Abstract
GeneDB (http://www.genedb.org/) is a genome database for prokaryotic and eukaryotic organisms. The resource provides a portal through which data generated by the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute and other collaborating sequencing centres can be made publicly available. It combines data from finished and ongoing genome and expressed sequence tag (EST) projects with curated annotation, that can be searched, sorted and downloaded, using a single web based resource. The current release stores 11 datasets of which six are curated and maintained by biologists, who review and incorporate information from the scientific literature, public databases and the respective research communities.
INTRODUCTION
The Pathogen Sequencing Unit (PSU) at the Wellcome Trust Sanger Institute sequences a large number of diverse prokaryotic and eukaryotic genomes (http://www.sanger.ac.uk/Projects/). In recent years, new sequencing and assembly technologies and collaborations between sequencing institutes have dramatically increased both the output and quality of sequence data. Maintenance and dissemination of such data required the development of an integrated, publicly accessible database.
During GeneDB’s development, four key points have been taken into consideration. GeneDB must be capable of storing and frequently updating sequences and annotations, irrespective of the status of the sequencing project. The resource should therefore support both the mining of preliminary datasets for gene discovery and the viewing of finished sequence data. Secondly, an intuitive user interface, which provides rapid access, visualization, searching and downloading of data, must be shared between the datasets. Thirdly, the database architecture should allow integration of diverse biological datasets with the sequence. Lastly, the use of structured vocabularies would ensure standardization, facilitating querying and comparison between species.
Currently, GeneDB houses the sequences and associated annotation of 11 organisms, including members of the bacteria, fungi, protozoa and arthropods. Of these, six are finished genomes and five are ongoing sequencing projects (Table 1).
Table 1. Details of the genomes either currently available in GeneDB or in preparation for addition to the database.
| Species | Genera | Genome size (haploid) (kb) | Status | Curated | Data type |
|---|---|---|---|---|---|
| Salmonella typhi | Bacteria | 4800 | Finished (15) | No | WGS |
| Schizosaccharomyces pombe | Fungi | 14 000 | Finished (16) | Yes | Clone based |
| Saccharomyces cerevisiae | 12 069 | Finished (17) | No | Clone based | |
| Aspergillus fumigatus | 35 000 | In progress | No | WGS | |
| Plasmodium falciparum | Protozoa | 22 900 | Completed (18) | Yes | WCS |
| Plasmodium chabaudi | 30 000 | In progress | Yes | WGS | |
| Plasmodium berghei | 26 000 | In progress | Yes | WGS | |
| Leishmania major | 33 600 | Completed | Yes | WCS | |
| Leishmania infantuma | ∼34 000 | In progress | No | WGS | |
| Trypanosoma brucei | 35 000 | In progress | Yes | WCS/B | |
| Trypanosoma congolensea | ∼35 000 | In progress | No | WGS | |
| Trypanosoma vivaxa | ∼35 000 | In progress | No | WGS | |
| Trypanosoma cruzia | ∼43 000 | Completed | No | WGS | |
| Dictyostelium discoideum | 34 000 | Completed | No | WCS/Y | |
| Glossina morsitans | Arthropoda | Unknown | In progress | No | EST |
In the ‘Status’ column, ‘Finished’ refers to published genomes without sequencing gaps and ‘Completed’ refers to genomes that are shotgun complete but still require gap closure. WGS = whole genome shotgun; clone based = sequenced on a clone by clone basis using physical maps; WCS/B = whole chromosome and BAC shotgun; WCS/Y = whole chromosome and YAC shotgun; EST = expressed sequence tag.
aDatasets to be added shortly.
DATA ANALYSES AND SYSTEM ARCHITECTURE
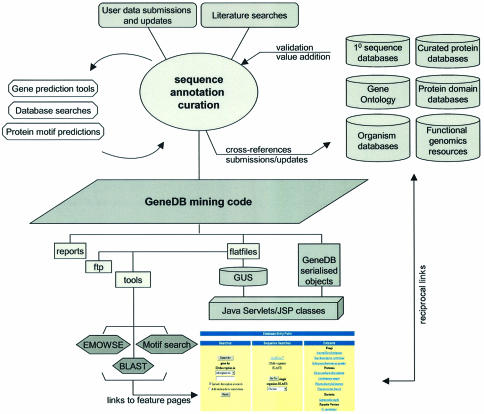
GeneDB stores sequence data and analyses generated via automated annotation pipelines, prior to manual annotation. The analysis pipelines include gene finding algorithms, protein feature predictions, BLAST and/or FASTA searches against nucleotide, protein and customized databases, protein domain and/or family search results and electronically inferred and manually revised gene ontology associations (GO) (1). Search results are continually reviewed during the curation process and complemented by additional datasets (Fig. 1).
Figure 1.
Analysis pipelines complemented by user-feedback, information gained from the literature and other public resources, generate data for GeneDB. Data are parsed through the mining code and serialized in binary files as well as stored in the GUS relational database (see accompanying text for further information).
The sequence and annotation files are processed by the GeneDB mining code, generating both the GeneDB Java objects and standardized files used to populate a Genomics Unified Schema (GUS) database (http://www.gusdb.org/). A set of data files including FASTA sequence files for third-party tools, such as BLAST, is also produced. Both the mining code and the GeneDB object layer take advantage of available code from the BioJava project. Access to the GeneDB data through the GeneDB website is provided by a set of servlets and Java Server Pages (JSP).
DATA CONTENT AND DISPLAY
The GeneDB homepage supplies links to the individual organism homepages. From these, researchers can take advantage of numerous ways to retrieve data and construct searches according to individual preferences and requirements. Clickable chromosome and contig maps, searchable text indices and browsable catalogues [GO assignments (1), descriptions, products, domains] provide fast and easy access. An additional query interface supports a wide range of queries on sequences and (curated) annotations stored in GUS, with the ability to combine searches with the Boolean operators AND and OR. For example, users can select all proteins of a specified length range with a specified number of introns. Other query options include GO assignments, keywords, chromosome, protein domains and predicted protein sequence features. The queries in each session are tracked via a history page, allowing further refinement of searches and downloading of results as a nucleotide or amino acid FASTA file. Furthermore, a variety of sequence similarity search facilities are available through GeneDB. In addition to WU-BLAST, GeneDB also supports omniBLAST, which permits searching across a set of selectable databases. An iterative BLAST (PSI-BLAST) search suited to the identification of distant homologues is envisaged to be available shortly. Peptide sequences can be searched with either user-specified motifs or using the peptide mass identification tool EMOWSE, part of the suit of EMBOSS open-source software tools (2). An alternative approach for accessing genes of interest is to use the official browser of the GO consortium, AmiGO. Several different methods are available for querying the data both externally (http://www.godatabase.org/) and internally via GeneDB, all of which include direct links to the gene pages.
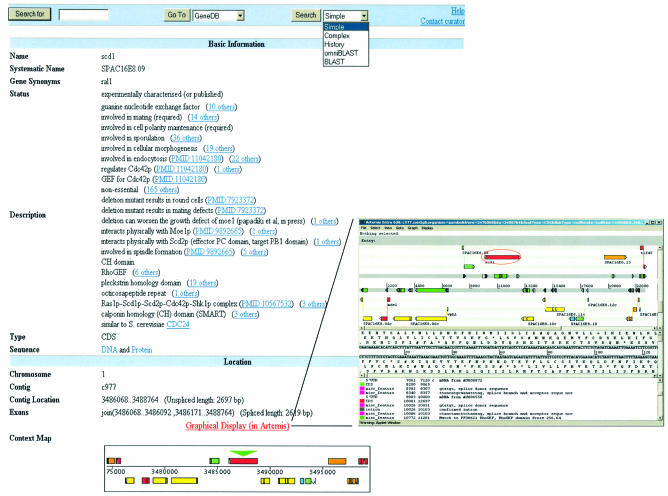
Feature pages, generated for coding sequences, display basic location information and a context map. The results of protein feature prediction algorithms [SignalP V2.0 (3), TMHMM v2.0 (4), GPI anchor predictions (http://129.194.185.165/dgpi/index_en.html)] and the manual annotation and curation processes are provided in both a graphical display and text format (Fig. 2). This information is complemented by the results of similarity searches, including the display of predicted and experimentally characterized orthologues and paralogues. Additional sequence features, both at the DNA level (e.g. polymorphisms, introns, UTRs, splice donor and acceptor sequences) and protein level (e.g. peptide domains), can be viewed in the context of the annotated sequence via an Artemis applet (5) (Fig. 2). The selected region can also be downloaded either in FASTA or annotated EMBL file format. Sequence data, either of the predicted coding sequence or the clustered ESTs, are accessible via a secondary page.
Figure 2.
The upper half of the S.pombe scd1 GeneDB feature page (http://www.genedb.org/genedb/Search?name=SPAC16E8.09&organism=pombe). Tools, a glossary and a feedback form, inviting comments and updates on a particular gene’s annotation, are accessible via the navigation bar. During the curation process, statements, extracted from public resources and the research community, are captured in structured syntax, referenced to the relevant database. Additional sequence features and annotation can be viewed in Artemis, accessible via the link above the gene neighbourhood map.
Extensive cross-referencing supports retrieval of related information from external resources, allowing rapid transfer between databases. This includes reciprocal links to numerous databases housing nucleotide and protein sequences [e.g. EMBL (6), Swiss-Prot/TrEMBL (7)], pathways [KEGG (8)], protein families [e.g. SCOP (9), Pfam (10), InterPro (11)], ontologies [e.g. GO (1)], expression data [e.g. microarray (http://www.sanger.ac.uk/perl/SPGE/geexview)], strain information [FYSSION (http://pombe.biols.susx.ac.uk)] and phenotype data [e.g. the Trypanosoma brucei RNAi project (http://www.TrypanoFAN.org/)]. Links to databases housing the same genome at different sites [e.g. SGD (12), TGAD (http://www.tigr.org/tdb/e2k1/tba1/tba1.shtml)] are also provided. These links to external resources are validated and updated on a monthly basis by the GeneDB mining code. Annotators and curators are automatically alerted to inconsistencies in the datasets and changed GO identifiers.
DATA CURATION
Experienced biologists curate data for six of the organisms in GeneDB (Table 1). Such curation involves several aspects, all aiming to facilitate data querying and retrieval. First as a number of organisms are sequenced by more than one sequencing centre, it ensures consistent annotation across the whole of the respective genome. Secondly, sequences and their annotation are updated according to new submissions to public databases, publications and contributions by the wider scientific community (Fig. 1). Public information is used not only to verify existing gene models and annotations but also to add value, enabling users to retrieve groups of genes/proteins not possible by purely computational methodologies. Wherever possible, controlled vocabularies such as GO (1) are used. In the absence of such vocabularies, statements are captured in structured syntax either in the description lines (Fig. 2) or in a dedicated curation field, providing a concise summary of the major aspects of a gene’s biology. Links are provided to PubMed records and other resources used to compile the statements. Text indices point to other products sharing the same description line.
Curators regularly exchange information and updates with the public databases, aiming to synchronize these datasets globally. Finally, the curation of related species (e.g. Plasmodium species, the Kinetoplastida) at one site enables extensive cross-referencing and comparative analyses between species, such as the inclusion of experimentally verified and predicted orthologues.
For three organisms (Schizosaccharomyces pombe, Leishmania major and Trypanosoma brucei), GeneDB curators are also involved in implementing nomenclature guidelines (13) and resolving nomenclature conflicts, ensuring accurate and complete retrieval of information.
FUTURE DEVELOPMENTS
GeneDB code development will continue to concentrate on integrating GeneDB with the GUS schema. Part of this development is a collaboration with the GUS team at the Computational Biology and Informatics Laboratory (CBIL, University of Pennsylvania) to design a common web interface architecture, permitting the creation of customized web pages to suit individual database requirements [e.g. GeneDB, PlasmoDB (14), AllGenes (http://www.allgenes.org/)].
Curation will be extended to integrate expression, phenotypic and interaction data. To this extent, GeneDB curators and developers have collaborated with the GUS team to implement modifications to the GUS schema in preparation for the incorporation of these large-scale biological data. Also, with the increasing emphasis on genomics projects of related organisms, the GeneDB team are already designing tools for comparative analyses that can be readily displayed via the web.
Furthermore, it is intended to substantially expand the available bacterial datasets to include all the bacterial genomes completed and published by the PSU (see http://www.sanger.ac.uk/Projects/Microbes/ for a comprehensive list).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the CBIL team, in particular Jonathan Crabtree, Steve Fischer, Jonathan Schug and Chris Stoeckert. We would also like to thank collaborating sequencing centres, in particular, Najib El-Sayed at The Institute for Genomic Research, Peter Myler and Ken Stuart at the Seattle Biomedical Research Institute, Adam Kuspa at Baylor College of Medicine, Angelika Noegel at the University of Cologne, Michel Veron at the Institut Pasteur, and Mike Lehane at the University of Wales, Bangor for sharing unpublished data as well as the numerous researchers who have contributed to the annotation of datasets in GeneDB. GeneDB is funded by the Wellcome Trust through its support of the Sanger Institute. GUS was developed by CBIL. GeneDB and the Centre for Tropical and Emerging Global Diseases, University of Georgia have made significant contributions to it as part of an ongoing collaborative effort with CBIL to further develop the schema.
REFERENCES
- 1.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen H. and Krogh,A. (1998) Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol., 6, 122–130. [PubMed] [Google Scholar]
- 4.Sonnhammer E.L., von Heijne,G. and Krogh,A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol., 6, 175–182. [PubMed] [Google Scholar]
- 5.Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.A. and Barrell,B. (2000) Artemis: sequence visualization and annotation. Bioinformatics, 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 6.Stoesser G., Baker,W., Van Den Broek,A., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V., Lopez,R. et al. (2003) The EMBL Nucleotide Sequence Database: major new developments. Nucleic Acids Res., 31, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M., Goto,S., Kawashima,S. and Nakaya,A. (2002) The KEGG databases at GenomeNet. Nucleic Acids Res., 30, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gough J., Karplus,K., Hughey,R. and Chothia,C. (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol., 313, 903–919. [DOI] [PubMed] [Google Scholar]
- 10.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng S., Dong,Q., Balakrishnan,R., Christie,K., Costanzo,M., Dolinski,K., Dwight,S.S., Engel,S., Fisk,D.G., Hong,E. et al. (2003) Saccharomyces Genome Database (SGD) provides biochemical and structural information for budding yeast proteins. Nucleic Acids Res., 31, 216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clayton C., Adams,M., Almeida,R., Baltz,T., Barrett,M., Bastien,P., Belli,S., Beverley,S., Biteau,N., Blackwell,J. et al. (1998) Genetic nomenclature for Trypanosoma and Leishmania. Mol. Biochem. Parasitol., 97, 221–224. [DOI] [PubMed] [Google Scholar]
- 14.Bahl A., Brunk,B., Crabtree,J., Fraunholz,M.J., Gajria,B., Grant,G.R., Ginsburg,H., Gupta,D., Kissinger,J.C., Labo,P. et al. (2003) PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res., 31, 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhill J., Dougan,G., James,K.D., Thomson,N.R., Pickard,D., Wain,J., Churcher,C., Mungall,K.L., Bentley,S.D., Holden,M.T. et al. (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature, 413, 848–852. [DOI] [PubMed] [Google Scholar]
- 16.Wood V., Gwilliam,R., Rajandream,M.A., Lyne,M., Lyne,R., Stewart,A., Sgouros,J., Peat,N., Hayles,J., Baker,S. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- 17.Goffeau A., Aert,R., Agostini-Carbone,M.L., Ahmed,A., Aigle,M., Alberghina,L., Albermann,K., Albers,M., Aldea,M., Alexandraki,D. et al. (1997) The Yeast Genome Directory. Nature, 387, 1–105. [Google Scholar]
- 18.Gardner M.J., Hall,N., Fung,E., White,O., Berriman,M., Hyman,R.W., Carlton,J.M., Pain,A., Nelson,K.E., Bowman,S. et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature, 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]