Abstract
Background
Low-risk breast cancer susceptibility alleles or SNPs confer only modest breast cancer risks ranging from just over 1.0 to1.3 fold. Yet, they are common among most populations and therefore are involved in the development of essentially all breast cancers. The mechanism by which the low-risk SNPs confer breast cancer risks is currently unclear. The breast cancer association consortium BCAC has hypothesized that the low-risk SNPs modulate expression levels of nearby located genes.
Methods
Genotypes of five low-risk SNPs were determined for 40 human breast cancer cell lines, by direct sequencing of PCR-amplified genomic templates. We have analyzed expression of the four genes that are located nearby the low-risk SNPs, by using real-time RT-PCR and Human Exon microarrays.
Results
The SNP genotypes and additional phenotypic data on the breast cancer cell lines are presented. We did not detect any effect of the SNP genotypes on expression levels of the nearby-located genes MAP3K1, FGFR2, TNRC9 and LSP1.
Conclusion
The SNP genotypes provide a base line for functional studies in a well-characterized cohort of 40 human breast cancer cell lines. Our expression analyses suggest that a putative disease mechanism through gene expression modulation is not operative in breast cancer cell lines.
Background
About ten percent of breast cancer patients have a history of multiple breast cancer cases in their family, suggesting the inheritance of breast cancer susceptibility alleles in these families. Germline mutations in the BRCA1 and BRCA2 genes are identified in about one quarter of the families with breast cancer. Female carriers of BRCA1 and BRCA2 mutations have an estimated 50–90% life-time risk to develop breast cancer, classifying both genes as high-risk susceptibility genes [1,2]. Other high-risk breast cancer genes include the p53, PTEN and STK11 genes, but mutations in these genes account for only few familial breast cancers. CHEK2 was the first moderate-risk breast cancer gene being identified [3-5]. Germline mutations in CHEK2 are identified in up to 5% of breast cancer families, albeit that their prevalence varies widely among populations. Female carriers of CHEK2 mutations have a moderate two to three fold increased risk to develop breast cancer. By now, several other moderate-risk breast cancer genes have been identified, including ATM, BRIP1 and PALB2 [6-9]. Mutations in these genes all confer increased breast cancer risks of two to three fold and mutations in each of these genes are identified in about 1% of the familial breast cancers. Recently, the international breast cancer association consortium (BCAC) has conducted a large genome-wide association study and identified five single nucleotide polymorphisms (SNPs) that associated with breast cancer [10]. Four of these SNPs were within haplotype blocks that contained genes: SNP rs2981582 locates in intron 2 of the FGFR2 gene at chromosome 10q; SNP rs889312 locates near MAP3K1 at 5q; SNP rs3803662 locates between TNRC9 and the LOC643714 gene at 16q; and SNP rs3817198 locates intronic in LSP1 at 11p. SNP rs13281615 locates at 8q24 in a region without any annotated genes. Importantly, independent genome-wide association studies have associated other SNPs in FGFR2 with breast cancer [11,12]. As FGFR2 had already been implicated in breast cancer [13-20], the significance of the FGFR2 SNPs as susceptibility alleles seemed evident. The TNRC9 SNP had also been associated with breast cancer in another study [21]. Lastly, the 8q24 SNP was of particular interest because other SNPs at 8q24 had been associated with increased risks of prostate cancer and colorectal cancer [22-26]. BCAC estimated that each of the five identified SNPs associated with rather small increased breast cancer risks, ranging from just over 1.0 to 1.3 fold, classifying them as low-risk susceptibility alleles [10]. However, these low-risk SNPs are very common and their impact is therefore still substantial, together accounting for almost 5% of the familial breast cancers.
The mechanism by which the low-risk susceptibility alleles confer breast cancer risks was obscure [10]. In analogy with the high-risk and moderate-risk breast cancer genes, it had been anticipated that the identified SNPs associated with disease-causing alleles in the coding sequences of nearby located genes. However, extensive sequencing efforts have not identified such alleles in the SNP-associated haplotype blocks, suggesting that the SNPs themselves might be the disease-causing susceptibility alleles [10]. BCAC therefore proposed an alternative disease mechanism that involves expression modulation of genes located in the vicinity of the identified SNPs, thereby conferring low breast cancer risks. Here, we have evaluated expression modulation in a well-characterized cohort of 40 human breast cancer cell lines, allowing us to specifically address whether this mechanism might operate in breast cancer cells.
Methods
Breast cancer cell lines
The 40 human breast cancer cell lines used in this study are listed in Table 1 and have been described in detail elsewhere [27]. Microsatellite analysis with nearly 150 polymorphic markers had shown that all cell lines are unique and monoclonal [28].
Table 1.
Genotypes of five low-risk SNPs in 40 human breast cancer cell lines
| Breast cancer cell lines | SNP genotypes and allelic losses | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 8q24 | Loss 8q | MAP3K1 | Loss 5q | FGFR2 | Loss 10q | TNRC9 | Loss 16q | LSP1 | Loss 11p | |
| SUM185PE | Het | No | Het | No | Maj H | nd | Min H | nd | Maj H | nd |
| BT483 | Het | No | Maj H | No | Min H | Yes | Maj H | No | Maj H | No |
| MDA-MB-134VI | Het | No | Het | No | Het | No | Het | No | Maj H | No |
| MDA-MB-175VII | Het | No | Min H | No | Het | No | Het | No | Maj H | No |
| MDA-MB-415 | Het | No | Het | No | Min H | Yes | Het | No | Het | No |
| MPE600 | Het | No | Maj H | nd | Het | No | Maj H | nd | Het | No |
| SUM52PE | Maj H | nd | Maj H | nd | Maj H | nd | Maj H | nd | Maj H | nd |
| CAMA-1 | Het | No | Het | No | Min H | No | Maj H | Yes | Maj H | Yes |
| MCF-7 | Maj H | No | Het | No | Maj H | No | Het | No | Maj H | Yes |
| ZR75-1 | Het | No | Het | No | Min H | Yes | Maj H | Yes | Het | No |
| SUM44PE | Het | No | Min H | nd | Maj H | nd | Het | No | Min H | nd |
| T47D | Het | No | Het | No | Maj H | No | Min H | Yes | Min H | Yes |
| MDA-MB-361 | Het | No | Het | No | Maj H | No | Het | No | Min H | No |
| BT474 | Het | No | Maj H | No | Maj H | Yes | Maj H | No | Maj H | No |
| UACC812 | Min H | No | Min H | No | Het | No | Het | No | Min H | Yes |
| ZR75-30 | Maj H | nd | Min H | nd | Min H | nd | Min H | nd | Maj H | nd |
| OCUB-F | Maj H | nd | Maj H | nd | Min H | nd | Min H | nd | Maj H | nd |
| SK-BR-5 | Het | No | Maj H | nd | Min H | nd | Min H | nd | Maj H | nd |
| SUM190PT | Min H | nd | Min H | nd | Maj H | nd | Min H | nd | Maj H | nd |
| SUM225CWN | Het | No | Maj H | nd | Maj H | nd | Maj H | nd | Het | No |
| MDA-MB-330 | Het | No | Min H | Yes | Min H | Yes | Het | No | Maj H | No |
| MDA-MB-453 | Het | No | Het | No | Min H | Yes | Maj H | Yes | Maj H | Yes |
| SK-BR-3 | Min H | Yes | Het | No | Het | No | Maj H | Yes | Min H | No |
| EVSA-T | Min H | nd | Min H | nd | Maj H | nd | Maj H | nd | Min H | nd |
| UACC893 | Maj H | No | Maj H | No | Maj H | Yes | Het | No | Maj H | No |
| BT20 | Min H | No | Maj H | Yes | Maj H | Yes | Maj H | Yes | Het | No |
| HCC1937 | Het | No | Min H | nd | Het | No | Het | No | Maj H | nd |
| MDA-MB-468 | Maj H | Yes | Maj H | Yes | Maj H | Yes | Maj H | Yes | Maj H | Yes |
| SUM149PT | Min H | nd | Het | No | Min H | nd | Min H | nd | Maj H | Yes |
| SUM229PE | Maj H | nd | Maj H | nd | Maj H | nd | Maj H | nd | Min H | nd |
| BT549 | Maj H | No | Maj H | No | Maj H | Yes | Het | No | Min H | Yes |
| Hs578T | Het | No | Maj H | Yes | Min H | Yes | Min H | No | Maj H | No |
| MDA-MB-157 | Maj H | No | Maj H | Yes | Maj H | Yes | Min H | No | Maj H | Yes |
| MDA-MB-231 | Maj H | Yes | Het | No | Het | No | Maj H | Yes | Het | No |
| MDA-MB-436 | Maj H | Yes | Maj H | Yes | Maj H | No | Min H | No | Maj H | Yes |
| SK-BR-7 | Het | No | Het | No | Min H | nd | Maj H | nd | Het | No |
| SUM159PT | Maj H | nd | Het | No | Min H | nd | Het | No | Maj H | nd |
| SUM1315MO2 | Het | No | Maj H | nd | Min H | nd | Het | No | Maj H | nd |
| SUM102PT | Het | No | Het | No | Maj H | nd | Het | No | Maj H | nd |
| MDA-MB-435s | Maj H | No | Maj H | Yes | Maj H | Yes | Maj H | Yes | Maj H | No |
| Total major homozygotes | 13 | 17 | 19 | 16 | 25 | |||||
| Total heterozygotes | 21 | 15 | 7 | 14 | 7 | |||||
| Total minor homozygotes | 6 | 8 | 14 | 10 | 8 | |||||
| Percentage of allelic loss | 13 | 25 | 52 | 32 | 37 | |||||
Genotypes of five low-risk SNPs have been determined in the current study. Allelotype data have been reported elsewhere and involved microsatellite analysis. Loss: Yes, allelic loss at the indicated chromosomal region; and No, no allelic loss at the indicated chromosomal region. nd, not determined; Maj H, major homozygotes; Min H, minor homozygotes; Het, heterozygote allele carriers.
Genotyping
Genotypes of five low-risk susceptibility alleles have been determined: rs889312 (A>C) near the MAP3K1 gene; rs2981582 (C>T) in the FGFR2 gene; rs3803662 (C>T) near the TNRC9 gene; rs3817198 (T>C) in the LSP1 gene and rs13281615 (A>G) that located in a gene desert at chromosome 8q24 [10]. Genotyping was performed by direct sequencing of PCR-amplified genomic templates, using the BigDye Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems) and an ABI 3130xL Genetic Analyzer. Primer sequences are available upon request.
Allele frequencies of cases and controls reported by BCAC have been obtained by using their reported Odds Ratio data [10], and inferring allele frequencies by assuming that Odds Ratios reflect the ratio of minor allele carriers versus major allele carriers from the cases divided by the ratio of minor allele carriers versus major allele carriers from the controls.
Expression analysis
Transcript expression levels of four genes have been determined: MAP3K1, FGFR2, TNRC9 and LSP1. Quantitative real-time PCR (qPCR) was performed on cDNA templates that had been generated with oligo-dT and random hexamer primers from total RNA isolates, using Power SYBR Green PCR Master Mix (Applied Biosystems) and an ABI Prism 7700. Ct values were normalized according HPRT and HMBS housekeeper Ct values. Transcript expression had also been determined by Human Exon 1.0 ST microarrays (Affymetrix), as described elsewhere [29]. The exon array data have been deposited in NCBI's Gene Expression Omnibus [30] and are accessible through GEO Series accession number GSE16732.
Statistical analysis
Statistical analyses were performed with Statistical Package for the Social Sciences (SPSS) version 11.5, considering P-values of less than 0.05 significant. Fisher's exact test was used to determine association of the SNP genotypes with the breast cancer cell lines. The Kruskal Wallis test was used to compare gene expression levels among three SNP genotype groups (major homozygotes, heterozygotes, and minor homozygotes).
Results and discussion
Genotyping of low-risk susceptibility alleles in breast cancer cell lines
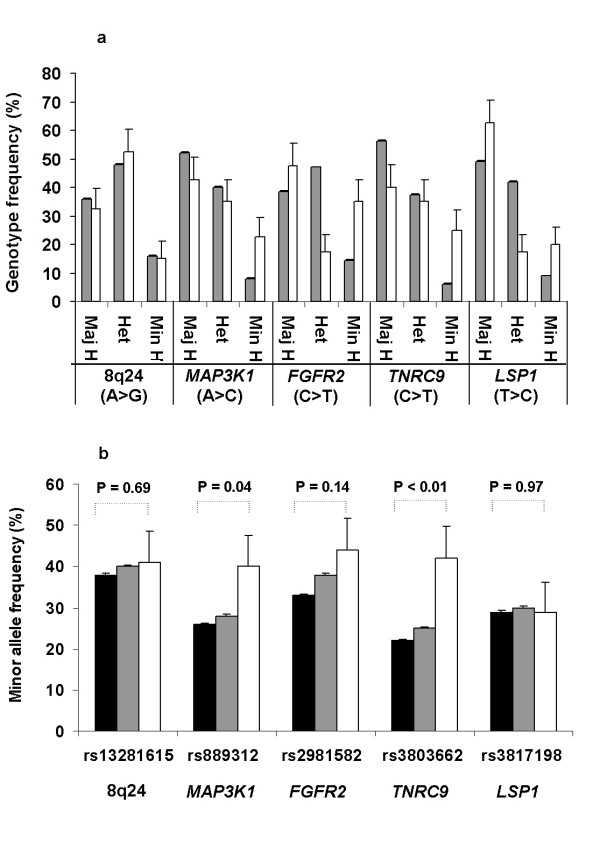
Genotypes of five low-risk susceptibility alleles [10] were determined in a cohort of 40 human breast cancer cell lines. For each SNP, frequencies of major homozygotes, heterozygotes and minor homozygotes are shown in Figure 1a and genotypes are detailed in Table 1. Frequencies of homozygote genotypes typically were higher than anticipated, likely related to allelic losses in the cell line samples (Figure 1a; [10]). For four SNPs (8q24, MAP3K1, FGFR2 and TNRC9), the minor allele frequencies among the cell lines were higher than among the 21,860 BCAC breast cancer cases and 22,578 population controls (Figure 1b; [10]). Fisher's exact testing indicated that the minor allele frequencies among the cell lines were significantly higher than the BCAC population controls for two SNPs: MAP3K1 and TNRC9 (Figure 1b). In Table 1 and 2, we also included previously-determined phenotypic and genotypic data on the breast cancer cell lines, including data on molecular subtyping and allelotyping (Hollestelle et al. submitted for publication; [28]). Together with the SNP genotypes, we provide a base line for functional studies in this cohort of breast cancer cell lines.
Figure 1.
Genotypes and minor allele frequencies of five low-risk breast cancer susceptibility alleles or SNPs in human breast cancer cell lines. 1a. Gray bars represent SNP genotype frequencies of 21,860 blood-derived samples from breast cancer cases reported by the breast cancer association consortium BCAC [10], and white bars represent genotype frequencies in 40 breast cancer cell lines. Maj H, major homozygotes; Min H, minor homozygotes; and Het, heterozygote allele carriers. The major and minor alleles of each allele are indicated between brackets. 1b. Black and gray bars represent minor allele frequencies in 22,578 population controls and 21,860 breast cancer cases, respectively, as reported by BCAC [10]. White bars represent frequencies identified in 40 breast cancer cell lines.
Table 2.
Molecular and phenotypic characterizations of 40 breast cancer cell lines
| Breast cancer cell lines | Breast cancer subtype | Intrinsic subtype | Protein expression | |||
|---|---|---|---|---|---|---|
| ER | PgR | ERBB2 | 3-neg | |||
| SUM185PE | Luminal-type | Luminal | - | - | - | + |
| BT483 | Luminal-type | Luminal | + | - | + | - |
| MDA-MB-134VI | Luminal-type | Luminal | + | - | - | - |
| MDA-MB-175VII | Luminal-type | Luminal | + | - | - | - |
| MDA-MB-415 | Luminal-type | Luminal | + | - | - | - |
| MPE600 | Luminal-type | Luminal | + | - | + | - |
| SUM52PE | Lumina-type | Luminal | + | - | + | - |
| CAMA-1 | Lumina-type | Luminal | + | + | + | - |
| MCF-7 | Luminal-type | Luminal | + | + | - | - |
| ZR75-1 | Luminal-type | Luminal | + | + | + | - |
| SUM44PE | Luminal-type | Luminal | + | + | - | - |
| T47D | Luminal-type | Luminal | + | + | - | - |
| MDA-MB-361 | Luminal-type | Luminal | + | + | ++ | - |
| BT474 | Luminal-type | Luminal | - | + | ++ | - |
| UACC812 | Luminal-type | Luminal | - | + | ++ | - |
| ZR75-30 | Luminal-type | Luminal | + | - | ++ | - |
| OCUB-F | Luminal-type | Luminal | - | - | ++ | - |
| SK-BR-5 | Luminal-type | Luminal | - | - | ++ | - |
| SUM190PT | Luminal-type | nd | - | - | ++ | - |
| SUM225CWN | Luminal-type | nd | - | - | ++ | - |
| MDA-MB-330 | Luminal-type | ERBB2 | + | - | ++ | - |
| MDA-MB-453 | Luminal-type | ERBB2 | - | - | ++ | - |
| SK-BR-3 | Luminal-type | ERBB2 | - | - | ++ | - |
| EVSA-T | Luminal-type | ERBB2 | - | - | ++ | - |
| UACC893 | Luminal-type | ERBB2 | - | - | ++ | - |
| BT20 | Basal-type | Basal-like | - | - | - | + |
| HCC1937 | Basal-type | Basal-like | - | - | - | + |
| MDA-MB-468 | Basal-type | Basal-like | - | - | - | + |
| SUM149PT | Basal-type | Basal-like | - | - | - | + |
| SUM229PE | Basal-type | Basal-like | - | - | - | + |
| BT549 | Basal-type | Normal-like | - | - | - | + |
| Hs578T | Basal-type | Normal-like | - | - | - | + |
| MDA-MB-157 | Basal-type | Normal-like | - | - | - | + |
| MDA-MB-231 | Basal-type | Normal-like | - | - | - | + |
| MDA-MB-436 | Basal-type | Normal-like | - | - | - | + |
| SK-BR-7 | Basal-type | Normal-like | - | - | - | + |
| SUM159PT | Basal-type | Normal-like | - | - | - | + |
| SUM1315MO2 | Basal-type | Normal-like | - | - | - | + |
| SUM102PT | Basal-type | Normal-like | nd | nd | nd | nd |
| MDA-MB-435s | Basal-type | Normal-like | - | - | - | + |
| Total phenotype positives | 14 | 8 | 13 | 16 | ||
Phenotypic characterizations have been reported elsewhere and involved protein expression patterns of the cell lines for the breast cancer subtyping (cytokeratins, ER, PgR and ERBB2) and expression of the intrinsic gene set for the intrinsic subtyping (Hollestelle et al. submitted for publication). Protein expression: +, expressed; ++, over expressed; and -, not detectable; 3-neg, triple-negative.
Expression levels of nearby located genes in breast cancer cell lines do not correlate with their SNP genotype
Surprisingly, BCAC had not identified disease-causing gene variants within the haplotype blocks of the five low-risk SNPs [10]. They proposed an alternative disease mechanism, in which SNP genotypes modulate expression levels of nearby located genes. Such disease mechanism was conceivable because the minor SNP alleles confer only low risks for breast cancer. Here, we have evaluated whether gene expression modulation is operative in breast cancer cell lines, by associating SNP genotypes of the breast cancer cell lines with the expression levels of nearby located genes.
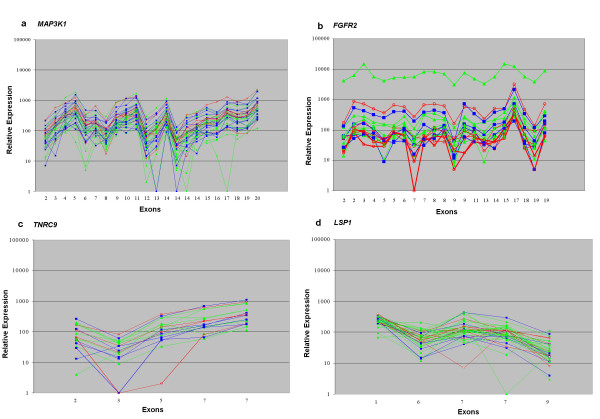
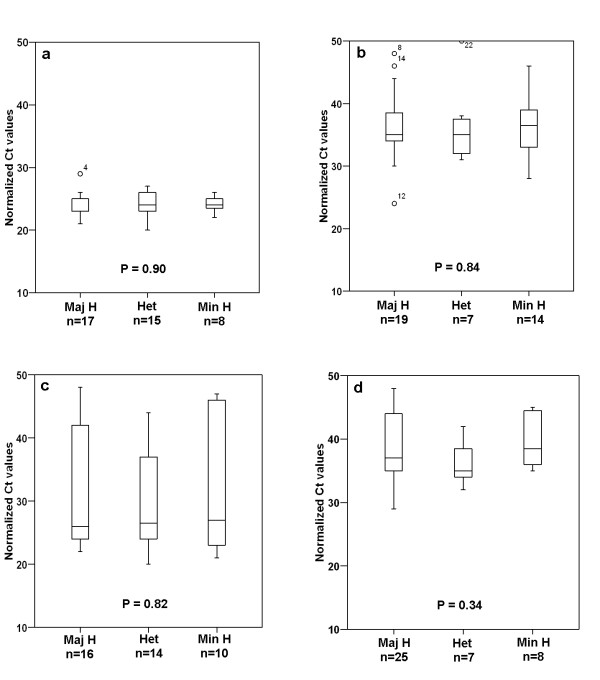
Gene expression data of the four genes physically nearest to the SNPs were obtained by Affymetrix Human Exon 1.0 ST microarray profiling and by qPCR analysis. Both transcript expression analysis methods revealed similar expression levels for each of the four genes: MAP3K1, FGFR2, TNRC9 and LSP1, with Spearman correlation coefficients of -0.6, -0.7, -0.8 and -0.4, respectively, among the 40 breast cancer cell lines (Table 3 and Figures 2 and 3). Because BCAC had shown that the low-risk SNPs confer breast cancer risks in a dose-dependent manner, with the highest risks for the minor homozygotes [10], association between gene expression levels and SNP genotypes was performed by three-group comparisons. Exon array data are shown in Figure 2, with cell lines from each genotype group depicted in a different color. Unique outliers typically represented decreased expression of one or more probes sets, such as exon 17 of MAP3K1 or exons 3–5 of TNRC9, possibly related to the presence of SNPs in probe sequences, alternative splicing or genomic deletions [29]. Expression of recurrent isoforms as reported by NCBI was detected only for the FGFR2 gene, with two cell lines expressing the isoform that lacked exon 9. Both cell lines were minor homozygotes for the FGFR2 SNP. Overall, there was no apparent association between the exon array expression level of each of the four genes and their SNP genotypes (Figure 2). The qPCR Ct-values are detailed in Table 3 and the three-group comparisons are shown in Figure 3. Again, we did not detect any association between gene expression levels with SNP genotypes for the four genes. It is possible that gene expression levels are affected by allelic loss of the gene loci. We therefore also have compared gene expression levels in major and minor homozygotes with allelic loss to the gene expression levels in cell lines without allelic loss, but gene expression levels did not correlate with allelic losses either (Table 4). Altogether, these results strongly suggest that a putative disease mechanism by expression modulation does not operate via cancer cells. Yet, recent studies have shown that expression levels of the FGFR2, MAP3K1 and TNRC9 genes associated with their SNP genotype in clinical breast cancer samples [31,32]. It may be that expression modulation is operative in non-neoplastic stromal or epithelial cells and perhaps only early in carcinogenesis. Alternatively, it may be that expression modulation of these genes was operative in invasive breast cancer cells but was lost upon in vitro propagation of the cell lines. Expression analysis of carefully dissected tumor cells and non-neoplastic epithelial and stromal cells from clinical breast cancer samples should resolve this issue and may determine the precise mechanism of expression modulation by low-risk breast cancer susceptibility alleles.
Table 3.
Gene expression analysis of MAP3K1, FGFR2, TNRC9 and LSP1 in 40 human breast cancer cell lines by quantitative RT-PCR, represented by normalized Ct values
| Breast cancer cell lines | Transcript expression (normalized Ct values) | |||
|---|---|---|---|---|
| MAP3K1 | FGFR2 | TNRC9 | LSP1 | |
| BT20 | 23 | 30 | 45 | 38 |
| BT474 | 24 | 35 | 22 | 35 |
| BT483 | 22 | 31 | 25 | 36 |
| BT549 | 29 | 32 | 39 | 40 |
| CAMA-1 | 24 | 33 | 26 | 45 |
| EVSA-T | 25 | 33 | 25 | 45 |
| HCC1937 | 26 | 32 | 37 | 45 |
| Hs578T | 26 | 45 | 45 | 34 |
| MCF-7 | 25 | 34 | 28 | 39 |
| MDA-MB-134VI | 22 | 37 | 24 | 35 |
| MDA-MB-157 | 25 | 44 | 33 | 29 |
| MDA-MB-175VII | 24 | 35 | 23 | 36 |
| MDA-MB-231 | 26 | 45 | 45 | 35 |
| MDA-MB-330 | 26 | 32 | 27 | 33 |
| MDA-MB-361 | 23 | 34 | 23 | 44 |
| MDA-MB-415 | 23 | 28 | 20 | 33 |
| MDA-MB-435s | 25 | 45 | 45 | 45 |
| MDA-MB-436 | 26 | 35 | 31 | 37 |
| MDA-MB-453 | 24 | 36 | 29 | 45 |
| MDA-MB-468 | 25 | 35 | 37 | 37 |
| MPE600 | 23 | 31 | 22 | 42 |
| OCUB-F | 23 | 39 | 23 | 42 |
| SK-BR-3 | 25 | 32 | 26 | 37 |
| SK-BR-5 | 23 | 37 | 21 | 43 |
| SK-BR-7 | 26 | 39 | 45 | 35 |
| SUM102PT | 25 | 35 | 32 | 29 |
| SUM1315M02 | 26 | 43 | 44 | 35 |
| SUM149PT | 27 | 38 | 45 | 37 |
| SUM159PT | 27 | 45 | 43 | 34 |
| SUM185PE | 23 | 38 | 23 | 45 |
| SUM190PT | 24 | 45 | 23 | 45 |
| SUM225CWN | 24 | 36 | 23 | 39 |
| SUM229PE | 25 | 39 | 33 | 36 |
| SUM44PE | 22 | 41 | 26 | 36 |
| SUM52PE | 24 | 24 | 24 | 37 |
| T47D | 20 | 36 | 45 | 35 |
| UACC812 | 24 | 38 | 24 | 45 |
| UACC893 | 21 | 36 | 25 | 36 |
| ZR75-1 | 24 | 36 | 24 | 32 |
| ZR75-30 | 23 | 33 | 23 | 43 |
| Total high expressers (Ct <20) | 1 | 0 | 1 | 0 |
| Total moderate expressers (Ct 20–30) | 39 | 2 | 23 | 2 |
| Total low expressers (Ct >30–35) | 0 | 12 | 4 | 5 |
| Total no expressers (Ct >35) | 0 | 26 | 12 | 33 |
Figure 2.
Normalized expression levels from Affymetrix Human Exon 1.0 ST microarrays of 2a. MAP3K1; 2b. FGFR2; 2c. TNRC9; and 2d. LSP1; from 40 human breast cancer cell lines. Kruskal Wallis testing using the average expression among all probe sets for each gene did not reveal significant associations between gene expression and SNP genotypes. Each line represents a cell line, with the color-coding according the genotype groups: green, major homozygotes; red, minor homozygotes; and blue, heterozygotes. Two cell lines with the delEx9 isoform of FGFR2 are indicated with bold red lines. Probe sets for each gene were ordered by physical location and indicated by exon, where probe sets that were not unique for that gene were omitted. Probe sets with expression values less than the background of 50 were also omitted, unless more than 3 cell lines had expression levels higher than 100.
Figure 3.
Correlation of gene expression levels of 3a. MAP3K1; 3b. FGFR2; 3c. TNRC9; and 3d. LSP1; with the SNP genotypes in 40 human breast cancer cell lines. Kruskal Wallis testing did not reveal any significant association between gene expression and SNP genotypes. Maj H, major homozygotes; Min H, minor homozygotes; and Het, heterozygote allele carriers. The number of cell lines in each genotype group is indicated under the genotypes and data are detailed in Table 1.
Table 4.
Gene expression of MAP3K1, FGFR2, TNRC9 and LSP1 in human breast cancer cell lines according to their allelic loss status at the gene locus
| Gene | Genotype (Number of cell lines) |
Average expression level (Normalized Ct values) |
|---|---|---|
| MAP3K1 | Het (n = 14) | 24 ± 2 |
| Maj H no loss (n = 4) | 24 ± 4 | |
| Min H no loss (n = 2) | 24 ± 0 | |
| Maj H allelic loss (n = 1) | 26 | |
| Min H allelic loss (n = 6) | 25 ± 1 | |
| FGFR2 | Het (n = 7) | 36 ± 5 |
| Maj H no loss (n = 4) | 31 ± 7 | |
| Min H no loss (n = 1) | 33 | |
| Maj H allelic loss (n = 6) | 35 ± 6 | |
| Min H allelic loss (n = 6) | 38 ± 6 | |
| TNRC9 | Het (n = 14) | 30 ± 8 |
| Maj H no loss (n = 2) | 24 ± 2 | |
| Min H no loss (n = 3) | 36 ± 8 | |
| Maj H allelic loss (n = 1) | 45 | |
| Min H allelic loss (n = 7) | 36 ± 9 | |
| LSP1 | Het (n = 7) | 37 ± 4 |
| Maj H no loss (n = 8) | 37 ± 4 | |
| Min H no loss (n = 2) | 41 ± 5 | |
| Maj H allelic loss (n = 3) | 40 ± 5 | |
| Min H allelic loss (n = 7) | 38 ± 6 | |
Although numbers are small for some sample groups, there are no apparent differences in gene expression levels related to allelic loss status. Allelic loss data and qPCR expression data are detailed in Table 1 and Table 3, respectively. Maj H, major homozygotes; Min H, minor homozygotes; Het, heterozygote allele carriers.
Conclusion
We present the genotypes of five low-risk susceptibility alleles or SNPs of 40 human breast cancer cell lines. Using this cell line model, we have evaluated the BCAC hypothesis that low-risk SNPs confer breast cancer risks by modulation of expression levels of nearby located genes. We found no evidence for expression modulation in the breast cancer cell lines, suggesting that such disease mechanism is more likely to operate in non-neoplastic epithelial or stromal cells or has been lost during in vitro propagation of the cell lines.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MR and FE carried out genotyping and transcript expression analyses, AH carried out protein expression analyses, and MR and AD performed statistical analyses. MR, JGMK and MS designed the study, and MR and MS wrote the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Muhammad Riaz, Email: m.riaz@erasmusmc.nl.
Fons Elstrodt, Email: f.elstrodt@bioke.com.
Antoinette Hollestelle, Email: a.hollestelle@erasmusmc.nl.
Abbas Dehghan, Email: a.dehghan@erasmusmc.nl.
Jan GM Klijn, Email: j.g.m.klijn@erasmusmc.nl.
Mieke Schutte, Email: a.schutte@erasmusmc.nl.
Acknowledgements
MR is funded by the Higher Education Commission (HEC) of the Government of Pakistan. Financial support was provided by grants from the Susan G. Komen Breast Cancer Foundation (BCT R0601309) and Erasmus MC Mrace 2007.
References
- Thompson D, Easton D. The genetic epidemiology of breast cancer genes. J Mammary Gland Biol Neoplasia. 2004;9(3):221–236. doi: 10.1023/B:JOMG.0000048770.90334.3b. [DOI] [PubMed] [Google Scholar]
- Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11(2):103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Meijers-Heijboer H, Ouweland A van den, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, van Veghel-Plandsoen M. et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31(1):55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomaki K, Heikkila P. et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71(2):432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium CBCC-C. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74(6):1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K. et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38(8):873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K. et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38(11):1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T. et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40(1):17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Kirchhoff T, Stefanov S, Lautenberger J, Viale A, Garber J, Friedman E, Narod S, Olshen AB, Gregersen P. et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA. 2008;105(11):4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnane J, Gaudray P, Dionne CA, Crumley G, Jaye M, Schlessinger J, Jeanteur P, Birnbaum D, Theillet C. BEK and FLG, two receptors to members of the FGF family, are amplified in subsets of human breast cancers. Oncogene. 1991;6(4):659–663. [PubMed] [Google Scholar]
- Luqmani YA, Graham M, Coombes RC. Expression of basic fibroblast growth factor, FGFR1 and FGFR2 in normal and malignant human breast, and comparison with other normal tissues. Br J Cancer. 1992;66(2):273–280. doi: 10.1038/bjc.1992.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penault-Llorca F, Bertucci F, Adelaide J, Parc P, Coulier F, Jacquemier J, Birnbaum D, deLapeyriere O. Expression of FGF and FGF receptor genes in human breast cancer. Int J Cancer. 1995;61(2):170–176. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- Dickson C, Spencer-Dene B, Dillon C, Fantl V. Tyrosine kinase signalling in breast cancer: fibroblast growth factors and their receptors. Breast Cancer Res. 2000;2(3):191–196. doi: 10.1186/bcr53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiskanen M, Kononen J, Barlund M, Torhorst J, Sauter G, Kallioniemi A, Kallioniemi O. CGH, cDNA and tissue microarray analyses implicate FGFR2 amplification in a small subset of breast tumors. Anal Cell Pathol. 2001;22(4):229–234. doi: 10.1155/2001/981218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffa AB, Tannheimer SL, Ethier SP. Transforming potential of alternatively spliced variants of fibroblast growth factor receptor 2 in human mammary epithelial cells. Mol Cancer Res. 2004;2(11):643–652. [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C. et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, Masson G, Jakobsdottir M, Thorlacius S, Helgason A. et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A. et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ. et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39(5):638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39(8):954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W. et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39(8):984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E. et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39(8):989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- Elstrodt F, Hollestelle A, Nagel JH, Gorin M, Wasielewski M, Ouweland A van den, Merajver SD, Ethier SP, Schutte M. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66(1):41–45. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- Harkes IC, Elstrodt F, Dinjens WN, Molier M, Klijn JG, Berns EM, Schutte M. Allelotype of 28 human breast cancer cell lines and xenografts. Br J Cancer. 2003;89(12):2289–2292. doi: 10.1038/sj.bjc.6601448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte M, Elstrodt F, Bralten LB, Nagel JH, Duijm E, Hollestelle A, Vuerhard MJ, Wasielewski M, Peeters JK, Spek P van der. et al. Exon expression arrays as a tool to identify new cancer genes. PLoS ONE. 2008;3(8):e3007. doi: 10.1371/journal.pone.0003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgard SH, Johansen FE, Alnaes GI, Naume B, Borresen-Dale AL, Kristensen VN. Genes harbouring susceptibility SNPs are differentially expressed in the breast cancer subtypes. Breast Cancer Res. 2007;9(6):113. doi: 10.1186/bcr1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KB, Maia AT, O'Reilly M, Teschendorff AE, Chin SF, Caldas C, Ponder BA. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008;6(5):e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]