of the glycerol backbone, these cross-peaks showed scrambled label patterns, i.e. for a given pair of protons, there were four 13C isotopomer species. Using the 2 and 3-CH2 correlation as an example, the four isotopomers were 12C212C3, 13C212C3, 12C213C3, and 13C213C3. These patterns are to be expected if the precursor acetyl CoA pool for fatty acid biosynthesis was both unlabeled and uniformly labeled, which in turn means that the citrate precursor to acetyl CoA synthesis were both unlabeled and doubly labeled at C1 and C2 or C5 and C6 positions (cf. Figs. 4 and 7). The 13C1,13C2-citrate species was discerned from the HAC-ACO analysis (cf. Fig. 4), while unlabeled citrate was detected by GC–MS (data not shown). It is interesting to note that there were at least three separate sets of unsaturated proton resonances, presumably reflecting different classes of unsaturated lipids.
Fig. 10.
13C enrichment in PL isotopomers determined by 1H–1H TOCSY and high-resolution 1H{13C}- HSQC. The A549 cells were grown in [U–13C]-glucose for 24 h before extraction with pure methanol as described in the “Methods”. The TOCSY spectrum was recorded at 14.1 T with acquisition times of 0.341 s in t2 and 0.043 s in t1 respectively and a recycle time of 2 s. The total experiment time was 10.9 h. The high resolution HSQC spectrum was recorded with acquisition times of 0.12 s and 0.034 s in t1 and t2, respectively with a recycle time of 2.1 s. Total experiment time was 16.7 h. Processing was as described in Figs. 2 and 9. a, b 1-D proton spectrum and TOCSY of the aliphatic region of the spectrum. Assignments were based on lipid standards recorded under the same conditions (Lane et al. 2009). c, d high resolution HSQC spectrum (d) and the 1-D 13C projection (c). Metabolite nomenclature and numbering are as described in (Fan and Lane 2008)
The 13C labeling pattern of lipids obtained from the TOCSY experiment was complemented and verified by high-resolution HSQC analysis, as shown in Fig. 10. The doublet splitting pattern of Cω and C2 suggests the dominance of acyl chain isotopomers containing the 13CH3–13CH2–12CH2– fragment while the doublet pattern for C3 and C1, and triplet pattern for C2 of the glycerol backbone is consistent with 13C labeling in all three carbons. This information is crucial to interpreting the corresponding FT-ICR-MS data, in terms of assigning the glycerolipid isotopologues of m + 3 (monoisotopic mass plus 3 neutrons) and m + odd number of neutrons to 13C labeling in the glycerol backbone (Lane et al. 2009). Together, the NMR and FT-ICR-MS data provide both the number and position of 13C labels in hundreds of lipid species directly in crude cell or tissue extracts, which is a feat unimaginable to achieve in past research.
Conclusions
The unique spectral editing capabilities of NMR can be used to select and quantify chemical classes such as phosphorylated metabolites, or enriched atoms at specific positions. These capabilities make NMR a technique of choice for reconstruction of metabolic network and quantitative flux analysis, especially when combined with high resolution MS. Both general and specific labeled tracers can be employed to probe different aspects of metabolic networks and how they are perturbed by the environment. We have applied a small fraction of the vast array of possible experiments to determine metabolic labeling topologies, as described here for the A549 cells. Further developments in fast acquisition techniques can increase the sample throughput as well as the information content of stable isotope-resolved metabolomic (SIRM) studies, particularly for the purpose of flux and network modeling in systems biology.
Acknowledgments
This work was supported in part by National Science Foundation EPSCoR grant # EPS-0447479; National Institutes of Health Grant numbers 5P20RR018733, 1R01CA118434-01A2, 3R01CA118434-02S1, R21CA133668-01; the Kentucky Challenge for Excellence, and the Brown Foundation. We thank Ms. JinLian Tan for expert technical assistance, and Dr. S. Arumugam for assistance with NMR experiments.
Contributor Information
Teresa W-M. Fan, Department of Chemistry, University of Louisville, Louisville, KY, USA Center for Regulatory Environmental Analytical Metabolomics, University of Louisville, Louisville, KY, USA; JG Brown Cancer Center, University of Louisville, Louisville, KY, USA.
Andrew N. Lane, Center for Regulatory Environmental Analytical Metabolomics, University of Louisville, Louisville, KY, USA JG Brown Cancer Center, University of Louisville, Louisville, KY, USA; Rm 217, CTRB, 505 S. Hancock St., Louisville, KY 40202, USA.
References
- Boren J, Cascante M, Marin S, Comin-Anduix B, Centelles JJ, Lim S, Bassilian S, Ahmed S, Lee WNP, Boros LG. Gleevec (ST1571) influences metabolic enzyme activities and glucose carbon flow toward nucleic acid and fatty acid synthesis in myeloid tumor cells. Journal of Biological Chemistry. 2001;276(41):37747–37753. doi: 10.1074/jbc.M105796200. [DOI] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Elsevier Academic Press; San Diego: 2007. [Google Scholar]
- Clore GM, Gronenborn AM. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998a;16(1):22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM. NMR structure determination of proteins and protein complexes larger than 20 kDa. Current Opinion in Chemical Biology. 1998b;2(5):564–570. doi: 10.1016/s1367-5931(98)80084-7. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: Molecular and biochemical studies. Int J Cancer. 2007;120(8):1721–1730. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- Emmerling M, Dauner M, Ponti A, Fiaux J, Hochuli M, Szyperski T, Wuthrich K, Bailey JE, Sauer U. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J Bacteriol. 2002;184(1):152–164. doi: 10.1128/JB.184.1.152-164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RR, Bodenhausen G, Wokaun A. Principles of nuclear magnetic resonance in one and two dimensions. Clarendon Press; Oxford: 1990. [Google Scholar]
- Evans DR, Guy HI. Mammalian pyrimidine biosynthesis: Fresh insights into an ancient pathway. Journal of Biological Chemistry. 2004;279(32):33035–33038. doi: 10.1074/jbc.R400007200. [DOI] [PubMed] [Google Scholar]
- Fan TW-M. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog Nucl Magn Reson Spectrosc. 1996;28:161–219. [Google Scholar]
- Fan TW-M. Comprehensive Toxicology. Vol. 2. Academic Press; McQueen, C.A. Oxford: 2010. Metabolomics-Edited Transcriptomics Analysis (Meta). pp. 685–706. [Google Scholar]
- Fan TWM, Lane AN. Identification of Glycerophosphorylcholine in Mussel Ovarian Extracts by 2-Dimensional Nuclear-Magnetic-Resonance. Anal Biochem. 1992;206(2):251–255. doi: 10.1016/0003-2697(92)90362-b. [DOI] [PubMed] [Google Scholar]
- Fan TW, Lane AN. Structure-based profiling of Metabolites and Isotopomers by NMR. Progress in NMR Spectroscopy. 2008;52:69–117. [Google Scholar]
- Fan TWM, Higashi RM, Lane AN, Jardetzky O. Combined use of proton NMR and gas chromatography-mass spectra for metabolite monitoring and in vivo proton NMR assignments. Biochim Biophys Acta. 1986;882(2):154–167. doi: 10.1016/0304-4165(86)90150-9. [DOI] [PubMed] [Google Scholar]
- Fan TWM, Higashi RM, Frenkiel TA, Lane AN. Anaerobic nitrate and ammonium metabolism in flood-tolerant rice coleoptiles. J Exp Bot. 1997;48(314):1655–1666. [Google Scholar]
- Fan T, Bandura L, Higashi R, Lane A. Metabolomics-edited transcriptomics analysis of Se anticancer action in human lung cancer cells. Metabolomics Journal. 2005;1(4):325–339. [Google Scholar]
- Fan T, Kucia M, Jankowski K, Higashi R, Ratajczak J, Ratajczak M, Lane A. Rhabdomyosarcoma cells show an energy producing anabolic metabolic phenotype compared with primary myocytes. Molecular Cancer. 2008;7(1):79. doi: 10.1186/1476-4598-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T, Bird J, Brodie E, Lane A. 13C-Isotopomer-based metabolomics of microbial groups isolated from two forest soils. Metabolomics. 2009a;5(1):108–122. [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol Cancer. 2009b;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW-M, Yuan P, Lane AN, Higashi RM, Wang Y, Hamidi A, Zhou R, Guitart X, Chen G, Manji HKM, Kaddurah-Daouk R. Stable Isotope-Resolved Metabolomic Analysis of Lithium Effects on Glial-Neuronal Metabolism and Interactions. Metabolomics. 2010;6(2):165–179. doi: 10.1007/s11306-010-0208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. Structure and mechanism in protein science. W. H. Freeman & Co; New York: 1999. [Google Scholar]
- Gardner KH, Kay LE. The use of H-2, C-13, N-15 multidimensional NMR to study the structure and dynamics of proteins. Annu Rev Biophys Biomol Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64(12):4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- Gradwell MJ, Fan TWM, Lane AN. Analysis of phosphorylated metabolites in crayfish extracts by two-dimensional 1H–31P NMR heteronuclear total correlation spectroscopy (hetero TOCSY). Anal Biochem. 1998;263(2):139–149. doi: 10.1006/abio.1998.2789. [DOI] [PubMed] [Google Scholar]
- Ippel J, Wijmenga S, de Jong B, Heus H, Hilbers C, Vroom E, van de Marel C, van Boom JH. Heteronuclear scalar couplings in the bases and sugar rings of nucleic acids: their determination and applicaiotn in assignment and conformational analysis. Magnetic Resoannce in Chemistry. 1996;34:S156–S176. [Google Scholar]
- Jeng MF, Dyson HJ. Direct measurement of the aspartic acid 26 pK(a) for reduced Escherichia coli thioredoxin by C-13 NMR. Biochemistry. 1996;35(1):1–6. doi: 10.1021/bi952404n. [DOI] [PubMed] [Google Scholar]
- King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25(34):4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- Lane AN, Fan TW. Quantification and identification of isotopomer distributions of metabolites in crude cell extracts using 1H TOCSY. Metabolomics. 2007;3:79–86. [Google Scholar]
- Lane AN, Fan TW, Higashi RM. Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Biophysical Tools for Biologists. 2008;84:541–588. doi: 10.1016/S0091-679X(07)84018-0. [DOI] [PubMed] [Google Scholar]
- Lane AN, Fan TW-M, Xie X, Moseley HN, Higashi RM. Stable isotope analysis of lipid biosynthesis by high resolution mass spectrometry and NMR Anal. Chim. Acta. 2009;651:201–208. doi: 10.1016/j.aca.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-NP, Boros LG, Puigjaner J, Bassilian S, Lim S, Cascante M. Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1, 2–13C2]glucose. Am J Physiol Endocrinol Metab. 1998;274(5):E843–E851. doi: 10.1152/ajpendo.1998.274.5.E843. [DOI] [PubMed] [Google Scholar]
- Lindon JC, Holmes E, Nicholson JK. Metabonomics: Systems biology in pharmaceutical research and development. Current Opinion in Molecular Therapeutics. 2004;6(3):265–272. [PubMed] [Google Scholar]
- Lu D, Mulder H, Zhao P, Burgess SC, Jensen MV, Kamzolova S, Newgard CB, Sherry AD. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion (GSIS). Proc Natl Acad Sci USA. 2002;99(5):2708–2713. doi: 10.1073/pnas.052005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom P, Teilum K, Carstensen T, Bezsonova I, Wiesner S, Hansen DF, Religa TL, Akke M, Kay LE. Fractional C-13 enrichment of isolated carbons using [1-C-13]- or [2-C-13]-glucose facilitates the accurate measurement of dynamics at backbone C-alpha and side-chain methyl positions in proteins. J Biomol NMR. 2007;38(3):199–212. doi: 10.1007/s10858-007-9158-6. [DOI] [PubMed] [Google Scholar]
- Maaheimo H, Fiaux J, Cakar ZP, Bailey JE, Sauer U, Szyperski T. Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional C-13 labeling of common amino acids. Eur J Biochem. 2001;268(8):2464–2479. doi: 10.1046/j.1432-1327.2001.02126.x. [DOI] [PubMed] [Google Scholar]
- Nagy M, Lacroute F, Thomas D. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc Natl Acad Sci U S A. 1992;89(19):8966–8970. doi: 10.1073/pnas.89.19.8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portais JC, Voisin P, Merle M, Canioni P. Glucose and glutamine metabolism in C6 glioma cells studied by carbon 13 NMR. Biochimie. 1996;78(3):155–164. doi: 10.1016/0300-9084(96)89500-9. [DOI] [PubMed] [Google Scholar]
- Qin J, Clore GM, Gronenborn AM. Ionization equilibria for side-chain carboxyl groups in oxidized and reduced human thioredoxin and in the complex with its target peptide from the transcription factor NF kappa B. Biochemistry. 1996;35(1):7–13. doi: 10.1021/bi952299h. [DOI] [PubMed] [Google Scholar]
- Rittenberg D, Schoenheimer R. Deuterium as an indicator in study of intermediatory metabolism. VIII. Hydrogenation of fatty acids in the animals organism. J. Biol. Chem. 1937;117:485–490. [Google Scholar]
- Sherry AD, Jeffrey FMH, Malloy CR. Analytical solutions for C-13 isotopomer analysis of complex metabolic conditions: substrate oxidation, multiple pyruvate cycles, and gluconeogenesis. Metabolic Engineering. 2004;6(1):12–24. doi: 10.1016/j.ymben.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6(2):469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- Vizan P, Alcarraz-Vizan G, Diaz-Moralli S, Rodriguez-Prados JC, Zanuy M, Centelles JJ, Jauregui O, Cascante M. Quantification of intracellular phosphorylated carbohydrates in HT29 human colon adenocarcinoma cell line using liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2007;79(13):5000–5005. doi: 10.1021/ac070170v. [DOI] [PubMed] [Google Scholar]
- Yuneva M. Finding an “Achilles’ heel” of cancer: the role of glucose and glutamine metabolism in the survival of transformed cells. Cell Cycle. 2008;7:2083–2089. doi: 10.4161/cc.7.14.6256. [DOI] [PubMed] [Google Scholar]