Abstract
This paper examines the benefits of Michaelis–Menten model reduction techniques in stochastic tau-leaping simulations. Results show that although the conditions for the validity of the reductions for tau-leaping remain the same as those for the stochastic simulation algorithm (SSA), the reductions result in a substantial speed-up for tau-leaping under a different range of conditions than they do for SSA. The reason of this discrepancy is that the time steps for SSA and for tau-leaping are determined by different properties of system dynamics.
INTRODUCTION
Biochemical systems typically involve complex networks with many reactions and many molecular species. While studying such systems, reduced models can not only benefit simulation speed but also aid in the understanding of complex models. The Michaelis–Menten (M–M) approximation1, 2 of enzyme–substrate reactions, which replaces the set of three reactions
| (1) |
with the single reaction
| (2) |
is a widely used model reduction technique for ordinary differential equation (ODE) chemical kinetics models, and has been the subject of refinements over the years.3, 4, 5
Recent studies have shown that the M–M approximation can also benefit the simulation of stochastic chemical kinetics models. With regard to the use of the M–M approximation in the stochastic simulation algorithm (SSA),6 Rao and Arkin7 derived a stochastic M–M approximation from the quasi-steady-state assumption (QSSA); and Mastny et al.8 verified the QSSA using perturbation analysis. Gillespie et al.9 examined the validity of the same abridgment for a similar but simpler reaction set
| (3) |
abridged to
| (4) |
They showed that the abridgment is valid for SSA under four sets of conditions, and that for only one of these cases does it result in a substantial speed-up for SSA. Sanft et al.10 extended the analysis to the M–M approximation and obtained similar results.
However, little attention has been paid to the abridgment when it is used in the context of tau-leaping. The tau-leaping algorithm11 is an approximate strategy to accelerate SSA simulations. This paper examines the conditions for validity and for speed-up of the abridgment applied to both reaction set 3 and reaction set 1 in the context of tau-leaping, and shows that the abridgment results in a substantial speed-up for tau-leaping under a different range of conditions than is the case for SSA.
BACKGROUND
Consider a well-stirred chemical reaction system with n molecular species S1, …, Sn and m reaction channels R1, …, Rm. Let xi(t) denote the population of species Si at time t, and x(t)=(x1(t),...,xn(t))T the state vector of the system at time t. Let each reaction Rj be characterized by its propensity function aj(x) and stoichiometry vector νj: the probability that one Rj reaction will occur in the next infinitesimal time interval [t, t + dt), given x(t)=x, is aj(x)dt, and the change to the system's state vector induced by one Rj reaction is νj. Then the dynamics of the whole system can be described by the chemical master equation.12 The SSA is an exact method to numerically solve the chemical master equation by simulating a large number of trajectories of the system.
The tau-leaping algorithm is an approximate method to accelerate SSA. Instead of simulating one reaction at a time, tau-leaping steps the system by a selected time interval, τ, during which many reactions may fire. The idea is that if the propensity functions are nearly constant during the interval, the number of times that each reaction fires can be approximated by a Poisson random number P(aj(x)τ). Then the state of the system can be advanced by the formula
| (5) |
The requirement that the propensities are nearly constant during the time interval [t, t + τ] is called the leap condition: for some ε ≪ 1,
| (6) |
The step size τ is dictated by the need to satisfy the leap condition.
It is easy to see that the tau-selection strategy is crucial to the accuracy and speed of tau-leaping simulations. Several strategies have been proposed. The most widely used strategy for mass action reactions is due to Cao et al.13 In that strategy, the leap condition is written in terms of the changes in species populations rather than the changes in propensities, and the expression for τ is given by
| (7) |
where ε ≪ 1 is the preset accuracy control parameter, νij are the stoichiometric terms, and gi is the highest order of reaction in which species Si appears as a reactant. (For further details, see Ref. 13.)
RESULTS
Abridgment of reaction set 3: Conditions for validity
Both SSA and tau-leaping are based on the same assumption that the dynamics of the chemical reaction system is governed by the propensity functions as defined above. In the case of the unimolecular reaction R3 in reaction set 3, given the population x2 of species S2, the probability for reaction R3 to fire in the next infinitesimal time dt is given by the product of the corresponding propensity function a3 and dt: a3dt = c3x2dt. Gillespie et al.9 showed that this condition is mathematically equivalent to the following condition: in the absence of competing reactions, the time to the next firing of reaction R3 is an exponential random variable with mean 1∕(c3x2). Therefore, the validity condition of the abridgment is that the time to the next firing of reaction R3 (the event of a S3 molecule being generated) must be approximately an exponential random variable with mean 1∕aa(x12), where x12 = x1 + x2 and aa(x12) is the propensity function of the abridged reaction. Note that aa is a function of x12 = x1 + x2, rather than a3 being a function of x2. Since both SSA and tau-leaping share the same assumption above of how the system dynamics is governed by propensity functions, this validity condition for the abridgment holds for both SSA and tau-leaping. The additional assumption in tau-leaping that the propensity functions are nearly constant during each time step does not affect the validity condition.
For reaction system 3, Gillespie et al.9 showed that the validity condition can be satisfied if and only if
| (8) |
which is satisfied if and only if at least one of the four conditions holds:
and the propensity function of the abridged model is given by
| (10) |
where x12 = x1 + x2.
Because SSA and tau-leaping share the same validity condition, 8 is also the condition for the validity of the abridgment for tau-leaping, and the propensity function is given by Eq. 10.
Abridgment of reaction set 3: Conditions for speed-up
Gillespie et al.9 also showed that under only one condition will the abridgment speed up the SSA simulation significantly: c2 ≫ c3. This is because the SSA simulates every reaction event of the system; thus the speed of an SSA simulation is determined by the number of reaction firings. When c2 ≫ c3, many fewer reaction firings take place in the abridged system than in the original system.
For tau-leaping, since the speed of the simulation is determined by the size of the leap step τ, we must re-examine the conditions for speed-up. The leap condition 7 requires that all propensity functions remain almost constant during a time step. Thus, the time step of tau-leaping is restricted by how fast the propensity functions change and is implicitly restricted by how fast the populations of the reactants change. The key to a significant acceleration of tau-leaping simulation by abridgment is to remove the intermediate highly reactive species with the following properties:
-
1
Their populations change rapidly, thus the step size of tau-leaping is restricted by those species.
-
2
The distributions of their populations remain almost constant for a much longer time than their populations do, thus the average values of the corresponding propensity functions over time during this new longer step can be approximated by the expectations of those propensity functions over the distributions.
For the abridgment of reaction set 3 to reaction set 4, the conditions for validity of the abridgment are given in Eqs. 8 or 9. To examine the extent of speed-up, we compare the time step τf of the full model 3 with the time step τa of the abridged model 4 under different conditions in the Appendix. Several interesting conclusions can be summarized from the analysis:
-
1
The abridgment always speeds up tau-leaping, as shown in A9, A18.
-
2
The conditions corresponding to a substantial speed-up from the abridgment for both tau-leaping and SSA are compared in Table 1. The condition εx12 ≫ 1 is assumed in all the conditions for tau-leaping, as this is the situation where tau-leaping is advantageous over SSA. It is easy to see that for most situations (except for when c2 ≫ c3), tau-leaping simulation benefits from the abridgment under a wider range of conditions than is the case for SSA simulation.
Table 1.
Conditions for a substantial speed-up from the abridgment, for tau-leaping and SSA, applied to reaction set 3.
|
c2≫̸︀c3 |
||||
|---|---|---|---|---|
| c2 ≫ c1 | c3 ≫ c1 | c1 ≫ c3 | c2 ≫ c3 | |
| Tau-leaping | c2 ≫ c1 or | |||
| SSA | ∅ | x12 > 0 | ||
Abridgment of enzyme-substrate system
For the enzyme–substrate system 1, Sanft et al.10 showed that the condition for validity of the stochastic M–M approximation is the same as that of the deterministic case,3 namely,
| (11) |
where ET = E(t) + ES(t), S0 = S(0), and Km = (c2 + c3)∕c1. The propensity function of the abridged system is also the same as the deterministic M–M rate:
| (12) |
where Vmax=c3ET. And, under only one condition will the abridgment speed up the SSA simulation significantly: c2 ≫ c3.
Applying the same arguments in Sec. 3A, the condition for validity and the stochastic M–M rate are the same for tau-leaping as for SSA.
On the other hand, for tau-leaping, the abridged system is no longer a mass-action system. However, it is easy to see that the propensity function 12 depends on the population of S, and
| (13) |
Thus the τ selection strategy in Eq. 7 with g = 1 can be used to calculate the tau-leaping step size of the abridged system:
| (14) |
To calculate the characteristic tau-leaping step size of the original system, we can make use of the corresponding ODE results as estimates of the expected populations:
| (15a) |
| (15b) |
Applying the τ selection strategy 7, we can easily see that τf < τa by examining the contribution to τ of the change in S. Then by examining the contribution to τ of the changes in E and ES, we have
| (16) |
When or , which is equivalent to
| (17) |
we obtain
| (18) |
| (19) |
Again, we are only interested in the case εS ≫ 1, where tau-leaping is advantageous over SSA. In this case, τf ≪ τa, and the speed gain grows as c2∕c3 increases. Note that since Km + S ≫ ET and ε ≪ 1, condition 17 is actually a very loose condition.
When and , which is equivalent to
| (20) |
we obtain
| (21) |
| (22) |
The condition for the abridgment to gain a substantial speed-up in this case is given by
| (23) |
Either , or c2 ≫ c3 and , or if the values of Km and S are widely separated will there be a large speed-up.
The conditions corresponding to a substantial speed-up from the M–M abridgment for both tau-leaping and SSA are compared in Table 2. It is easily seen that the M–M abridgment yields a significant speed-up for tau-leaping under a different range of conditions than it does for SSA, for the enzyme–substrate system.
Table 2.
Conditions for a substantial speed-up from the M–M abridgment, for tau-leaping and SSA, applied to the enzyme–substrate system 1.
| c2≫̸︀c3 | c2 ≫ c3 | |
|---|---|---|
| Tau-leaping | or | or |
| SSA | ∅ | S > 0 |
NUMERICAL EXAMPLES
Based on the analysis in Sec. 3, we tested and timed the abridgment to different reaction systems under different conditions using the adaptive tau-leaping code in STOCHKIT 2.0.14 The tests were carried on a personal computer with an Intel Core 2 Quad Q9300 2.5 GHz central processing unit and 4 GB RAM.
Speed-up of reaction set 3
For the abridgment of reaction set 3, we choose two sets of parameters to show the different speed-up behavior of SSA and tau-leaping. Parameter set I was chosen as follows: c1 = 1, c2 = 100, c3 = 100. This corresponds to the condition c2 + c3 ≫ c1. To show how condition A16 factors into the speed-up, we set x2(0) = x3(0) = 0, and varied x1(0) = x12(0). The simulation time and speed-up of SSA and tau-leaping under different initial conditions or to the different system end time tend, all with 10 000 runs, is shown in Table 3. ε was set to 0.01 in the τ selection. We can see from the table that: (a) the speed-up gain of SSA from the abridgment does not change as the initial condition changes. This agrees with the results of Gillispie et al.9 because the speed-up gain of SSA is determined solely by the c2∕c3 ratio; (b) tau-leaping gains a considerable speed-up when condition A16 is satisfied; and (c) the speed-up of tau-leaping is not large when condition A16 is not satisfied, but if tend is large enough the system will go into the region of condition A16 eventually and tau-leaping will benefit substantially from the abridgment. All of these results agree with the analysis.
Table 3.
Simulation time and speed-up of SSA and tau-leaping under different initial conditions, for parameter set I, applied to reaction set 3.
| SSA |
Tau-leaping |
||||||
|---|---|---|---|---|---|---|---|
| x1(0) | tend | Original(s) | Abridged(s) | Speed-up | Original (s) | Abridged (s) | Speed-up |
| 1 × 105 | 1 | 58 | 9 | 6 | 50 | 1 | 50 |
| 1 × 106 | 1 | 650 | 110 | 6 | 5 | 1 | 5 |
| 1 × 107 | 1 | 5800 | 900 | 6 | 3 | 1 | 3 |
| 1 × 106 | 10 | 1660 | 290 | 6 | 220 | 2 | 110 |
Parameter set II was chosen so that c1 ≫ c3 ≫ c2 to illustrate the only case for which tau-leaping would not gain a substantial speed-up, regardless of the system end time tend. The speed-up of tau-leaping under different initial conditions or to different tend is shown in Table 4. The other parameters and values are set to: c1 = 100, c2 = 0.01, c3 = 1, x2(0) = x3(0) = 0, ε = 0.01. Noting that c3∕(2εc2) = 5000 and , we can see that the results are consistent with condition A26: tau-leaping will benefit substantially from the abridgment when . A quick test of SSA shows that the speed-up for SSA under this set of parameters is between 3 and 4. Tau-leaping benefits from the abridgment more significantly than SSA when condition A26 is satisfied.
Table 4.
Speed-up of tau-leaping (abridged system vs original system) for different initial conditions or system end times, for parameter set II, applied to reaction set 3.
| x1(0) | 1 × 104 | 1 × 106 | 1 × 108 |
|---|---|---|---|
| tend=1 | 4 | 20 | 3 |
| tend=10 | 3 | 17 | 34 |
Speed-up of enzyme–substrate system
For the abridgment of enzyme–substrate system 1, we chose three sets of parameters and initial conditions to show the different speed-up behavior of SSA and tau-leaping:
-
1
c1 = 1, c2 = 10, c3 = 10, S(0) = 1 × 105, E(0) = 100, ES(0) = P(0) = 0, tend=10. This set corresponds to the condition c2≫̸︀c3 and . According to the analysis, only tau-leaping should be able to gain a substantial speed-up from the abridgment;
-
2
c1 = 1, c2 = 100, c3 = 1, S(0) = 1 × 105, E(0) = 100, ES(0) = P(0) = 0, tend=10. This set corresponds to the condition c2 ≫ c3 and . Under this condition, both SSA and tau-leaping should be able to gain a substantial speed-up; and
-
3
c1 = 1, c2 = 1 × 1010, c3 = 1 × 108, S(0) = 1 × 1010, E(0) = 1 × 108, ES(0) = P(0) = 0, tend=1×10−12. This set corresponds to the condition that c2 ≫ c3 holds but neither Eq. 17 nor Eq. 23 holds. The analysis suggests that only SSA should be able to gain a substantial speed-up from the abridgment.
The simulation time and speed-up of SSA and tau-leaping under different parameters and initial conditions, all with 10 000 runs, are shown in Table 5. It is easy to see that the results agree with the analysis.
Table 5.
Simulation time and speed-up of SSA and tau-leaping under different parameters and initial conditions, applied to enzyme–substrate system 1.
| SSA |
Tau-leaping |
|||||
|---|---|---|---|---|---|---|
| Parameters set | Original (s) | Abridged (s) | Speed-up | Original (s) | Abridged (s) | Speed-up |
| I | 28 | 8 | 4 | 39 | 0.08 | 488 |
| II | 139 | 0.8 | 174 | 221 | 0.02 | 1105 |
| III | 695 | 3 | 232 | 0.03 | 0.02 | 2 |
Accuracy of the M–M abridgment
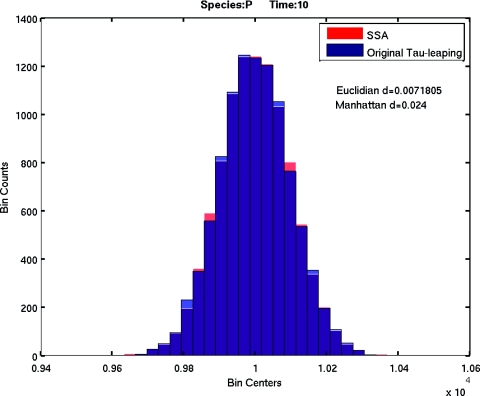
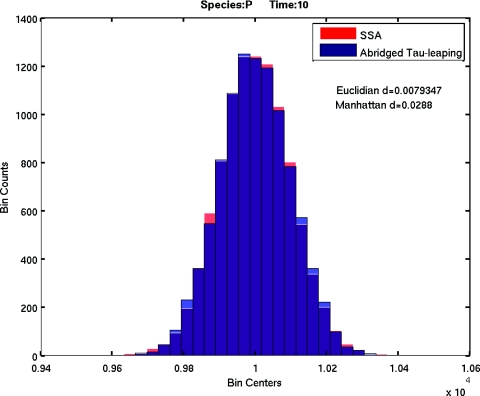
Next, we tested the accuracy of the M–M abridgment for tau-leaping. We chose an enyzme substrate reaction set with the following set of parameters: c1 = 1, c2 = 10, c3 = 10. Initial conditions were set to S(0) = 105, E(0) = 100, ES(0) = P(0) = 0. The system end time was set to 10. Thus, the validity condition for the M–M approximation 11 will be satisfied all the time. The baseline is the histogram of product P at the system end time of 10 000 SSA simulations of the original enzyme substrate model. 10 000 tau-leaping simulations were performed for both the original model and the abridged M–M model. The comparison of the histograms of the product P for both SSA simulation and tau-leaping simulation of the original model is shown in Fig. 1, while the comparison of the histograms of P for SSA simulation and tau-leaping simulation of the abridged model is shown in Fig. 2. The Euclidian distance and Manhattan distance in the figures are respectively L2 norm and L1 norm of the histogram distance:15 suppose X and Y are two groups of samples with N samples in X and M samples in Y, and all the sample values are bounded in the interval I=[xmin,xmax). Let L=xmax−xmin. Divide the interval I into K subintervals Ii=[xmin+(i−1)L∕K,xmin+iL∕K). Then the histogram distance is given by
| (24) |
where the characteristic function χ(x, Ii) is defined as
| (25) |
The histogram distance results show that the M–M abridgment is accurate and valid under this condition. Different conditions are also tested and all of them show that the M–M abridgment is valid when the validity condition 11 is satisfied.
Figure 1.
The comparison of histograms of product P at the system end time of SSA simulation and tau-leaping simulation of the original model.
Figure 2.
The comparison of histograms of product P at the system end time of SSA simulation and tau-leaping simulation of the abridged model.
Circadian oscillation model
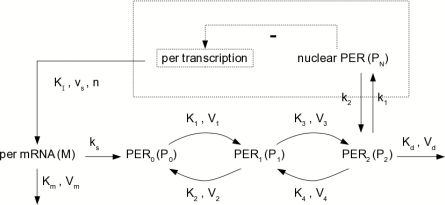
Next, we consider a more complicated model: a circadian oscillation model in the Drosophila period protein (PER).16 The scheme of the model is shown in Fig. 3. In this model, per mRNA (M) is synthesized in the nucleus and transfers to the cytosol at a maximum rate vs. It is also degraded by an enzyme there with a maximum rate Vm and Michaelis constant Km. The PER protein ( PER 0) is synthesized at a rate proportional to M by a first-order rate constant ks. The reversible phosphorylations of the PER proteins, between P0 and P1 and between P1 and P2, are governed by a set of Michaelis–Menten reactions at maximum rates Vi, with Michaelis constants Ki (i = 1, 2, 3, 4). The bisphosphorylated form P2 is degraded by an enzyme with a maximum rate Vd and Michaelis constant Kd, and transported into the nucleus with a first-order rate constant k1. The nuclear bisphosphorylated form of PER PN is transported into the cytosol with a first-order rate constant k2, while it also exerts a negative feedback on per transcription described by a Hill equation with repression threshold constant KI and Hill coefficient n. The detailed kinetic laws, parameter values, and initial conditions can be found in Goldbeter.16
Figure 3.
Scheme of the model for circadian oscillations in PER. Six out of the ten reactions are Michaelis–Menten reactions.
To compare the results with the original M–M model to a comparable mass-action model, we converted this 5 species, 10 reaction model into a full 17 species, 22 reaction stochastic model by replacing all the 6 Michaelis–Menten reactions with enzyme–substrate reactions. The system volume was chosen to be the characteristic size of a cell nucleus, 1000 μm3. Then we varied the ratios of backward dissociation rates [c2 in system 1] to forward dissociation rates [c3 in system 1] of the enzyme–subsrate compounds, to see the different speed-up behavior of the Michaelis–Menten abridgment applied to SSA and tau-leaping. To simplify the problem, we took all of the six ratios (corresponding to c2∕c3) to be the same. The simulation time and speed-up of SSA and tau-leaping with different c2∕c3 ratios, all with 1000 runs and tend=1, is shown in Table 6. The result shows that for this model, SSA will gain a large speed-up from the M–M abridgment only when c2 ≫ c3 is satisfied, while tau-leaping can benefit substantially from the M–M abridgment under a wider range of conditions.
Table 6.
Simulation time and speed-up of SSA and tau-leaping with different c2∕c3 ratios, applied to the circadian oscillation model.
| SSA |
Tau-leaping |
|||||
|---|---|---|---|---|---|---|
| c2∕c3 | Original (s) | Abridged (s) | Speed-up | Original (s) | Abridged (s) | Speed-up |
| 0.01 | 1042 | 674 | 2 | 1271 | 3 | 424 |
| 1 | 1701 | 674 | 3 | 2103 | 3 | 701 |
| 100 | 78 200 | 674 | 116 | 83 800 | 3 | 2.8 × 104 |
ACKNOWLEDGMENTS
The authors gratefully acknowledge financial support from the National Institute of Biomedical Imaging and Bioengineering through Grant No. R01EB007511, the U.S. Department of Energy (DOE) through Grant No. DEFG02-04ER25621, and the Institute for Collaborative Biotechnologies through Grant No. DFR3A-8-447850-23002 from the U.S. Army Research Office (USARO). The work of Yang Cao has also been supported by grants from National Science Foundation (NSF) under Grant Nos. CCF-0726763 and CCF-0953590 and from National of Institutes of Health (NIH) under Grant No. 2R01GM078989.
APPENDIX: ANALYSIS OF SPEED-UP OF THE ABRIDGMENT OF REACTION SET 3
For this analysis, we assume that εx12 ≫ 1, as this is the situation where tau-leaping is advantageous over SSA.
For the abridged model 4, applying the τ selection strategy 7, we get
| (A1) |
On the other hand, the step size τf of the original model 3 is restricted by the minimum of
Since τf varies with x1 and x2, the expectations of x1 and x2 are used to calculate the characteristic time step of the original system. Because it is a linear system, the expectations of x1 and x2 are the same as the results of the corresponding ODE solutions. Let and denote the expectation of x1 and x2, and x′ = dx∕dt. Then
To evaluate the τ values in A2, we are concerned with the ratio r between and :
| (A4) |
Substituting A4 into A3, we have
| (A5) |
Applying the QSSA (r′ = 0) and requiring that r > 0, we obtain
| (A6) |
Now consider the different conditions under which the abridgment is valid:
-
1
In this case,
and εx1 ≈ εx12 ≫ 1 as assumed. Thus the τf is at most(A8) (A9) Further calculation reveals that τ21 = ∞ and τ12 > τ22. Thus τf = min {τ11, τ22}.
When εx2 ⩽ 1, we have
thus (recalling that εx12 ≫ 1)(A10) (A11) In this case a significant speed up is ensured. Condition εx2 ⩽ 1 is equivalent to
When εx2 > 1, we have(A12) (A13) (A14) To yield a substantial speed-up in this case, the following condition must be satisfied:(A15) As a consequence, when c2 + c3 ≫ c1, the condition for the abridgment to gain a significant speed up in tau-leaping is A12 or A15:
Since c2 + c3 ≫ c1 and ε ≪ 1, this is actually a very loose condition. If the end time of a simulation is long enough, the simulation will always gain a large speed up when there are enough S1 and S2 consumed and the system goes into the region of condition A16.(A16) -
2
In this case, x1 ≈ c2x12∕(c1 + c2), x2 ≈ c1x12∕(c1 + c2), and the τf is at most
(A18) Further calculation reveals that τ11 = ∞. Thus τf = min {τ21, τ12, τ22}.
If c2 ≫ c1, then τ21 ≪ τa, thus τf < τ12 ≪ τa.
If c2 ∼ c1, then , both εx1 ≫ 1 and εx2 ≫ 1 hold as assumed, thus
The condition to yield a substantial speed gain in this case is(A19)
If c2 ≪ c1, then ,(A20) (A21)
whence(A22) (A23)
To yield a major speed-up, at least one of A23 or A24 should be much less than 1, which is equivalent to(A24) (A25) Consequently, when c1 + c2 ≫ c3, the condition of a substantial speed up in tau-leaping is
Notice c1 + c2 ≫ c3, this is also a very loose condition. If the end time of a simulation is long enough, the simulation can always gain a large speed up except for the case c2 ≪ c3 and x12≫̸︀c3∕(2ɛc2).(A26)
References
- Michaelis L. and Menten M. L., Biochem. Z. 49, 333 (1913). [Google Scholar]
- Nelson D. and Cox M. M., Lehninger Principles of Biochemistry, 4th ed. (Freeman, New York, 2005). [Google Scholar]
- Segel L. A. and Slemrod M., SIAM Rev. 31, 446 (1989). 10.1137/1031091 [DOI] [Google Scholar]
- Tzafriri A. R. and Edelman E. R., J. Theor. Biol. 226, 303 (2004). 10.1016/j.jtbi.2003.09.006 [DOI] [PubMed] [Google Scholar]
- Pedersen M. G., Bersani A. M., and Bersani E., Bull. Math. Biol. 69, 433 (2005). 10.1007/s11538-006-9136-2 [DOI] [PubMed] [Google Scholar]
- Gillespie D. T., J. Phys. Chem. 81, 2340 (1977). 10.1021/j100540a008 [DOI] [Google Scholar]
- Rao C. and Arkin A. P., J. Chem. Phys. 118, 4999 (2003). 10.1063/1.1545446 [DOI] [Google Scholar]
- Mastny E. A., Haseltine E. L., and Rawlings J. B., J. Chem. Phys. 127, 094106 (2007). 10.1063/1.2764480 [DOI] [PubMed] [Google Scholar]
- Gillespie D. T., Cao Y., Sanft K. R., and Petzold L. R., J. Chem. Phys. 130, 064103 (2009). 10.1063/1.3072704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanft K. R., Gillespie D. T., and Petzold L. R., IET Syst. Biol. 5, 58 (2011). 10.1049/iet-syb.2009.0057 [DOI] [PubMed] [Google Scholar]
- Gillespie D. T., J. Chem. Phys. 115, 1716 (2001). 10.1063/1.1378322 [DOI] [Google Scholar]
- Gillespie D. T., Annu. Rev. Phys. Chem. 58, 35 (2007). 10.1146/annurev.physchem.58.032806.104637 [DOI] [PubMed] [Google Scholar]
- Cao Y., Gillespie D. T., and Petzold L. R., J. Chem. Phys. 124, 044109 (2006). 10.1063/1.2159468 [DOI] [PubMed] [Google Scholar]
- See http://engineering.ucsb.edu/~cse/StochKit for details of the STOCHKIT 2.0 software.
- Cao Y. and Petzold L. R., J. Comp. Phys. 212, 6 (2006). 10.1016/j.jcp.2005.06.012 [DOI] [Google Scholar]
- Goldbeter A., Proc. Biol. Sci. 261, 319 (1995). 10.1098/rspb.1995.0153 [DOI] [PubMed] [Google Scholar]