Abstract
The SWISS-MODEL Repository is a database of annotated three-dimensional comparative protein structure models generated by the fully automated homology-modelling pipeline SWISS-MODEL. The Repository currently contains about 300 000 three-dimensional models for sequences from the Swiss-Prot and TrEMBL databases. The content of the Repository is updated on a regular basis incorporating new sequences, taking advantage of new template structures becoming available and reflecting improvements in the underlying modelling algorithms. Each entry consists of one or more three-dimensional protein models, the superposed template structures, the alignments on which the models are based, a summary of the modelling process and a force field based quality assessment. The SWISS-MODEL Repository can be queried via an interactive website at http://swissmodel.expasy.org/repository/. Annotation and cross-linking of the models with other databases, e.g. Swiss-Prot on the ExPASy server, allow for seamless navigation between protein sequence and structure information. The aim of the SWISS-MODEL Repository is to provide access to an up-to-date collection of annotated three-dimensional protein models generated by automated homology modelling, bridging the gap between sequence and structure databases.
INTRODUCTION
Three-dimensional protein structures are key to a detailed understanding of the molecular basis of protein function. Combining sequence information with 3D structure gives invaluable insights for the development of effective rational strategies for experiments such as site-directed mutagenesis, studies of disease related mutations, or the structure based design of specific inhibitors. Techniques for experimental structure solution by X-ray crystallography and nuclear magnetic resonance spectroscopy have made great progress in recent years and currently more than 22 000 experimental protein structures have been deposited in the Protein Data Bank PDB (1). However, experimental protein structure determination is still a time-consuming process without guaranteed success. This is reflected by the fact that the number of structurally characterized proteins is about two orders of magnitude smaller than the number of known protein sequences in the Swiss-Prot and TrEMBL (2) databases, which hold more than one million entries. Thus, no experimental structural information is available for the vast majority of protein sequences. Therefore, theoretical methods for protein structure prediction aiming to bridge this structure knowledge gap have gained much interest in recent years. Among all current computational approaches, homology modelling is the only method that can reliably generate a three-dimensional model for a protein (3). If a target protein shares significant amino acid sequence similarity to at least one experimentally solved three-dimensional structure (template), homology or comparative modelling can be applied to construct a three-dimensional model for the new protein. During the past few years, several structural genomics initiatives (4) were started with the goal to speed up the experimental elucidation of new protein folds. Protein structure determination and comparative modelling complement one another in the exploration of the protein structure space (5).
Information from three-dimensional comparative protein models is used routinely in a wide variety of applications (6,7). The usefulness of homology models for specific applications is strongly dependent on their quality. The accuracy of a protein model can be evaluated by assessing the deviation of the model from its actual experimentally determined structure. Manual assessment of prediction methods, e.g. during the biannual CASP experiments (8), is a good means to evaluate new algorithmic developments based on a small number of examples. Likewise, automated blind assessment of modelling servers provides statistically meaningful estimates for the expected accuracy and stability of automated prediction methods (9,10). Several attempts at automated evaluation of modelling methods have been developed during recent years (11–13). SWISS-MODEL was among the first modelling servers to join the EVA (12) project, which continuously and automatically monitors the accuracy and reliability of the participating protein structure prediction pipelines. Applications for high quality models are manifold, and include planning site-directed mutagenesis experiments and rationalizing the effect of mutations (14–16), characterization of molecular functions (17,18) and structure based drug design (19,20). Although medium accuracy models are prone to significant errors (7,21), often such inaccuracies are located in the variable surface and loop regions, while the conserved core and active sites are modelled correctly. These protein models can still provide a valuable basis for identifying functionally relevant residues for site-directed mutagenesis experiments, or for the validation of sequence based functional annotations (22).
Homology modelling of protein structures consists of four steps: template selection, target-template alignment, model building, and model evaluation. Each of these individual steps usually requires expertise in structural biology and the use of specialized computer programs. A huge and constantly growing number of structurally uncharacterized protein sequences together with the increasing number of available template structures motivated the development of automated, stable and reliable modelling methods (23,24). The idea of an internet based automated modelling facility with integrated expert knowledge was first implemented 10 years ago by Peitsch and co-workers (23,25) and formed the starting point for the SWISS-MODEL server. With the presently available computing power, it is possible to apply comparative modelling on a large scale to whole genomes, e.g. Escherichia coli (26), Saccharomyces cerevisiae (24), or entire sequence databases (27,28). It was proposed by Sanchez et al. (29) that the availability of structural information for whole protein families, organisms, or metabolic pathways will encourage new types of applications. For example, the development of drugs with higher selectivity for a given target protein would be facilitated by the availability of structural models for all proteins sharing similar ligand binding sites. Structural comparisons would allow screening for drug candidates with better specificity at an earlier stage of drug development.
Storing and organizing results of large-scale automated modelling in a database makes better use of the available computing resources, and gives instant and queryable access to models without having to wait for a computation to complete. The easy access to pre-computed and annotated comparative models through a model repository helps to enrich other database projects with structural information, e.g. sequence knowledge bases like Swiss-Prot (2), or databases dedicated to specific cellular functions, e.g. the meiosis specific database GermOnline (30). In this paper, we describe the SWISS-MODEL Repository, a database of annotated three-dimensional protein models created by the SWISS-MODEL server pipeline (31).
SWISS-MODEL REPOSITORY
Web access
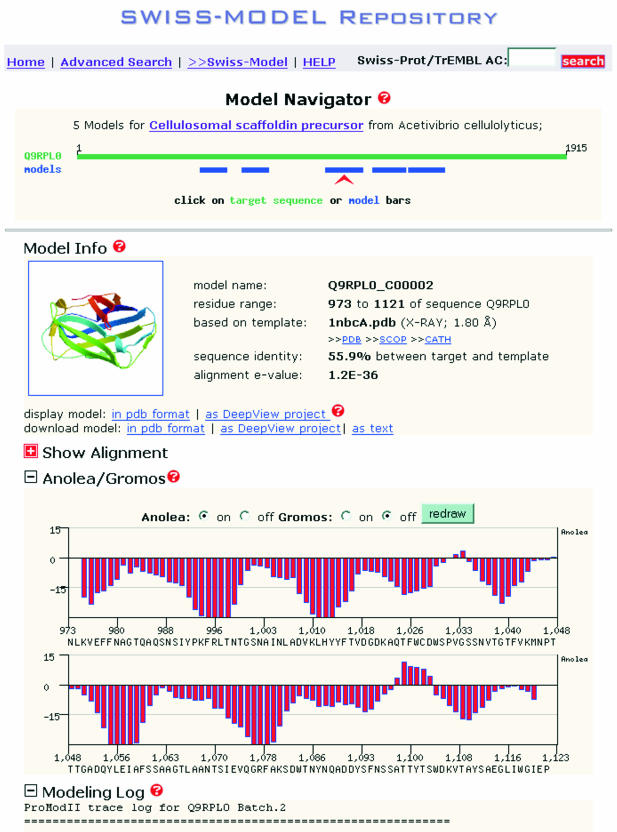
The aim of the SWISS-MODEL Repository is to provide access to an up-to-date collection of annotated models generated by automated homology modelling, bridging the gap between sequence and structure databases. All models in the Repository are publicly accessible via our interactive website at http://swissmodel.expasy.org/repository/. The design of the web page is printer-friendly, so that all information can be printed in one step from any standard web browser (Fig. 1). A graphical ‘model navigator’ provides an overview of the models that have been generated for a selected sequence, allowing fast and easy navigation for the different regions in the protein, for which three-dimensional models are available. The ‘model info’ section contains information about the template structure and target-template sequence alignment on which the modelling has been based. The interactive display allows expanding detailed views of the target-template sequence alignment, the force field based assessment of the model and the modelling log files. The model assessment is presented as a diagram of Gromos96 (32) force field energies and Anolea (33) mean force potentials on a per residue basis. These allow visual inspection of the model quality to identify unreliable regions, e.g. caused by errors in the target-template alignment. A small ribbon representation is included to obtain a first impression of the model structure. Model coordinates can be downloaded in PDB format or as DeepView projects. Protein models can be displayed directly from within the web browser using any molecular viewer application, e.g. DeepView (25), Dino (http://www.dino3d.org), or Rasmol (34). Moreover, complete DeepView (Swiss-PdbViewer) modelling projects can be exported. These project files contain the final model superposed on the template structure. DeepView is used to visualize the model and analyse certain structural features, e.g. Ramachandran plots or electrostatic properties. It allows manual adjustment of the placement of insertions and deletions in the alignment on which the initial modelling process was based. The project with the modified alignment can then be re-submitted to the SWISS-MODEL server for further model building.
Figure 1.
Typical view of a SWISS-MODEL Repository entry. The ‘Model Navigator’ allows toggling between the different models available (blue bars) for the given protein sequence (green bar).
The Repository can be queried for protein or gene name, Swiss-Prot accession codes, protein description key words, E.C. numbers and organism names. The search interface allows combining all these different descriptors to complex queries, e.g. searching the Repository for all models of a certain enzyme in several organisms. For each model, the Repository provides links to the target sequence entry in Swiss-Prot (2), the template structure entries in PDB (1), SCOP (35) and CATH (36), and domain organization in InterPro (37). Cross-linking individual repository entries to and from other databases, e.g. ExPASy (38), allows navigation between protein sequence and structure information.
Content and update
As of August 2003, the Swiss-Model Repository contained 317 616 models for 282 096 different Swiss-Prot/TrEMBL sequence entries, i.e. for 34% of the 132 244 Swiss-Prot (Release 41.19) and 25% of 941 322 trEMBL (Release 24.6) entries, a significant part of the sequence could be modelled. The length of the models varies from 45 to 1524 residues (ferredoxin-dependent glutamate synthase from Oryza sativa), with an average model size of 200, which corresponds well with the expected size of individual protein domains; 47% of the models in the Repository correspond to eukaryotic proteins, 4.7% to human protein sequences, and 20% are of prokaryotic origin. The Repository is updated regularly to take into account new sequences, modifications of existing sequence entries, and new template structures released by the PDB that might allow the construction of models for previously unmodelled proteins, or might provide a better template for already existing model entries. Also, fundamental changes and improvements of the modelling pipeline initiate a new update cycle. An individual checksum is assigned to each model entry to ensure the consistency of the data with the information in other databases.
The SWISS-MODEL Repository has been implemented using relational database technology. During the modelling process, it communicates with the SWISS-MODEL server pipeline and keeps track of the workflow for individual target sequences. The models in the SWISS-MODEL Repository are computed by a modified version of the SWISS-MODEL server pipeline (31). Since no manual intervention takes place during the model building process, care must be taken to assess the quality of models generated to minimize the number of erroneous models in the database. We have defined criteria for entry of models to the Repository based on the EVA evaluation of the SWISS-MODEL server during a period of 150 weeks comprising 12 100 models built for 9125 individual proteins, and the evaluation during the 3D Crunch experiment comprising 1200 model/control structure comparisons (21,39). Additionally, each model is assessed using a partial Gromos96 force field implementation (32) and the empirical Anolea mean force potential (33). Protein models with a minimum length of 45 residues sharing at least 40% sequence identity with their template structure are entered into the database if their Anolea mean force potential is below 200 kJ/mol. The chosen threshold values for models to enter the Repository are specific for the current implementation of the modelling pipeline and will be adjusted with improvements of the modelling algorithms.
OUTLOOK
The number of models in the Repository is expected to grow rapidly as a result of ongoing genome sequencing and structural genomics efforts. We will continue to develop the SWISS-MODEL Repository as a resource connecting sequence to structure information. Integrating InterPro domain information (37) in our data schema will provide functional annotation mapped onto three-dimensional structural models. The SWISS-MODEL Repository will widen its spectrum of the provided biological information by adding species-specific views and cross-linking with other knowledge bases.
CITATION
Users of the SWISS-MODEL Repository are requested to cite this article in their publications.
Acknowledgments
ACKNOWLEDGEMENTS
We are deeply indebted to Manuel C. Peitsch (Novartis AG, Basel) and to Nicolas Guex (GSK, Raleigh, NC) for their pioneering work on large-scale protein structure modelling. We would like to thank Jozef Aerts (Biozentrum and SIB, Basel) for excellent technical help on the Anolea mean force potentials. We are grateful to Nicola Mulder, Rolf Apweiler (EBI Hinxton, UK) and Lorenza Bordoli (EMBnet and Biozentrum and SIB, Basel) for very fruitful and encouraging discussions. We would like to acknowledge the financial support by the Swiss National Science Foundation (SNF) and Novartis Pharma AG, Basel.
REFERENCES
- 1.Westbrook J., Feng,Z., Chen,L., Yang,H. and Berman,H.M. (2003) The Protein Data Bank and structural genomics. Nucleic Acids Res., 31, 489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tramontano A., Leplae,R. and Morea,V. (2001) Analysis and assessment of comparative modeling predictions in CASP4. Proteins, 45 (Suppl. 5), 22–38. [DOI] [PubMed] [Google Scholar]
- 4.Brenner S.E. (2001) A tour of structural genomics. Nature Rev. Genet., 2, 801–809. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez R., Pieper,U., Melo,F., Eswar,N., Marti-Renom,M.A., Madhusudhan,M.S., Mirkovic,N. and Sali,A. (2000) Protein structure modeling for structural genomics. Nature Struct. Biol., 7 (Suppl.), 986–990. [DOI] [PubMed] [Google Scholar]
- 6.Kopp J., Peitsch,M.C. and Schwede,T. (2003) Protein structure modeling. In Galperin,M.Y. and Koonin,E.V. (eds), Frontiers in Computational Genomics. Caister Academic Press, Norfolk, Vol. 3, pp. 89–121. [Google Scholar]
- 7.Marti-Renom M.A., Stuart,A.C., Fiser,A., Sanchez,R., Melo,F. and Sali,A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct., 29, 291–325. [DOI] [PubMed] [Google Scholar]
- 8.Moult J., Fidelis,K., Zemla,A. and Hubbard,T. (2001) Critical assessment of methods of protein structure prediction (CASP): round IV. Proteins, 45 (Suppl. 5), 2–7. [PubMed] [Google Scholar]
- 9.Moult J., Fidelis,K., Zemla,A., Hubbard,T. and Tramontano,A. (2002) The significance of performance ranking in CASP—response to Marti-Renom et al. Structure, 10, 291–293. [DOI] [PubMed] [Google Scholar]
- 10.Marti-Renom M.A., Madhusudhan,M.S., Fiser,A., Rost,B. and Sali,A. (2002) Reliability of assessment of protein structure prediction methods. Structure, 10, 435–440. [DOI] [PubMed] [Google Scholar]
- 11.Bujnicki J.M., Elofsson,A., Fischer,D. and Rychlewski,L. (2001) LiveBench-2: large-scale automated evaluation of protein structure prediction servers. Proteins, 45 (Suppl. 5), 184–191. [DOI] [PubMed] [Google Scholar]
- 12.Eyrich V.A., Marti Renom,M.A., Przybylski,D., Madhusudhan,M.S., Fiser,A., Pazos,F., Valencia,A., Sali,A. and Rost,B. (2001) EVA: continuous automatic evaluation of protein structure prediction servers. Bioinformatics, 17, 1242–1243. [DOI] [PubMed] [Google Scholar]
- 13.Fischer D., Barret,C., Bryson,K., Elofsson,A., Godzik,A., Jones,D., Karplus,K.J., Kelley,L.A., MacCallum,R.M., Pawowski,K. et al. (1999) CAFASP-1: critical assessment of fully automated structure prediction methods. Proteins, 37 (Suppl. 3), 209–217. [DOI] [PubMed] [Google Scholar]
- 14.Bajorath J. and Aruffo,A. (1997) Construction and analysis of a detailed three-dimensional model of the ligand binding domain of the human B cell receptor CD40. Proteins, 27, 59–70. [PubMed] [Google Scholar]
- 15.Srinivasan N., Antonelli,M., Jacob,G., Korn,I., Romero,F., Jedlicki,A., Dhanaraj,V., Sayed,M.F., Blundell,T.L., Allende,C.C. et al. (1999) Structural interpretation of site-directed mutagenesis and specificity of the catalytic subunit of protein kinase CK2 using comparative modelling. Protein Eng., 12, 119–127. [DOI] [PubMed] [Google Scholar]
- 16.Wattenhofer M., Di Iorio,M.V., Rabionet,R., Dougherty,L., Pampanos,A., Schwede,T., Montserrat-Sentis,B., Arbones,M., Iliades,T., Pasquadibisceglie,A. et al. (2002) Mutations in the TMPRSS3 gene are a rare cause of childhood non-syndromic deafness in Caucasian patients. J. Mol. Med., 80, 124–131. [DOI] [PubMed] [Google Scholar]
- 17.Vaidehi N., Floriano,W.B., Trabanino,R., Hall,S.E., Freddolino,P., Choi,E.J., Zamanakos,G. and Goddard,W.A.,III (2002) Prediction of structure and function of G protein-coupled receptors. Proc. Natl Acad. Sci. USA, 99, 12622–12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray D. and Honig,B. (2002) Electrostatic control of the membrane targeting of C2 domains. Mol. Cell, 9, 145–154. [DOI] [PubMed] [Google Scholar]
- 19.Schafferhans A. and Klebe,G. (2001) Docking ligands onto binding site representations derived from proteins built by homology modelling. J. Mol. Biol., 307, 407–427. [DOI] [PubMed] [Google Scholar]
- 20.Schapira M., Raaka,B.M., Das,S., Fan,L., Totrov,M., Zhou,Z., Wilson,S.R., Abagyan,R. and Samuels,H.H. (2003) Discovery of diverse thyroid hormone receptor antagonists by high-throughput docking. Proc. Natl Acad. Sci. USA, 100, 7354–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwede T., Diemand,A., Guex,N. and Peitsch,M.C. (2000) Protein structure computing in the genomic era. Res. Microbiol., 151, 107–112. [DOI] [PubMed] [Google Scholar]
- 22.Duret L., Guex,N., Peitsch,M.C. and Bairoch,A. (1998) New insulin-like proteins with atypical disulfide bond pattern characterized in Caenorhabditis elegans by comparative sequence analysis and homology modeling. Genome Res., 8, 348–353. [DOI] [PubMed] [Google Scholar]
- 23.Peitsch M.C. (1996) ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans., 24, 274–279. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez R. and Sali,A. (1998) Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl Acad. Sci. USA, 95, 13597–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 26.Peitsch M.C., Wilkins,M.R., Tonella,L., Sanchez,J.C., Appel,R.D. and Hochstrasser,D.F. (1997) Large-scale protein modelling and integration with the SWISS-PROT and SWISS-2DPAGE databases: the example of Escherichia coli. Electrophoresis, 18, 498–501. [DOI] [PubMed] [Google Scholar]
- 27.Peitsch M.C. (1997) Large scale protein modelling and model repository. Proc. Int. Conf. Intell. Syst. Mol. Biol., 5, 234–236. [PubMed] [Google Scholar]
- 28.Pieper U., Eswar,N., Stuart,A.C., Ilyin,V.A. and Sali,A. (2002) MODBASE, a database of annotated comparative protein structure models. Nucleic Acids Res., 30, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez R., Pieper,U., Melo,F., Eswar,N., Marti-Renom,M.A., Madhusudhan,M.S., Mirkovic,N. and Sali,A. (2000) Protein structure modeling for structural genomics. Nature Struct. Biol., 7 (Suppl.), 986–990. [DOI] [PubMed] [Google Scholar]
- 30.Wiederkehr C., Basavaraj,R., Sarrauste de Menthière,C., Hermida,L., Koch,R., Schlecht,U., Amon,A., Brachat,S., Breitenbach,M., Briza,P. et al. (2004) GermOnline, a cross-species community knowledgebase on germ cell growth and development. Nucleic Acids Res., 32, D509–D511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwede T., Kopp,J., Guex,N. and Peitsch,M.C. (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res., 31, 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Gunsteren W.F., Billeter,S.R., Eising,A.A., Hünenberger,P.H., Krüger,P., Mark,A.E., Scott,W.R.P. and Tironi,I.G. (1996) Biomolecular Simulations: The GROMOS96 Manual and User Guide. VdF Hochschulverlag ETHZ, Zurich, Switzerland. [Google Scholar]
- 33.Melo F. and Feytmans,E. (1998) Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol., 277, 1141–1152. [DOI] [PubMed] [Google Scholar]
- 34.Sayle R.A. and Milner-White,E.J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci., 20, 374. [DOI] [PubMed] [Google Scholar]
- 35.LoConte L., Brenner,S.E., Hubbard,T.J.P., Chothia,C. and Murzin,A.G. (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orengo C.A., Pearl,F.M.G., Bray,J.E., Todd,A.E., Martin,A.C., Lo,C.L. and Thornton,J.M. (1999) The CATH Database provides insights into protein structure/function relationships. Nucleic Acids Res., 27, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gasteiger E., Jung,E. and Bairoch,A. (2001) Swiss-Prot: connecting biological knowledge via a protein database. Curr. Issues Mol. Biol., 3, 47–55. [PubMed] [Google Scholar]
- 39.Peitsch M.C., Schwede,T. and Guex,N. (2000) Automated protein modelling—the proteome in 3D. Pharmacogenomics, 1, 257–266. [DOI] [PubMed] [Google Scholar]