Abstract
Metastasis is a defining feature of malignant tumors and is the most common cause of cancer-related death, yet the genetics of metastasis are poorly understood. We used massively parallel exome sequencing coupled with Sanger re-sequencing to search for metastasis-related mutations in highly metastatic uveal melanomas of the eye. Inactivating somatic mutations were identified in the gene encoding BRCA1-associated protein 1 (BAP1) on chromosome 3p21.1 in 26 of 31 (84%) metastasizing tumors, including 15 mutations causing premature protein termination, and six affecting its ubiquitin carboxy-terminal hydrolase (UCH) domains. One tumor harbored a frameshift mutation that was germline in origin, thus representing a susceptibility allele. These findings implicate loss of BAP1 in uveal melanoma metastasis and suggest the BAP1 pathway as a therapeutic target.
Uveal melanoma (UM) is the most common primary cancer of the eye and has a strong propensity for fatal metastasis (1). UMs are divided into class 1 (low metastatic risk) and class 2 (high metastatic risk) based on a validated multi-gene clinical prognostic assay included in the TNM classification system (2, 3). However, the genetic basis of metastasis remains unclear. Oncogenic mutations in the Gαq stimulatory subunit GNAQ are common in UM (4), but these mutations occur early in tumorigenesis and are not correlated with molecular class or metastasis (5, 6). On the other hand, class 2 tumors are strongly associated with monosomy 3 (7), suggesting that loss of one copy of chromosome 3 may unmask a mutant gene on the remaining copy which promotes metastasis.
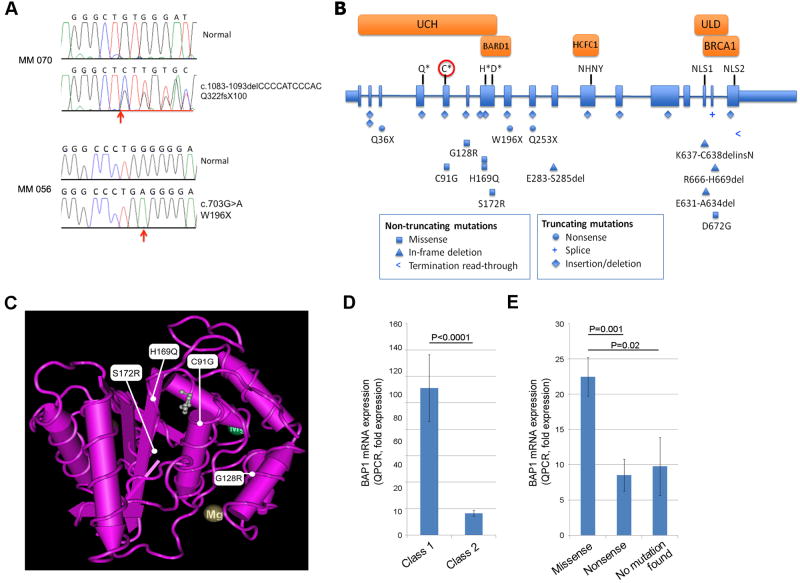
Using exome capture followed by massively parallel sequencing (8-10), we analyzed two class 2 tumors that were monosomic for chromosome 3 (MM56 and MM70) and matching normal DNA from peripheral blood lymphocytes. Both tumors contained inactivating mutations in BAP1, located at chromosome 3p21.1 (Fig. 1A) (10). MM56 contained a C/G to T/A transition that created a premature termination codon (p.W196X). MM70 contained a deletion of 11bp in exon 11, leading to a frameshift and premature termination of the BAP1 protein (p.Q322fsX100). The matched normal DNA samples did not contain these mutations, indicating that they were likely to be somatic in origin. No gene on chromosome 3 other than BAP1 contained deleterious somatic mutations that were present in both tumors (table S3).
Fig. 1.
Inactivating mutations in BAP1 occur frequently in uveal melanomas. (A) Sanger sequence traces of MM 056 and MM 070 at the sites of the mutations. Location of mutated base in MM 056 and the start of the deletion of MM 070 are indicated (arrows). The non-coding BAP1 strand is shown for MM 070. (B) Map of BAP1 gene and location of BAP1 mutations. BAP1 contains 17 exons (shaded boxes) that encode a 728 amino acid protein. Introns are not to scale. Mutations are shown below the gene figure as indicated. The UCH domain (aa. 1-188) and UCH37-like domain (ULD) (aa. 635-693) are indicated (12, 13). The critical Q, C, H and D residues of the active site (Gln85, Cys91, His169 and Asp184) are indicated with asterisks. The catalytic cysteine is indicated with a circle. Also shown are: the NHNY consensus sequence for interaction with HCFC1 (aa. 363-365, exon 11), nuclear localization signals (NLS) at aa. 656-661 (exon 15) and aa. 717-722 (exon 17), the BARD1 binding domain within the region bounded by aa. 182-240 (13), and the BRCA1 binding domain within aa. 598-729 (11). (C) Location of BAP1 missense mutations in the UCH domain aligned to the crystal structure of UCH-L3 (21). Three-dimensional structure of UCH-L3 was visualized with MMDB software (22). The small molecule near C91G, H169Q and S172R represents a suicide inhibitor, illustrating the critical location of these mutations for catalytic activity. (D) BAP1 mRNA levels measured by quantitative RT-PCR in 9 non-metastasizing class 1 UMs and 28 metastasizing class 2 UMs. (E) Relationship between BAP1 mRNA levels (measured by quantitative RT-PCR) and type of BAP1 mutation in 9 UMs with nonsense mutations, 10 UMs with missense mutations (including small in-frame deletions, splice acceptor, and stop codon read-through mutations), and 4 class 2 UMs in which no BAP1 mutations were detected.
BAP1 encodes a nuclear ubiquitin carboxy-terminal hydrolase (UCH), one of several classes of deubiquitinating enzymes (11). In addition to the UCH catalytic domain, BAP1 contains a UCH37-like domain (ULD) (12), binding domains for BRCA1 and BARD1, which form a tumor suppressor heterodimeric complex (13), and a binding domain for HCFC1, which interacts with histone-modifying complexes during cell division (12, 14, 15). BAP1 also interacts with ASXL1 to form the Polycomb group repressive deubiquitinase complex (PR-DUB), which is involved in stem cell pluripotency and other developmental processes (16, 17). BAP1 exhibits tumor suppressor activity in cancer cells (11, 13), and BAP1 mutations have been reported in a small number of breast and lung cancer samples (11, 18).
To further investigate BAP1, genomic DNA from 29 additional class 2 UMs, and 26 class 1 UMs were subjected to Sanger re-sequencing of all BAP1 exons. Altogether, BAP1 mutations were identified in 26 of 31 (84%) class 2 tumors, including 13 out-of-frame deletions and two nonsense mutation leading to premature protein termination, six missense mutations, four in-frame deletions, and one mutation predicted to produce an abnormally extended BAP1 polypeptide (Fig. 1A-C) (10). Three of the missense mutations affected catalytic residues of the UCH active site (C91 and H169), two occurred elsewhere in the UCH domain, and one affected the ULD (Fig. 1B-C). All BAP1 missense mutations and in-frame deletions affected phylogenetically conserved amino acids (fig. S1). Only one of 26 class 1 tumors contained a BAP1 mutation (NB101) (10). This case may represent a transition state in which the tumor has sustained a BAP1 mutation but has not yet converted to class 2, suggesting that BAP1 mutations may precede the emergence of the class 2 signature. Somatic BAP1 mutations were also detected in two of three metastatic tumors (10).
One copy of chromosome 3 was missing in all 17 BAP1-mutant class 2 tumors for which cytogenetic data were available, consistent with chromosome 3 loss uncovering recessive BAP1 mutations (10). Normal DNA from 20 patients with BAP1-mutant class 2 primary tumors and the two with metastatic tumors was available and did not contain a BAP1 mutation, indicating that the mutations were somatic in origin. However, we detected one germline mutation (p.E402fsX2; c.1318-1319insA) in the patient with tumor MM 087 (table S2) suggesting that germline alterations in BAP1 can predispose to UM. GNAQ mutation status was available in 15 cases. GNAQ mutations were present in 4/9 BAP1 mutant tumors and 3/6 BAP1 wildtype tumors, indicating that there was no correlation between GNAQ and BAP1 mutation status.
Quantitative RT-PCR showed that BAP1 mRNA levels were significantly lower in class 2 tumors compared to class 1 tumors (P<0.0001) (Fig. 1D). Truncating mutations were associated with significantly lower mRNA levels than missense mutations (P=0.001) (Fig. 1E), consistent with nonsense mediated mRNA decay in the former group. Class 2 tumors in which BAP1 mutations were not identified expressed very low levels of BAP1 mRNA (Fig. 1E).
To determine whether the low BAP1 mRNA levels in class 2 tumors without detectable BAP1 mutations may be explained by DNA methyation, we performed a preliminary analysis of DNA methylation of BAP1. This did not reveal a convincing difference between class 1 and class 2 tumors (10). However, analysis of the BAP1 promoter was limited by an unusually complex CpG island that will require further work to resolve. Thus, we cannot rule out a role for methylation in class 2 tumors in which BAP1 mutations were not found. However, with almost 85% of class 2 tumors harboring mutations, we do not expect that methylation will be a major mechanism of BAP1 inactivation. An alternative explanation is that these tumors may contain very large deletions of the BAP1 locus not detectable by our sequencing method.
Immunofluorescence revealed abundant nuclear BAP1 protein in two class 1 tumors but virtually none in four BAP1 mutant class 2 tumors (fig. S3). This was expected for the two tumors with mutations expected to cause premature protein terminations (MM 091 and MM 100), but it was surprising for the two tumors with missense mutations (MM 071 and MM 135) and suggests that these mutations lead to protein instability.
RNAi-mediated knock down of BAP1 in 92.1 UM cells, which did not harbor a detectable BAP1 mutation, recapitulated many characteristics of the de-differentiated class 2 UM phenotype (19). Cells transfected with control siRNA exhibited typical melanocytic morphology, including dendritic projections and cytoplasmic melanosomes (Fig. 2A), whereas cells transfected with BAP1 siRNA lost these features, developed a rounded epithelioid morphology and grew as multicellular non-adherent spheroids, strikingly similar to the features of class 2 clinical biopsy samples (Fig. 2A). Microarray gene expression profiling of 92.1 UM cells transfected with control versus BAP1 siRNA showed that most of the top genes that discriminate between class 1 and class 2 tumors shifted in the class 2 direction in BAP1 depleted cells compared to control cells (fig. S4). Similarly, depletion of BAP1 shifted the gene expression profile of the multi- gene clinical prognostic assay towards the class 2 signature (Fig. 2B). BAP1 depletion caused a reduction in mRNA levels of neural crest migration genes (ROBO1), melanocyte differentiation genes (CTNNB1, EDNRB and SOX10) and other genes that are down-regulated in class 2 tumors (LMCD1 and LTA4H) (19). In contrast, BAP1 depletion caused an increase in mRNA levels of CDH1 and the proto-oncogene KIT, which are highly expressed in class 2 tumors (20). Similar results were seen in other UM cell lines and with an independent BAP1 siRNA (10).
Fig. 2.

UM cells depleted of BAP1 acquire properties that are typical of metastasizing class 2 tumor cells. (A) Phase contrast photomicrographs of 92.1 uveal melanoma cells transfected with BAP1 or control siRNA at the indicated days. Bottom panels show representative examples of class 1 and class 2 uveal melanoma cells obtained from patient biopsy samples (Papanicolaou stain). Scale bars, 10 microns. (B) 92.1 cells transfected with BAP1 siRNA and evaluated after five days. BAP1 protein levels were efficiently depleted to less than 95% of control levels (see western blot). Upper panel depicts principal component analysis to show effect of BAP1 knockdown on gene expression signature. The small spheres represent the training set of known class 1 (blue) and class 2 (red) tumors. Large spheres represent the control-transfected (gray) and BAP1 siRNA transfected (red) cells. Lower panel depicts mRNA levels measured by quantitative RT-PCR of a panel of melanocyte lineage genes, presented as fold change in BAP1 siRNA/control siRNA transfected cells. Results are representative of three independent experiments.
GNAQ mutations occur early in UM and are not sufficient for malignant transformation (4), but they may create a dependency of the tumor cells on constitutive GNAQ activity. In contrast, BAP1 mutations occur later in UM progression and coincide with the onset of metastatic behavior. Thus, simultaneous targeting of both genetic alterations might have synergistic therapeutic effects. One potential strategy to counteract the effects of BAP1 mutation would be to inhibit the RING1 ubiquinating activity that normally opposes the deubiquinating activity BAP1 (16). Our findings strongly implicate mutational inactivation of BAP1 as a key event in the acquisition of metastatic competence in UM, and they dramatically expand the role of BAP1 and other deubiquitinating enzymes as potential therapeutic targets in cancer.
Supplementary Material
Acknowledgments
Supported by grants to J.W.H. from the National Cancer Institute (R01 CA125970), Barnes-Jewish Hospital Foundation, Kling Family Foundation, Tumori Foundation, Horncrest Foundation, and a Research to Prevent Blindness David F. Weeks Professorship, and by awards to the Department of Ophthalmology and Visual Sciences at Washington University from a Research to Prevent Blindness, Inc. Unrestricted grant, and the NIH Vision Core Grant P30 EY02687c. E.D.O.R. was supported by NIH/NIAMS training grant AR007279-31A1. We thank Jess Hoisington-Lopez from the Center for Genome Sciences (Wash Univ. School of Medicine) for running Solexa paired end sequences, and Dr. Michael Lovett for comments on the manuscript. J.W.H. and Washington University may receive income based on a license of related technology by the University to Castle Biosciences, Inc.
Footnotes
- Materials and Methods
- Figs. S1 to S5
- Tables S1 to S3
- References
References and Notes
- 1.Landreville S, Agapova OA, Harbour JW. Future Oncol. 2008;4:629. doi: 10.2217/14796694.4.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onken MD, Worley LA, Tuscan MD, Harbour JW. J Mol Diagn. 2010;12:461. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finger PT. Arch Pathol Lab Med. 2009;133:1197. doi: 10.5858/133.8.1197. [DOI] [PubMed] [Google Scholar]
- 4.Van Raamsdonk CD, et al. Nature. 2009;457:599. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onken MD, et al. Invest Ophthalmol Vis Sci. 2008;49:5230. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer J, et al. Br J Cancer. 2009;101:813. doi: 10.1038/sj.bjc.6605226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worley LA, et al. Clin Cancer Res. 2007;13:1466. doi: 10.1158/1078-0432.CCR-06-2401. [DOI] [PubMed] [Google Scholar]
- 8.Bashiardes S, et al. Nat Methods. 2005;2:63. doi: 10.1038/nmeth0105-63. [DOI] [PubMed] [Google Scholar]
- 9.Ng SB, et al. Nat Genet. 2010;42:30. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.See supporting material on Science Online
- 11.Jensen DE, et al. Oncogene. 1998;16:1097. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 12.Misaghi S, et al. Mol Cell Biol. 2009;29:2181. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishikawa H, et al. Cancer Res. 2009;69:111. doi: 10.1158/0008-5472.CAN-08-3355. [DOI] [PubMed] [Google Scholar]
- 14.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. J Biol Chem. 2009;284:34179. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyagi S, Chabes AL, Wysocka J, Herr W. Mol Cell. 2007;27:107. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 16.Gaytan de Ayala Alonso A, et al. Genetics. 2007;176:2099. doi: 10.1534/genetics.107.075739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheuermann JC, et al. Nature. 2010;465:243. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood LD, et al. Science. 2007;318:1108. [Google Scholar]
- 19.Onken MD, et al. Cancer Res. 2006;66:4602. doi: 10.1158/0008-5472.CAN-05-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onken MD, Worley LA, Ehlers JP, Harbour JW. Cancer Res. 2004;64:7205. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misaghi S, et al. Journal of Biological Chemistry. 2005;280:1512. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Nucleic Acids Res. 2007;35:D298. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.