Abstract
As fundamental units of neuronal communication, chemical synapses are composed of presynaptic and postsynaptic specializations that form at specific locations with defined shape and size. Synaptic assembly must be tightly regulated to prevent overgrowth of the synapse size and number, but the molecular mechanisms that inhibit synapse assembly are poorly understood. We identified regulator of synaptogenesis–1 (RSY-1) as an evolutionarily conserved molecule that locally antagonized presynaptic assembly. The loss of RSY-1 in Caenorhabditis elegans led to formation of extra synapses and recruitment of excessive synaptic material to presynaptic sites. RSY-1 directly interacted with and negatively regulated SYD-2/liprin-alpha, a master assembly molecule that recruits numerous synaptic components to presynaptic sites. RSY-1 also bound and regulated SYD-1, a synaptic protein required for proper functioning of SYD-2. Thus, local inhibitory mechanisms govern synapse formation.
Synapse formation is a highly dynamic and regulated process. Although many positive factors that promote synaptogenesis have been identified (1–3), less is known about negative regulators of synapse formation and their mode of action (4–10). We investigated synapse development and its regulation in the hermaphrodite-specific neuron HSNL, one of a pair of motor neurons that controls egg-laying behavior in Caenorhabditis elegans (11). Presynaptic specializations in the HSNL neuron assemble within a spatially discrete location along the axon that is stereotyped between animals (Fig. 1, A and B) (12). These presynaptic sites were visualized by transgenically expressing fluorophore-tagged synaptic proteins such as synaptic vesicle component synaptobrevin (SNB-1) or active-zone components ERC/CAST/Bruchpilot (ELKS-1) and GIT-1 (Fig. 1B and fig. S1) (12, 13). SYD-1 and SYD-2 are essential for synapse development in the HSNL neuron; numerous synaptic components failed to localize to presynaptic sites in syd-1 and syd-2 mutants (Fig. 1, C and E, and fig. S1) (13, 14). However, increasing SYD-2 function in syd-1 null mutants, either by overexpressing SYD-2 or by introducing a gain-of-function (gf) mutation in syd-2, rescues the synaptic defects observed in syd-1 mutants (13, 14). Thus, under normal circumstances, SYD-1 is required for SYD-2 function. The existence of such positive regulators suggests that negative regulators of SYD-2 might counteract and balance the pro-synaptic function of SYD-1.
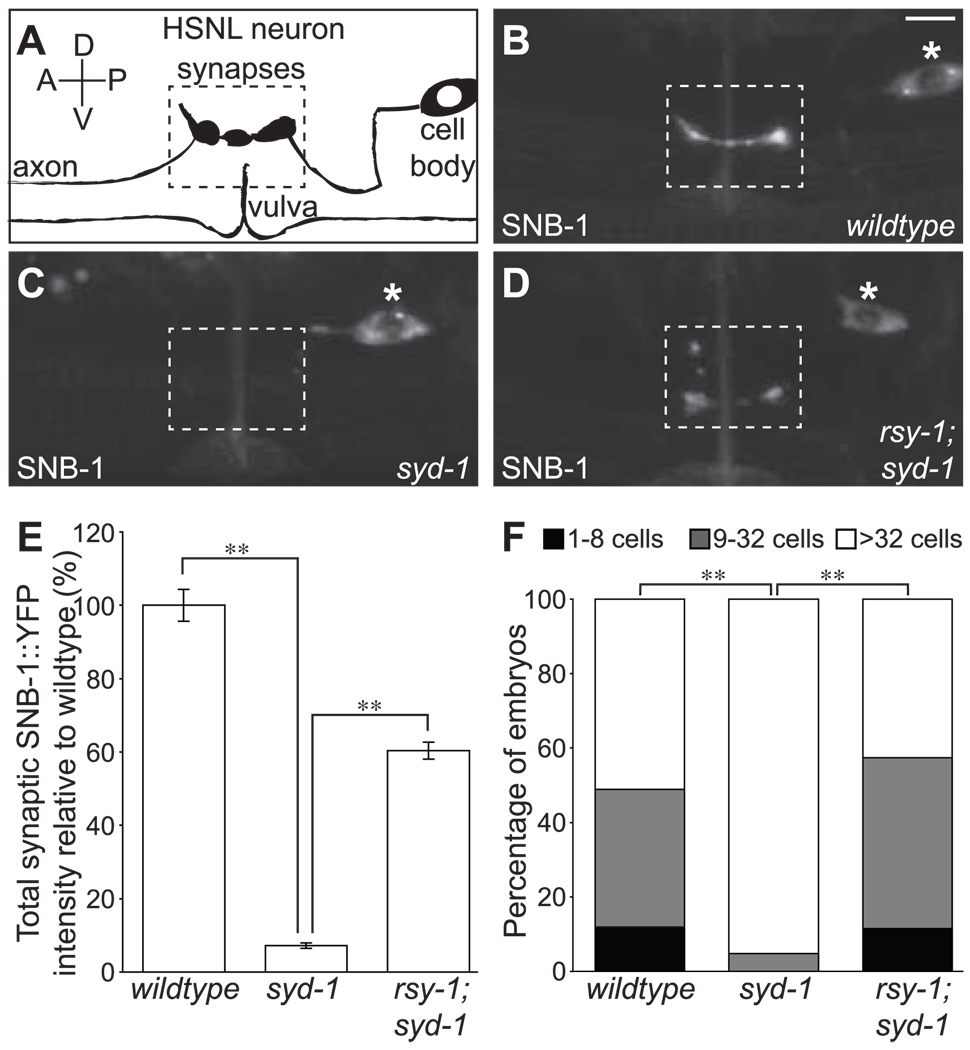
Fig. 1.
RSY-1 antagonizes the pro-synaptic function of SYD-1. (A) Schematic of the HSNL neuron. Synapses are shown in the dotted rectangle. Anterior, A; posterior, P; dorsal, D; ventral, V. (B) SNB-1::YFP is expressed in the HSNL in wildtype(N2), (C) syd-1(ju82), and (D) rsy-1(wy94);syd-1(ju82) mutants. All images are of adults. The dotted rectangle indicates the synaptic region, and the asterisk marks the HSNL cell body. Scale bar, 5 µm. (E) Total SNB-1::YFP intensity at the synaptic region normalized to wild type. **P < 0.01; Student’s t test; n > 20 per group. Error bars indicate SEM. (F) Proportion of eggs at a particular stage when laid. Scored double blind. **P < 0.01; Fisher’s exact test; n > 20 per group.
To isolate putative negative regulators of synaptogenesis, we performed a visual genetic screen formutants that suppress the synaptic defects in the HSNL of syd-1 mutants (15). We recovered two alleles of regulator of synaptogenesis–1 (rsy-1). In syd-1;rsy-1 double mutants, the accumulation of SNB-1::yellow fluorescent protein (YFP) in the synaptic region of the HSNL was significantly higher than in syd-1 single mutants (Fig. 1, C to E). The accumulation of SNB-1::YFP in these double mutants probably represents a restoration of functional synapses. First, active-zone components ELKS-1 and GIT-1 also localized at higher levels to presynaptic sites in rsy-1;syd-1 double mutants than in syd-1 single mutants (fig. S1). Second, defects in the egg-laying behavior in syd-1 mutants, which are a cell-autonomous consequence of the presynaptic assembly defects in the HSNL neuron, were rescued in rsy-1;syd-1 double mutants (Fig. 1F). Thus RSY-1 appears to counteract and balance the pro-synaptic function of SYD-1.
We mapped rsy-1 to the genetic locus Y53H1A.1, which encodes two isoforms. Isoform A encodes a 589–amino acid protein with a proline-rich region, a coiled-coil domain, and a serine/arginine–rich (SR) domain (fig. S2). The SR domain of RSY-1 contains multiple putative nuclear localization sequences (NLSs). Isoform B encodes a smaller 517–amino acid protein that lacks the SR domain, which is replaced by 22 amino acids with a single putative NLS that is unique to the B isoform (fig. S2). RSY-1 is well conserved in vertebrates with both a long and a short isoform in Mus musculus (fig. S2). The vertebrate homolog of RSY-1 interacts with pinin (16), a dual resident of the nucleus and the desmosome junction with proposed functions including cell adhesion, transcription, and splicing (17–20). Beyond its interaction with pinin, no function has yet been assigned to RSY-1.
Both alleles of rsy-1 that we isolated contained mutations that generate early stop codons, suggesting that they are likely to be null alleles (fig. S2). To determine where rsy-1 is expressed, we made transgenic animals with a synthetic operon in which expression of both RSY-1 and cytoplasmic green fluorescent protein (GFP) is under control of the rsy-1 promoter (fig. S3). RSY-1 was reproducibly expressed in the HSNL (Fig. 2A), as well as in other neurons and tissues (fig. S4). To determine whether RSY-1 functions cell-autonomously in the HSNL, we transgenically expressed isoform A of RSY-1 under the control of the unc-86 promoter, which only expresses in the HSNs in the vulva region (21). Expression of the Punc-86::rsy-1 transgene in rsy-1;syd-1 double mutants restored synaptic defects in the HSNL of syd-1 single mutants (Fig. 2B and fig. S5), consistent with a cell-autonomous role for RSY-1 in inhibiting presynaptic assembly.
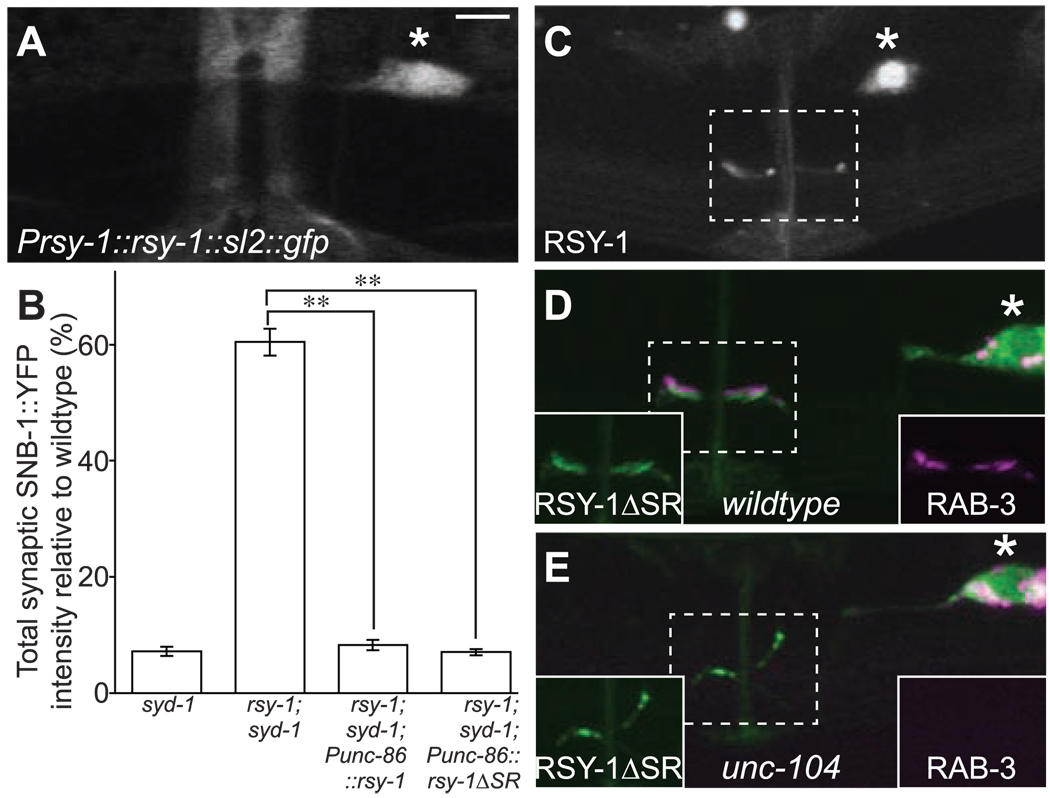
Fig. 2.
Molecular characterization of rsy-1. (A) rsy-1::sl2::gfp expressed under the rsy-1 promoter, which consists of 580 base pairs upstream of the rsy-1 start site. (B) Total SNB-1::YFP intensity at the synaptic region normalized to wild type. Punc-86::rsy-1 and Punc-86::rsy-1ΔSR denote transgenic expression of rsy-1 or rsy-1ΔSR cDNA (isoform A), respectively. **P < 0.01; Student’s t test; n > 20. Error bars indicate SEM. (C) GFP::RSY-1 (isoform A) expressed in the HSNL under the unc-86 promoter. (D) GFP::RSY-1ΔSR (RSY-1 without amino acids 503 to 589 of isoform A) (in green) and mCherry::RAB-3 (in magenta) coexpressed in the HSNL in wild-type (N2) and (E) in unc-104(e1265) mutants. Insets in the lower left-hand corner in (D) and (E) show GFP::RSY-1ΔSR (green) alone, and insets in the lower right-hand corner show mcherry::RAB-3 (magenta) alone. All images are of adults. The dotted rectangle indicates the synaptic region, and the asterisk marks the HSNL cell body. Scale bar, 5 µm.
Next, we sought to determine the subcellular localization of RSY-1. GFP-tagged RSY-1 (isoform A) expressed in the HSNL localized to presynaptic sites as well as to the nucleus (Fig. 2C and fig. S6). Because both isoforms of RSY-1 contain putative NLSs at the C terminus, we used a truncated version of RSY-1 (RSY-1ΔSR) that lacks all putative NLSs to determine whether the synaptic function of RSY-1 requires its localization to the nucleus. When expressed in the HSNL of rsy-1;syd-1 double mutants, RSY-1ΔSR completely restored the reduction of SNB-1::YFP accumulation at presynaptic sites to levels observed in syd-1 mutants (Fig. 2B). Furthermore, GFP-tagged RSY-1ΔSR was mostly excluded from the HSNL nucleus but robustly localized to the presynaptic sites, where it colocalized with RAB-3, a synaptic vesicle marker (Fig. 2D and fig. S6). Thus, localization of RSY-1 at the presynaptic sites (but not in the nucleus) is probably important for its function in inhibiting synaptogenesis.
To further characterize presynaptic localization of RSY-1, we examined RSY-1ΔSR localization in unc-104 mutants. UNC-104 is an ortholog of the vertebrate Kif1A, a kinesin motor essential for trafficking of synaptic vesicles (22). RSY-1ΔSR localization to presynaptic sites was not affected in unc-104 mutants (Fig. 2E), suggesting that RSY-1 is not associated with synaptic vesicles. Furthermore, RSY-1ΔSR tightly colocalized with active-zone component SYD-2 (fig. S7). RSY-1ΔSR also colocalized with synaptic protein GIT-1 and was juxtaposed to another active-zone protein, ELKS-1 (fig. S8). Thus, RSY-1 occupies a particular subdomain of the active zone and locally regulates synapse assembly.
To determine when RSY-1 acts during the synaptic maturation process, we examined the localization of RSY-1ΔSR at nascent synapses of HNSL in the early and mid-L4 stages of development (12). RSY-1 accumulated at the developing presynaptic sites in early and mid-L4 stages concomitantly with RAB-3 (fig. S9), suggesting that RSY-1 could regulate synaptogenesis from the very early stages of synapse development. Future localization study of endogenous RSY-1 protein will probably provide additional information.
If RSY-1 is a negative regulator of synapse formation, then rsy-1 single mutants should have elevated synaptogenic activity. SNB-1::YFP levels at presynaptic sites in the HSNL were increased in rsy-1 mutants (Fig. 3, A, B, and I), suggesting that excessive synaptic material is recruited to the presynaptic sites. During development, synapses form at secondary sites outside of the normal synaptic region of the HSNL, which are then gradually eliminated as animals reach adulthood (8). Elimination of these synapses is partially dependent on proteasomal degradation (8). We observed a partial failure in the elimination of SNB-1::YFP localized to these secondary sites in rsy-1 mutants (Fig. 3, A to D). These SNB-1::YFP accumulations probably represent presynaptic specializations because RAB-3 colocalized with the active-zone components ELKS-1 and GIT-1 at these sites (fig. S10). The persistence of synapses at secondary sites in rsy-1 mutants suggests that local inhibition of presynaptic assembly by RSY-1 also contributes to the elimination of synapses. Thus, RSY-1 plays a role in controlling the size and number of presynaptic sites.
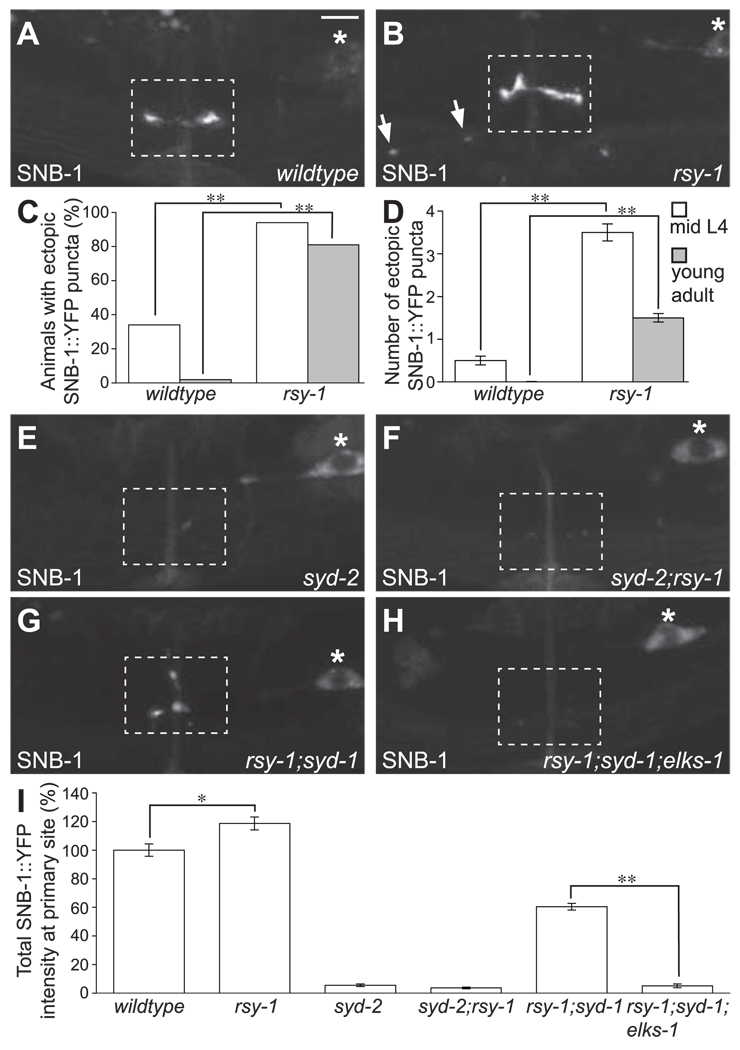
Fig. 3.
RSY-1 is a negative regulator of SYD-2–dependent synapse assembly. (A) SNB-1::YFP expressed in the HSNL wildtype(N2) and (B) rsy-1(wy94) mutants. Arrows denote ectopic SNB-1::YFP puncta. (C) Percentage of animals with ectopic SNB-1::YFP puncta in wildtype(N2) and rsy-1(wy94) mutants at mid-L4 and young adult stages. **P < 0.01; Fisher’s exact test; n = 100 per group. (D) Average number of ectopic SNB-1::YFP puncta in wildtype(N2) and rsy-1(wy94) mutants at mid-L4 and young adult stages. **P < 0.01; Student’s t test; n = 100 per group. Error bars indicate SEM. (E) SNB-1::YFP accumulation at synapses in syd-2(ju37), (F) syd-2(ju37);rsy-1(wy94), (G) rsy-1(wy94);syd-1(ju82), and (H) rsy-1(wy94);syd-1(ju82);elks-1(tm1233) mutants. All images are of adults. The dotted rectangle indicates the synaptic region, and the asterisk marks the HSNL cell body. Scale bar, 5 µm. (I) Total SNB-1::YFP intensity at the primary synaptic region in the HSNL relative to wild type. *P < 0.05; **P < 0.01; Student’s t test; n > 20 per group. Error bars indicate SEM.
RSY-1 can inhibit synaptogenesis by either negatively regulating SYD-2 function or antagonizing an unknown SYD-2–independent parallel assembly pathway. Similar to syd-1 suppression, if rsy-1 mutation is able to suppress synaptic defects in syd-2 mutants, then the data would be indicative of a model in which RSY-1 functions in parallel with SYD-2. However, we found that syd-2 is epistatic to rsy-1; synaptic defects in syd-2 mutants, as assayed by localization of synaptic proteins and the egg-laying behavior, were not rescued in syd-2;rsy-1 double mutants (Fig. 3, E, F, and I, and fig. S5). Thus, RSY-1 probably acts upstream of or in parallel with SYD-2. Both models suggest that RSY-1 is a negative regulator of SYD-2–dependent synapse assembly.
According to the linear model in which syd-2 acts downstream of rsy-1, the function of SYD-2 should increase in rsy-1 mutants. If this is the case, then the phenotype in rsy-1 mutants and syd-2(gf) animals should be similar. Indeed, more synaptic material was recruited to presynaptic sites in the HSNL in syd-2(gf) animals (14), similar to our observation in rsy-1 loss-of-function mutants (Fig. 3I). The linear model also predicts that the rescued synapse assembly in rsy-1;syd-1 mutants, like in syd-2(gf);syd-1 mutants, is due to an increase in SYD-2 function. If this is the case, then synapses in both double mutants should be similarly susceptible to various genetic manipulations. ELKS-1 is a presynaptic active-zone component shown to be important for the development of presynaptic terminals at the neuromuscular junction in Drosophila (23, 24). In C. elegans, although the loss of ELKS-1 function by itself does not result in any detectable defects in synapse assembly (13, 14, 25), synapse formation in the HSNL of syd-2(gf);syd-1 mutants is crucially dependent on ELKS-1 (14). Thus, ELKS-1 functions redundantly with SYD-1 to promote synapse assembly. Synapse formation in rsy-1;syd-1 double mutants was also dependent on elks-1 (Fig. 3, G to I, and fig. S5). Thus, the synapse assembly program deployed in rsy-1;syd-1 mutants is similar to the one in syd-2(gf);syd-1 mutants. In summary, the loss of RSY-1 function has similar consequences as the gain of SYD-2 function. Although these results do not rule out the possibility that RSY-1 functions in parallel with SYD-2, given that RSY-1 interacts with SYD-2 as described below, they are consistent with a genetic model in which RSY-1 is a negative regulator of SYD-2.
RSY-1 could interfere with the pro-synaptic function of SYD-2 directly or inhibit the function of SYD-2 indirectly by blocking SYD-1. To test these possibilities, we used a single-cell in situ protein-protein interaction assay (26, 27), in which translocation of the prey to the plasma membrane is tested in the presence of a membrane-tethered bait, to determine whether RSY-1 physically interacts with SYD-2 and SYD-1. RSY-1ΔSR bound to both SYD-2 [via the first two SAM domains] and SYD-1 (note the enrichment of SYD-2SAM1-2 and SYD-1 on the plasma membrane in the presence of membrane-targeted RSY-1ΔSR) (Fig. 4, A, B, and G). Furthermore, coimmunoprecipitation from worm lysates confirmed that RSY-1ΔSR interacted with both SYD-2 and SYD-1 in vivo (fig. S11).
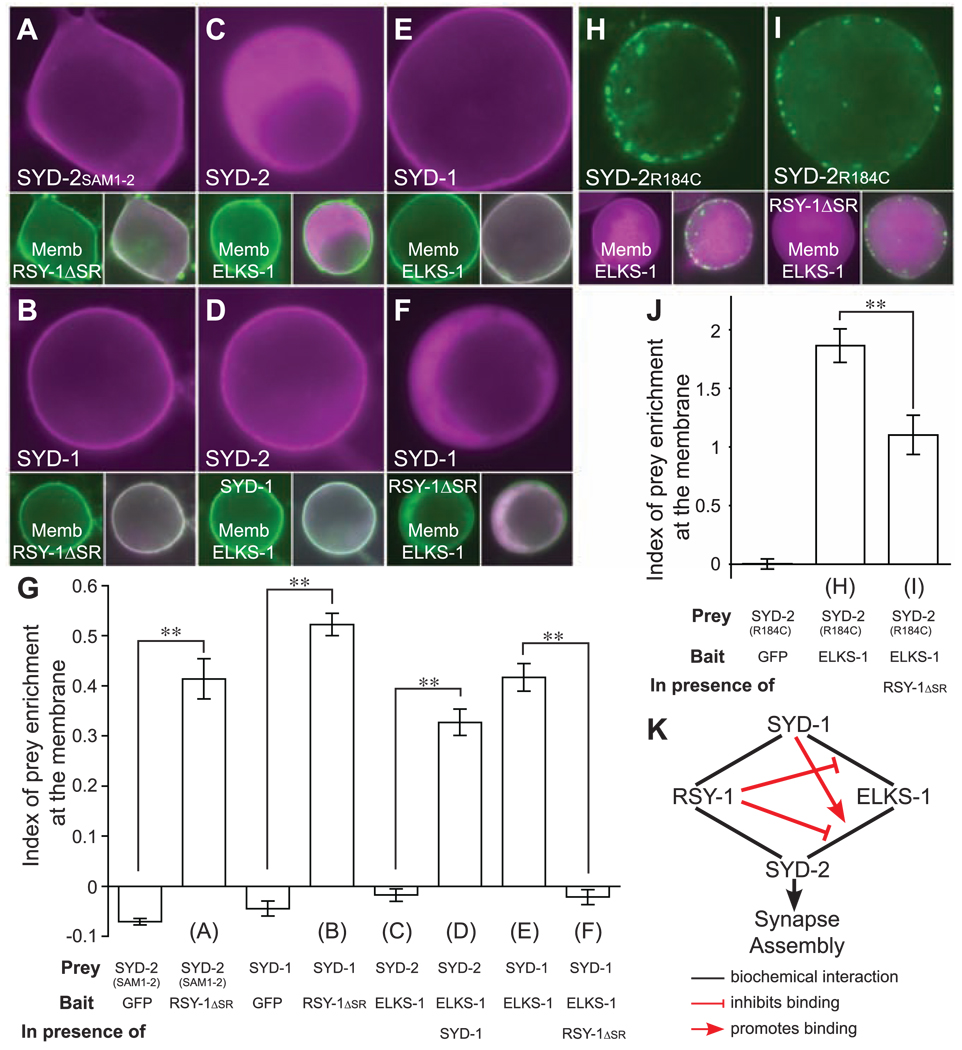
Fig. 4.
Molecular mechanisms of RSY-1 function. (A) mCherry::SYD-2SAM1-2 (amino acids 853 to 1085 of SYD-2) (magenta, main panel) coexpressed in Hek293T cells with membrane-targeted GFP::RSY-1ΔSR (green, lower left panel; lower right panel shows a merged image). (B) mCherry::SYD-1 coexpressed with membrane-targeted GFP::RSY-1ΔSR. (C) mCherry::SYD-2 coexpressed with membrane-targeted GFP::ELKS-1 in the absence of and (D) in the presence of cytoplasmic GFP::SYD-1. (E) mCherry::SYD-1 coexpressed with membrane-targeted GFP::ELKS-1 in the absence of and (F) in the presence of cytoplasmic GFP::RSY-1ΔSR. (G) Quantification of prey translocation to the plasma membrane. (H) eYFP::SYD-2R184C (arginine amino acid at position 184 switched to cysteine) (green, main panel) coexpressed with membrane-targeted mCerulean::ELKS-1 (magenta, lower left panel; lower right panel shows a merged image) in the absence of and (I) in the presence of cytoplasmic mCherry::RSY-1ΔSR. (J) Quantification of prey translocation to the plasma membrane. In (G) and (J), the index of prey enrichment at the membrane was calculated by the following expression: (prey fluorescence intensity at the cell membrane/cytoplasm) – 1. An index of 0 indicates equal fluorescence intensity at the membrane and in the cytoplasm. **P < 0.01; Student’s t test; n = 15 per group. Error bars indicate SEM. Letters corresponding to figure panels are shown below the appropriate bar. The prey, membrane-targeted bait, and third cytoplasmic protein present are indicated below each bar. (K) Summary of biochemical interactions and their consequences. Promotion of SYD-2/ELKS-1 interaction by SYD-1 supports the genetic data that syd-1 is normally required for syd-2 function. Inhibition of SYD-2/ELKS-1 and SYD-1/ELKS-1 interaction by RSY-1 supports the genetic role of rsy-1 in antagonizing syd-2 directly and indirectly via syd-1.
To study the molecular consequence of interaction of RSY-1 with SYD-2 and SYD-1, we first established a readout of SYD-2 function. Vertebrate homologs of ELKS-1 and SYD-2 directly bind in vitro (28), whereas in C. elegans, SYD-2 and ELKS-1 coimmunoprecipitate from worm lysate (14). Furthermore, syd-2 loss-of-function analysis suggests that SYD-2 is necessary for localizing ELKS-1 to presynaptic sites, whereas experiments with the syd-2(gf) allele suggest that SYD-2 is sufficient for recruiting ELKS-1 (13, 14). Given these data and the genetic evidence that ELKS-1 is an important component of the presynaptic assembly program, we used the ELKS-1/SYD-2 interaction as one of the readouts of SYD-2 function.
Although there was little detectible interaction between ELKS-1 and SYD-2 in our assay, the interaction was greatly enhanced in the presence of SYD-1 (Fig. 4, C, D, and G), suggesting that SYD-1 facilitates binding between ELKS-1 and SYD-2. Consistent with this result, SYD-1 directly interacted with ELKS-1 (Fig. 4, E and G), and this interaction was weakened in the presence of RSY-1ΔSR (Fig. 4, F and G). Thus, one way in which RSY-1 regulates SYD-2 function is indirectly by weakening the interaction of SYD-1 with ELKS-1 and thus potentially blocking the ability of SYD-1 to facilitate SYD-2 function (Fig. 4K).
Given that the ELKS-1/SYD-2 binding is very weak in the absence of SYD-1 in our assay, we could not test whether interaction of RSY-1 with SYD-2 inhibited ELKS-1/SYD-2 binding. However, the ELKS-1/SYD-2 interaction does increase when SYD-2 contains a gain-of-function mutation, Arg184 → Cys184 (R184C) (14), which was verified in our cell-based assay (Fig. 4, H and J). We then tested the effect of RSY-1 on this interaction and found that the interaction between ELKS-1 and SYD-2R184C was weakened in the presence of RSY-1ΔSR (Fig. 4, I and J), suggesting that, besides acting via SYD-1, RSY-1 can also directly antagonize the ability of SYD-2 to recruit ELKS-1 (Fig. 4K).
It is increasingly clear that positive and negative regulators control synapse development at multiple levels. For example, the transcription factor MEF2 globally regulates the number of excitatory synapses (7). Three ubiquitin ligase complexes also regulate presynaptic development (5, 8, 29). Here, RSY-1 was shown to act as a negative regulator of synaptogenesis by counteracting SYD-1 function to inhibit SYD-2–dependent presynaptic assembly in the HSNL neuron. RSY-1 controls the amount of synaptic material recruited to presynaptic sites. RSY-1 also plays a role in establishing a balance between synapse formation and synapse elimination. RSY-1 achieves these functions by interacting with integral components of the synapse assembly machinery and by regulating a dense network of protein-protein interactions between various active-zone molecules (Fig. 4K).
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/323/5920/1500/DC1
Materials and Methods
Figs. S1 to S11
References
References and Notes
- 1.Zhen M, Jin Y. Nature. 1999;401:371. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
- 2.Zhen M, Jin Y. Curr. Opin. Neurobiol. 2004;14:280. doi: 10.1016/j.conb.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Waites CL, Craig AM, Garner CC. Annu. Rev. Neurosci. 2005;28:251. doi: 10.1146/annurev.neuro.27.070203.144336. [DOI] [PubMed] [Google Scholar]
- 4.Chang Q, Balice-Gordon RJ. Neuron. 2000;26:287. doi: 10.1016/s0896-6273(00)81161-7. [DOI] [PubMed] [Google Scholar]
- 5.van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH. Cell. 2004;119:707. doi: 10.1016/j.cell.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 6.Nakata K, et al. Cell. 2005;127:407. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Flavell SW, et al. Science. 2006;311:1008. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 8.Ding M, Chao D, Wang G, Shen K. Science. 2007;317:947. doi: 10.1126/science.1145727. published online 11 July 2007 (10.1126/science.1145727) [DOI] [PubMed] [Google Scholar]
- 9.Klassen MP, Shen K. Cell. 2007;130:704. doi: 10.1016/j.cell.2007.06.046. [DOI] [PubMed] [Google Scholar]
- 10.Poon VY, Klassen MP, Shen K. Nature. 2008;455:669. doi: 10.1038/nature07291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai C, Horvitz HR. Genetics. 1989;121:703. doi: 10.1093/genetics/121.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen K, Bargmann CI. Cell. 2003;112:619. doi: 10.1016/s0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- 13.Patel MR, et al. Nat. Neurosci. 2006;9:1488. doi: 10.1038/nn1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai Y, et al. Nat. Neurosci. 2006;9:1479. doi: 10.1038/nn1808. [DOI] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supporting material on Science Online
- 16.Zimowska G, et al. Invest. Ophthalmol. Vis. Sci. 2003;44:4715. doi: 10.1167/iovs.03-0240. [DOI] [PubMed] [Google Scholar]
- 17.Ouyang P. Biochem. Biophys. Res. Commun. 1999;263:192. doi: 10.1006/bbrc.1999.1353. [DOI] [PubMed] [Google Scholar]
- 18.Joo JH, et al. Mol. Vis. 2005;11:133. [PubMed] [Google Scholar]
- 19.Alpatov R, et al. Mol. Cell. Biol. 2004;24:10223. doi: 10.1128/MCB.24.23.10223-10235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P, Lou PJ, Leu S, Ouyang P. Biochem. Biophys. Res. Commun. 2002;294:448. doi: 10.1016/S0006-291X(02)00495-3. [DOI] [PubMed] [Google Scholar]
- 21.Baumeister R, Liu Y, Ruvkun G. Genes Dev. 1996;10:1395. doi: 10.1101/gad.10.11.1395. [DOI] [PubMed] [Google Scholar]
- 22.Hall DH, Hedgecock EM. Cell. 1991;65:837. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- 23.Wagh DA, et al. Neuron. 2006;49:833. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Kittel RJ, et al. Science. 2006;312:1051. doi: 10.1126/science.1126308. published online 13 April 2006 (10.1126/science.1126308) [DOI] [PubMed] [Google Scholar]
- 25.Deken SL, et al. J. Neurosci. 2005;25:5975. doi: 10.1523/JNEUROSCI.0804-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchard D, Hutter H, Fleenor J, Fire A. Mol. Cell. Proteomics. 2006;5:2175. doi: 10.1074/mcp.T600025-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. Genesis. 2007;45:593. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 28.Ko J, Na M, Kim S, Lee JR, Kim E. J. Biol. Chem. 2003;278:42377. doi: 10.1074/jbc.M307561200. [DOI] [PubMed] [Google Scholar]
- 29.Fulga TA, Van Vactor D. Neuron. 2008;57:339. doi: 10.1016/j.neuron.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 30.We would like to thank the Caenorhabditis Genetics Center and the Japanese NBPR for strains; Y. Kohara (National Institute of Genetics, Japan) for Y53H1A.1 cDNAs; J. Audhya (University of California, San Diego) for C. elegans optimized mCherry cDNA; L. Luo (Stanford University) for vertebrate mCherry cDNA; T. Meyer (Stanford University) for eYFP-C3 plasmid; M. Park (Stanford University) for mCerulean-C1 plasmid; C. Gao for technical assistance; and members of the Shen lab, C. Bargmann, T. Clandinin, and L. Luo for critical comments on the manuscript. We would also like to thank B. Grill (Stanford University) for providing protocol and guidance for coimmunoprecipitation experiments. This work was funded by grants awarded to K.S. [from NIH (1R01NS048392), the Human Frontier Science Foundation, the Howard Hughes Medical Institute, and the W. M. Keck Foundation] and by a National Research Service Award predoctoral fellowship awarded to M.R.P. by NIH.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.