Abstract
HbVar (http://globin.cse.psu.edu/globin/hbvar/) is a relational database developed by a multi-center academic effort to provide up-to-date and high quality information on the genomic sequence changes leading to hemoglobin variants and all types of thalassemia and hemoglobinopathies. Extensive information is recorded for each variant and mutation, including sequence alterations, biochemical and hematological effects, associated pathology, ethnic occurrence and references. In addition to the regular updates to entries, we report two significant advances: (i) The frequencies for a large number of mutations causing β-thalassemia in at-risk populations have been extracted from the published literature and made available for the user to query upon. (ii) HbVar has been linked with the GALA (Genome Alignment and Annotation database, available at http://globin.cse.psu.edu/gala/) so that users can combine information on hemoglobin variants and thalassemia mutations with a wide spectrum of genomic data. It also expands the capacity to view and analyze the data, using tools within GALA and the University of California at Santa Cruz (UCSC) Genome Browser.
INTRODUCTION
Hemoglobinopathies resulting from mutations in the α- or β-like globin gene clusters are the most common inherited disorders in humans, with around 7% of the world population being carriers of a globin gene mutation [reviewed in (1)]. Single nucleotide substitutions can lead to amino acid replacements that cause hemolytic anemias, such as sickle cell disease, or hemoglobins that are unstable or have altered oxygen affinity. Molecular defects in either regulatory or coding regions of the human α-, β- or δ-globin genes can minimally or drastically reduce their expression, leading to α-, β- or δ-thalassemia, respectively. Other sequence changes have little or no effect on hemoglobin function, but are useful polymorphisms for genetic studies.
We recently developed HbVar as a publicly available database not only to store information from previous compilations (2,3), but also to allow regular updates and corrections, since new hemoglobin variants and thalassemias continue to be discovered. The query interface provides easy access to this information for the research community and for physicians as an aid in diagnosis. We also find that other interested individuals, such as patients and their parents, people involved in the provision of genetic services and counseling, pharmaceutical industries, etc. are using HbVar (4).
DATABASE DESCRIPTION AND STRUCTURE
The initial sources of information in HbVar were the books A Syllabus of Human Hemoglobin Variants (2nd Edn) (3) and A Syllabus of Thalassemia Mutations (2). This information has been expanded by more than 200 additional entries and corrections made by the database curators. Published information on pathology, hematology, clinical presentation and laboratory findings (range of hemoglobin levels, hematocrit, etc.) is included, while considerable biochemical data on the variants is also recorded, including techniques used to identify, isolate and determine their structure, stability, function, and qualitative distribution in ethnic groups and geographic locations. Controlled vocabularies are enforced, and entries include literature references. These data can be accessed through summary listings or user-generated queries, which can be highly specific.
The HbVar database and associated resources at the Globin Gene Server (http://globin.cse.psu.edu/), such as the online Syllabi, the GALA database, etc., are currently in use world-wide. Since January 2000, we have recorded 6372 accesses to the HbVar query page and 37 915 accesses to the online Syllabi, an almost 5-fold increase compared to when HbVar was initially described (4). Users frequently contact the curators and the rest of the HbVar team members in order to submit new hemoglobin variants and/or thalassemia mutations, report missing information for existing mutants and pinpoint inconsistencies and/or erroneous entries. This is particularly important, since the user input improves the quality and accuracy of the data. Therefore we urge the HbVar users to notify the curators of such errors or incomplete information (detailed contact information is at http://globin.cse.psu.edu/html/contact.html).
POPULATION FREQUENCY DATA
Since the initial launch of HbVar in mid 2000, users have requested information on the frequencies of thalassemia mutations in different populations. The β-thalassemias are the most common autosomal recessive disorder in populations of Mediterranean, Middle and Far Eastern, Asian/Indian and African descent with a history of malaria endemicity, but each at-risk population has its own spectrum of common mutations. Such information significantly simplifies mutation analysis and molecular diagnosis. Carrier and prenatal diagnoses, using a combined hematological and mutation analysis, have made it possible to screen women at childbearing age in several Mediterranean at-risk populations and, when combined with nondirective genetic counseling, the screening resulted in a consistent decline of the birth of affected homozygotes [(5) and references therein].
To provide the requisite information, we extracted from the published literature the frequency spectrum in 48 countries or ethnic backgrounds for 121 β-thalassemia mutations (Table 1). Now a user can query not only for all of the β-thalassemia mutations found in a given population, but also to specify their frequency range (Fig. 1a), and therefore to focus a search on common or rare alleles, depending on the study.
Table 1. Frequency of β-thalassemia mutations in different populations.
| Origin | Countries/ethnic backgroundsa |
|---|---|
| Arab | 13 |
| Mediterranean | 11 |
| European | 16 |
| Asian/Indian | 18 |
| African | 4 |
| American | 2 |
The total number of countries/ethnic backgrounds (categorized in different origin groups), for which information on the β-thalassemia mutation frequencies is made available in the HbVar database.
aA given country can be included in different ethnic origin groups (e.g. Italy can be found in both the Mediterranean and the European countries).
Figure 1.
Query on HbVar for mutation frequencies in different populations. (a) Construction of the query ‘Find all β-thalassemia mutants in the Greek Cypriot population’. Only parts of the query page are shown. The user needs to specify beta0 or beta+ or beta(0 or + unclear) in the ‘Type of Thalassemia’ field and Greek Cypriots in the ‘Ethnic background’ field (the selections required for such a query are highlighted). The query page also allows the user to insert the desired frequency range (5–100% for this example). (b and c) Output from the query. Four different mutations are displayed. The mutation names are hyperlinked to further information. Upon selection of a specific mutant (IVS-II-745 C → G β+ for this example), the frequencies of this mutation for different populations/ethnic backgrounds are displayed (depicted in c), together with detailed information on this hemoglobin variant/thalassemia mutation (not shown).
As an example, one may want to see only the β-thalassemia mutations that are common in Greek Cypriots. To do this, the user selects beta0, beta+ and beta(0 or + unclear) from the field Type of Thalassemia, Greek Cypriot as the Ethnic background and a frequency range of 5–100% to restrict the output to more frequent mutations (Fig. 1a). Four thalassemia mutations, listed in Figure 1b, are returned. The user can find more detailed information by following the hyperlinks to the individual entries. Such information could be, apart from the properties of this mutant, the frequencies of a specific mutation (the IVS-II-745 C → G β+ for this example) in different populations/ethnic backgrounds (Fig. 1c).
INTEGRATION WITH OTHER GENOMIC RESOURCES: LINKING HbVar WITH THE GALA DATABASE AND THE UCSC GENOME BROWSER
For many studies, the information in HbVar needs to be combined with the wealth of information about features of the genomic DNA, such as gene structures, conservation among species, repetitive elements, recombination frequencies and many others. The latter information is stored in genome browsers such as those at UCSC (6), Ensembl (7) and MapViewer at NCBI (8). Genomic DNA features (annotations) and much data about interspecies conservation recently were organized into a relational database, called GALA (9). This database allows a user to query across fields for different types of information from multiple locations. We have linked HbVar with GALA so that users can access information in both databases. The output from GALA can be examined in a variety of formats, including UCSC Genome Browser views, which facilitates some analyses.
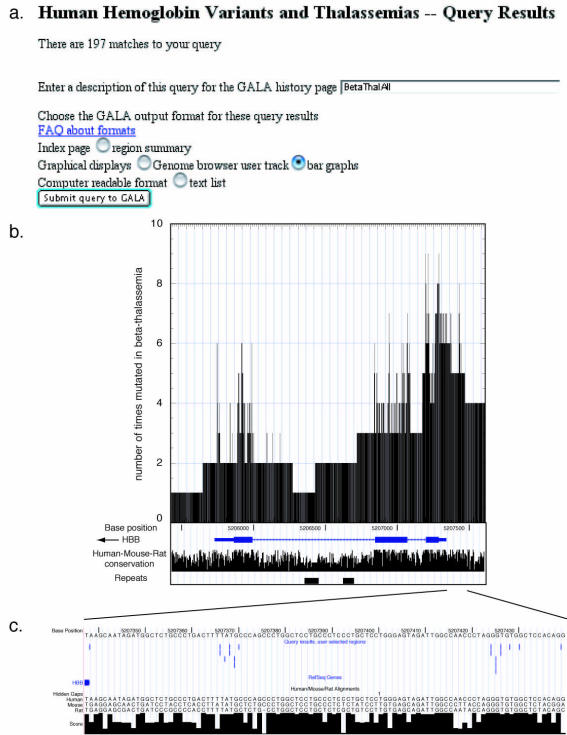
One example of the new capacities is to generate a mutation spectrum from the linked databases. A query to HbVar finds the 197 β-thalassemia mutations currently recorded (from the field ‘Type of Thalassemia’ as in Fig. 1a). After selecting a ‘GALA query’ as the output, the system automatically brings the user to an interface with GALA to select the output format desired. If the bar graph option under ‘Graphical displays’ is selected (Fig. 2a), the system generates a graph indicating how many times the query results (β-thalassemia mutations in this case) fall within a bin along a designated region [both the bin size (the number of nucleotides included in each vertical bar) and the region are specified by the user]. Using a bin size of one nucleotide for optimal resolution, we see that most β-thalassemia mutations fall within the promoter, exons 1 and 2 and intron 1 (Fig. 2b). Most of the gene regions and surrounding DNA have mutation frequencies of 1 to 4; these result from the large deletions that can cause β-thalassemia.
Figure 2.
Linking HbVar and GALA databases and the UCSC Genome Browser to examine the spectrum of mutations that cause β-thalassemia. (a) The set of all β-thalassemia mutations collected from HbVar can be exported to GALA, which is used to generate a bar graph (see also text). (b) The graphical output from the query. The number of mutations found at every nucleotide (bin size of 1) is shown on the vertical axis and the chromosomal position in the horizontal axis. The coordinates for the human β-globin gene (HBB) are based on the April 2003 human reference sequence and include 265 bp of the promoter region (bounded by a SnaBI restriction site) through the gene and extending 300 bp beyond exon 3. Note that HBB is transcribed from right to left in this display (CEN to TEL on the short arm of human chromosome 11). A view from the UCSC Genome Browser is added beneath the plot to show landmarks in HBB. (c) The β-thalassemia mutations in the promoter of HBB are exported to the UCSC Genome Browser and viewed along with alignments between human, mouse and rat sequences.
A more detailed view can be obtained by directing the output to the UCSC Genome Browser as a custom track, which GALA does automatically upon a user’s request. Figure 2c shows the point mutations in the HBB promoter that cause β-thalassemia, with additional tracks selected to show the human DNA sequence aligned with orthologous segments of mouse and rat. Nucleotides in the TATA and CAC (EKLF binding site) boxes have been mutated multiple times in different β-thalassemia mutations. The template strand is shown, i.e. the one that is complementary to the mRNA within the exons. These mutationally sensitive regions are highly conserved, especially the CAC box. Interestingly, other highly conserved regions, such as the CCAAT box, are not mutated in the known β-thalassemias. This intriguing observation is difficult to explain. It is unlikely that CCAAT box mutations are too severe since deletions of the entire gene have been found; these loss-of-function mutations are recessive as expected. Finding multiple mutations of the same nucleotide in other parts of the promoter is consistent with the current collection of β-thalassemia mutations being quite comprehensive. An alternative hypothesis is that the CCAAT box mutations have a dominant negative phenotype, thus removing them from the population soon after they occur.
Examining a mutation spectrum illustrates the power of combining HbVar with the analysis and display capacity of other databases. Additional examples illustrate combinations of data from different databases. Starting with the 96 β-thalassemia substitution mutations found by HbVar and collected as a simple query in GALA, we can use GALA to find those that are found in exons. We find that 51 of the β-thalassemia mutations caused by nucleotide substitutions intersect with the set of all exons (not shown). One may want to find the nucleotide substitutions that occur in highly conserved regions. Again, using GALA to combine information from HbVar with alignment data reveals that of the 96 nucleotide substitutions that cause β-thalassemia, 39 occur in highly conserved regions (defined as at least 70% identity in at least 100 bp ungapped alignment between human and mouse sequences). Users can easily access information about the mutations that fall in these categories.
FUTURE PROSPECTS
The recent assembly of the human reference sequence opens many opportunities for enhancing the accuracy and expanding the application of information on human sequence variants (10). One particular example illustrates this for the thalassemias. A substantial number of the β-thalassemia and HPFH mutations (over 40) are large deletions, removing one or more genes or in some cases the Locus Control Region (LCR). It has long been recognized that critical sequences controlling switch in expression from a fetal to an adult pattern can potentially be identified by comparing the endpoints of deletions that do or do not allow persistent expression of the γ-globin genes in adult erythropoiesis [reviewed in (11)]. However, rigorous identification of all such control sequences has not been completed and some remain controversial. One of the limitations to interpreting these data has been incomplete information about the deleted sequences. Fortunately, the DNA sequence for the HBB gene complex and surrounding regions is now complete. This will allow the precise identification of all deletion junctions and annotation of the DNA features affected by the deletions. Indeed, this information will be critical for interpreting all deletional mutants. Not only will the junction sequences allow better analysis and interpretation of the mutations, but they will also allow specialized screening strategies to be designed and implemented for each mutation.
The link between HbVar and GALA databases, coupled with the UCSC Genome Browser, was the first step towards integrating the available resources in the Globin Gene Server (12). Future work will explore integration of these resources with databases of experimental data on gene regulation, dbERGE (13). Anticipating many genomic DNA sequences to be determined from a wider variety of organisms, mechanisms are now in place for generating whole-genome pair wise (14) and multiple sequence alignments (15). We plan to integrate this wealth of information with HbVar and other resources by expanding the links among databases, following the example of the linkage of HbVar and GALA described here.
Acknowledgments
ACKNOWLEDGEMENTS
We thank all the HbVar users world-wide for their valuable comments and suggestions, which helped us to keep the information as updated and complete as possible and also contributed to the continuous improvement of the database profile and content. We will always be indebted to the late Prof. Titus H. J. Huisman and his colleagues for their detailed compilations of hemoglobin variants and thalassemia mutations. This work was supported by PHS grants HG02238 to W.M. and DK27635 to R.C.H. and by support from the Huck Institute of Life Sciences at Penn State University.
REFERENCES
- 1.Forget B.G., Higgs,D.R., Steinberg,M. and Nagel,R.L. (2001) Disorders of Hemoglobin: Genetics, Pathophysiology and Clinical Management. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 2.Huisman T.H.J., Carver,M.F.H. and Baysal,E. (1997) A Syllabus of Thalassemia Mutations. The Sickle Cell Anemia Foundation, Augusta, GA, USA. [Google Scholar]
- 3.Huisman T.H.J., Carver,M.F.H. and Efremov,G.D. (1998) A Syllabus of Human Hemoglobin Variants. 2nd edn. The Sickle Cell Anemia Foundation, Augusta, GA, USA. [Google Scholar]
- 4.Hardison R.C., Chui,D.H., Giardine,B., Riemer,C., Patrinos,G.P., Anagnou,N., Miller,W. and Wajcman,H. (2002) HbVar: A relational database of human hemoglobin variants and thalassemia mutations at the globin gene server. Hum. Mutat., 19, 225–233. [DOI] [PubMed] [Google Scholar]
- 5.Cao A. (2002) Carrier screening and genetic counseling in beta-thalassemia. Int. J. Hematol., 76 (Suppl. 2), 105–113. [DOI] [PubMed] [Google Scholar]
- 6.Karolchik D., Baertsch,R., Diekhans,M., Furey,T.S., Hinrichs,A., Lu,Y.T., Roskin,K.M., Schwartz,M., Sugnet,C.W., Thomas,D.J., Weber,R.J., Haussler,D. and Kent,W.J. (2003) The UCSC Genome Browser Database. Nucleic Acids Res., 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clamp M., Andrews,D., Barker,D., Bevan,P., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T., Durbin,R., Eyras,E., Gilbert,J., Hammond,M., Hubbard,T., Kasprzyk,A., Keefe,D., Lehvaslaiho,H., Iyer,V., Melsopp,C., Mongin,E., Pettett,R., Potter,S., Rust,A., Schmidt,E., Searle,S., Slater,G., Smith,J., Spooner,W., Stabenau,A., Stalker,J., Stupka,E., Ureta-Vidal,A., Vastrik,I. and Birney,E. (2003) Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res., 31, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. and Wagner,L. (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giardine B., Elnitski,L., Riemer,C., Makalowska,I., Schwartz,S., Miller,W. and Hardison,R.C. (2003) GALA, a database for genomic sequence alignments and annotations. Genome Res., 13, 732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins F.S., Green,E.D., Guttmacher,A.E. and Guyer,M.S. (2003) A vision for the future of genomics research. Nature, 422, 835–847. [DOI] [PubMed] [Google Scholar]
- 11.Stamatoyannopoulos G. and Grosveld,F. (2001) Hemoglobin switching. In Stamatoyannopoulos,G., Majerus,P.W., Perlmutter,R.M. and Varmus,H. (eds), The Molecular Basis of Blood Diseases. W.B. Saunders Company, Philadelphia, PA, pp. 135–182. [Google Scholar]
- 12.Hardison R., Chao,K.-M., Schwartz,S., Stojanovic,N., Ganetsky,M. and Miller,W. (1994) Globin gene server: A prototype e-mail database server featuring extensive multiple alignments and data compilation. Genomics, 21, 344–353. [DOI] [PubMed] [Google Scholar]
- 13.Riemer C., ElSherbini,A., Stojanovic,N., Schwartz,S., Kwitkin,P.B., Miller,W. and Hardison,R. (1998) A database of experimental results on globin gene expression. Genomics, 53, 324–337. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz S., Kent,W.J., Smit,A., Zhang,Z., Baertsch,R., Hardison,R.C., Haussler,D. and Miller,W. (2003) Human-mouse alignments with Blastz. Genome Res., 13, 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz S., Elnitski,L., Li,M., Weirauch,M., Riemer,C., Smit,A., Green,E.D., Hardison,R.C. and Miller,W., NISC_Comparative_Sequencing_Program (2003) MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res., 31, 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]