Abstract
CADRE is a public resource for housing and analysing genomic data extracted from species of Aspergillus. It arose to enable maintenance of the complete annotated genomic sequence of Aspergillus fumigatus and to provide tools for searching, analysing and visualizing features of fungal genomes. By implementing CADRE using Ensembl, a framework is in place for storing and comparing several genomes: the resource will thus expand by including other Aspergillus genomes (such as Aspergillus nidulans) as they become available. CADRE is accessible at http://www.cadre.man.ac.uk.
INTRODUCTION
Aspergillus is a genus of fungi found worldwide; over 180 species are officially recognized (1), some of which are of medical or industrial importance. Aspergillus fumigatus is the most common mould pathogen of humans, causing both life-threatening invasive disease in immunocompromised patients and allergic disease in patients with atopic immune systems (2). Aspergillus nidulans, an occasional human pathogen, is a model organism that has contributed to our understanding of genetics, gene regulation and cellular biology (3,4), while Aspergillus niger (5,6) and Aspergillus oryzae (7) are both used in industrial processes. Several other Aspergillus species are known to be significant allergens or to be responsible for mycotoxin production on stored food (8–10).
Interest in A.fumigatus has increased in recent years, not only because it is the most frequently isolated Aspergillus species from patients, but also because the incidence of invasive aspergillosis is rising (11). Moreover, it produces a number of toxins, such as fumagillin, which has been developed as a treatment for angiogenesis and microsporidiosis (12). To gain a better insight into the pathogenicity of this organism, an international consortium was established in 1998 to sequence the small (∼30 Mb) A.fumigatus genome (13). Sequencing is almost complete and first pass annotation is being carried out by the Wellcome Trust Sanger Institute (UK) and The Institute for Genomic Research (USA). A Central Aspergillus Data REpository (CADRE) was subsequently established in 2001 to manage the information produced by the sequencing effort, to contribute secondary annotation and to facilitate future comparative studies by incorporating genomic data from A.nidulans and other Aspergillus species as they become available.
The principal role of CADRE is to manage the complete annotated genomic sequence of A.fumigatus. Using a subset of these data as a test case, we have therefore implemented a database and Web-based software to facilitate searching and visualization of genomic features. These tools offer relatively simple displays for viewing gene and protein annotation, as well as more complex displays for viewing different gene predictions and other sequence features (e.g. RNA-encoding genes and repeats).
SOURCE DATA AND METHODS
Source data were provided by the A.fumigatus pilot sequence project. Using a bacterial artificial chromosome (BAC) genomic library, which was constructed using DNA from a clinical isolate of A.fumigatus (AF293), a bidirectional clone-by-clone walk was undertaken from niaD-positive BAC clones. Sixteen BAC clones were completely sequenced to yield an assembly of 922 kb, for which 360 protein-coding genes and eight tRNA genes have been predicted. This sequence and set of annotated genes were used to implement a database that will eventually house the full genome.
The infrastructure used to organize the genomic data was provided by the Ensembl system (14). This comprises: (i) a database schema that has been designed for storing annotated eukaryotic genomes; (ii) BioPerl and Ensembl object-oriented modules for describing biological entities; (iii) a series of Perl scripts that generate Web pages for viewing genomic data and (iv) an annotation pipeline for predicting genetic features.
As Ensembl was developed for the management and automated annotation of large eukaryotic genomes, we adapted this system both to handle smaller genomes (such as those of fungi), and to accommodate automatic and manual annotation provided by different research groups. For A.fumigatus, annotation has been provided in an XML format, which we hope will be adopted by the Aspergillus annotation community for data exchange.
The foundation of CADRE is Ensembl version 8.1. The database schema has been implemented using the MySQL relational database management system.
CONTENT OF CURRENT RELEASE
Release 1.0 (September 2003) contains information pertaining to the A.fumigatus pilot sequence project and includes 368 predicted genes. Each of these has been given a unique CADRE identifier and has been classified as known, putative or novel. Known genes are those corresponding to previously characterized A.fumigatus genes or orthologous genes from other Aspergillus species, whose protein sequences are available in the public databases. Putative genes are those found to be similar to known publicly available protein sequences. Novel genes are those predicted with no similarity to any known protein. Of the total number of predicted protein-coding genes, 35 have been classified as known genes, 149 as putative and 176 as novel.
DISPLAY AND SEARCH SOFTWARE
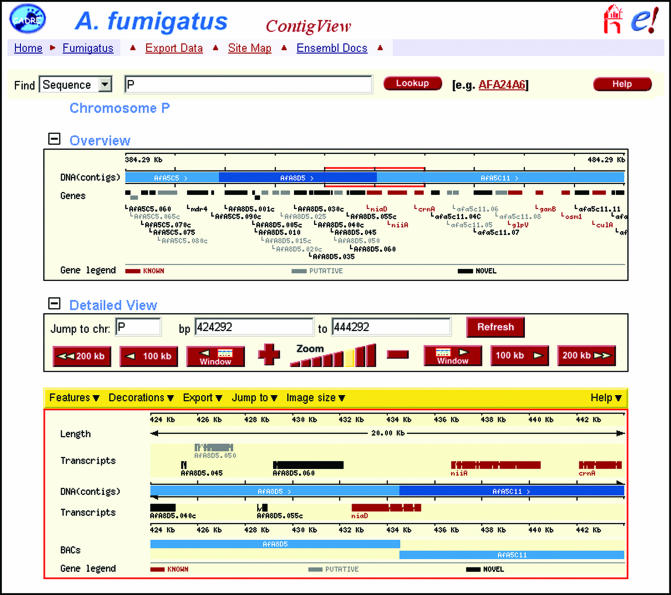
Several tools are provided for viewing genomic data within CADRE, the three main ones being ContigView, GeneView and ProtView. ContigView (Fig. 1) is the principal data visualization tool in the Ensembl system. It provides a high-level view of the contigs that make up a genome assembly, as well as the genomic features that have been mapped onto it. ContigView can also be customized: i.e. display colours can be changed and features can be added or removed to aid data assimilation. Within CADRE, this view is provided on two levels: (i) an ‘overview’, displaying predicted genes within a 100 kb region of the assembly, and (ii) a ‘detailed view’, showing a range of predicted features within a smaller region of the assembly (by default 10 kb is shown).
Figure 1.
Screenshot of ContigView. A region of 100 kb around the niaD locus is shown in the top panel (or ‘Overview’); names for all predicted genes within that region of the assembly are also displayed. A region of 20 kb around the niaD locus is shown in the bottom panel (or ‘Detailed View’); names and structures of all predicted transcripts within that region of the assembly are also displayed. In both panels, transcripts are colour-coded according to their classification: i.e. known, red; putative, grey; and novel, black. Each transcript is labelled with its standard name, if known; otherwise the systematic name is used.
The main features of ContigView are predicted transcripts, which are colour-coded according to our classification system and displayed parallel to the assembly in accordance with their position and strand orientation. Each transcript provides a pop-up menu of additional information (systematic feature name, CADRE transcript and gene identifiers) and hyperlinks to other views (GeneView, ProtView and ExportView). The position of other features, such as BAC clones, tRNAs and start/stop codons, can also be presented alongside the transcripts. The ability to integrate a range of features within a single view is a vital facility provided by Ensembl. As the resource expands, it will allow CADRE to provide results obtained from various prediction programs, as well as data from other research groups, thereby aiding functional assignment. In addition, it will facilitate genome comparison, as transcripts in other Aspergillus genomes found to be similar to those in the currently viewed genome can also be displayed. Ensembl provides two means of handling information gathered by other groups: data can be stored in-house, as an auxiliary database, or it can be dynamically imported using the Distributed Annotation System (15). ContigView is extensible and can act as a portal to other databases, providing the opportunity for collaborative genome analysis amongst research groups.
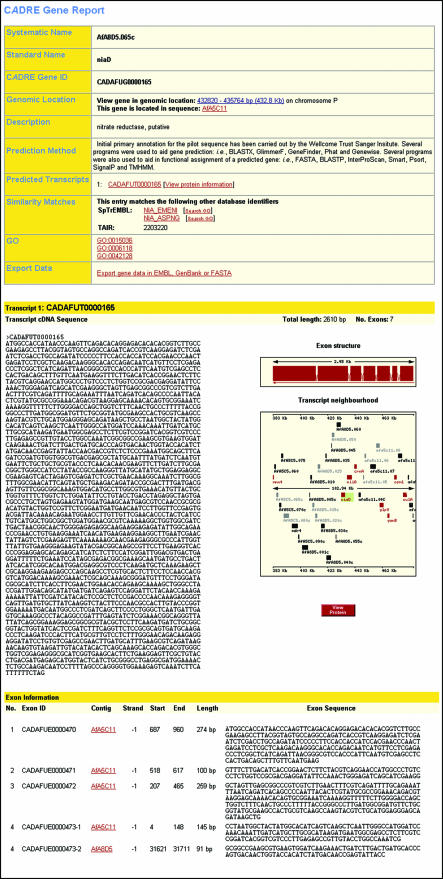
GeneView (Fig. 2) provides detailed information about a particular gene. The summary table at the top of the report provides: (i) the systematic feature name; (ii) the standard gene name, as represented in the literature; (iii) the CADRE gene identifier; (iv) the chromosomal location; (v) a short description of the gene, manually transferred from the external sequence database entry [e.g. SWISS-PROT (16)] to which the predicted gene mapped; (vi) how the gene was predicted; (vii) a list of predicted transcripts, each of which is hyperlinked to the sequence and its translation; (viii) a list of cross-references to similar sequences; (ix) GO terms that have been mapped to the gene and (x) a link to ExportView, for data download.
Figure 2.
Screenshot of GeneView. The report generated for the gene niaD is shown. The top section provides a summary of information concerning the gene of interest, whereas the bottom section provides further details pertaining to each predicted transcript. For each transcript, a clickable image of the transcript neighbourhood is provided. As in ContigView, each transcript in this image is colour-coded and provides a pop-up menu of further information and hyperlinks. In addition, exon (only some are shown here) and splice-site (omitted) information is provided for each transcript.
Below the summary table are reports describing each predicted transcript. Each report provides: (i) the cDNA sequence; (ii) an image of the exon structure; (iii) the transcript neighbourhood, highlighting the transcript of interest; (iv) exon information (CADRE exon identifier, contig identifier, strand orientation, contig coordinates and exon sequence) and (v) splice-site information (CADRE identifiers of adjacent exons and splice-site sequence). For (iv) and (v), an exon may lie across two contigs: in this event, the exon sub-sequences are distinguished by a numerical suffix, e.g. CADAFUE0000473-1 and CADAFUE0000473-2.
ProtView provides information about a particular protein. The summary table at the top of this report provides: (i) the CADRE protein identifier; (ii) the corresponding CADRE gene identifier; (iii) the name of the protein, manually transferred from the external sequence database entry to which the predicted protein mapped and (iv) how the protein was predicted. Below this table, the sequence is provided in FASTA format, with a link to the transcript within GeneView. ProtView is also able to provide information about any matches to family- or domain-based databases [e.g. Pfam (17) and PRINTS (18)] and structural features (e.g. transmembrane, low complexity and coiled-coil regions). However, this information has not yet been stored for the pilot sequence.
Other views available are CytoView and ExportView. CytoView allows navigation and display of much larger sections of an assembly than ContigView (i.e. up to 50 Mb can be shown). ExportView allows data to be downloaded as a FASTA sequence, a tab-delimited feature list or a flat file in EMBL or GenBank format.
For all of the above views, a search box is provided, allowing searches against any of the main features present in the database (i.e. sequence, gene, transcript and peptide) using identifiers or descriptions.
FUTURE DIRECTIONS
To address the need for continuing management and ongoing annotation of Aspergillus genomic data within CADRE, we are implementing an automated annotation pipeline. We will also establish a community annotation notice board to aid manual annotation. Our policy is to provide reusable code, which will be made available for other groups using Ensembl-based databases. For all stored genomes, we will eventually provide two sets of transcripts: (i) predicted transcripts—those originally annotated by the sequencing centres and (ii) revised transcripts—those annotated by the sequencing centres and edited over time to reflect current public sequence databases and literature.
Other areas of development will be ‘views’ that form part of the standard Ensembl system that have not yet been implemented in CADRE, e.g. BLASTView and SyntenyView. BLASTView will allow similarity searches against DNA and protein sequences within CADRE. SyntenyView will allow us to provide information pertaining to the conservation of large-scale gene order between any two stored Aspergillus genomes.
CONCLUSIONS
Using Ensembl as a foundation, we have provided the Aspergillus research community with a platform for collaboration. Through data integration and its range of analysis tools, CADRE will increasingly support comparative genomics and functional analyses of an important group of fungi, thereby establishing its role as a central Aspergillus data repository.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank the Pathogen Sequencing Unit (Sanger) for providing the primary annotation of the pilot sequence. We also wish to thank the Pathogen Sequencing Unit, the Ensembl Team (Sanger/European Bioinformatics Institute) and the Fungal Genomics Laboratory (North Carolina State University) for useful discussions on implementing CADRE. CADRE is funded by the Wellcome Trust (grant no. 062322).
REFERENCES
- 1.Pitt J.I., Samson,R.A. and Frisvad,J.C. (2000) List of accepted species and their synonyms in the family Trichocomaceae. In Samson,R.A. and Pitt,J.I. (eds), Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Harwood, Amsterdam, pp. 9–49. [Google Scholar]
- 2.Marr K.A., Patterson,T. and Denning,D. (2002) Aspergillosis. Pathogenesis, clinical manifestations, and therapy. Infect. Dis. Clin. North Am., 16, 875–894. [DOI] [PubMed] [Google Scholar]
- 3.Pontecorvo G., Roper,J.A., Hemmons,L.M., MacDonald,K.D. and Bufton,A.W.J. (1953) The genetics of Aspergillus nidulans. Adv. Genet., 5, 141–239. [DOI] [PubMed] [Google Scholar]
- 4.Martinelli S.D. and Kinghorn,J.R. (eds) (1994) Aspergillus: 50 Years On. Elsevier, Amsterdam. [Google Scholar]
- 5.Bennett J.W. and Klich,M.A. (1999) Aspergillus. In Flickinger,M.C. and Drew,S.W. (eds), Encyclopedia of Bioprocess Technology: Fermentation, Biocatalysis, and Bioseparation. John Wiley and Sons, Inc., New York, pp. 213–220. [Google Scholar]
- 6.Ruijter G.J.G., Kubicek,C.P. and Visser,J. (2002) Production of organic acids by fungi. In Osiewacz,H.D. (ed.), Industrial Applications. Springer-Verlag, Berlin, The Mycota, Vol. X, pp. 213–230. [Google Scholar]
- 7.Nout M.J. and Aidoo,K.E. (2002) Asian fungal fermented food. In Osiewacz,H.D. (ed.), Industrial Applications. Springer-Verlag, Berlin, The Mycota, Vol. X, pp. 23–47. [Google Scholar]
- 8.Blyth W., Grant,I.W., Blackadder,E.S. and Greenberg,M. (1977) Fungal antigens as a source of sensitization and respiratory disease in Scottish maltworkers. Clin. Allergy, 7, 549–562. [DOI] [PubMed] [Google Scholar]
- 9.Jarvis J.Q. and Morey,P.R. (2001) Allergic respiratory disease and fungal remediation in a building in a subtropical climate. Appl. Occup. Environ. Hyg., 16, 380–388. [DOI] [PubMed] [Google Scholar]
- 10.Pitt J.I. (2000) Toxigenic fungi: which are important? Med. Mycol., 38 (Suppl. 1), 17–22. [PubMed] [Google Scholar]
- 11.Groll A.H., Shah,P.M., Mentzel,C., Schneider,M., Just-Nuebling,G. and Huebner,K. (1996) Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J. Infect., 33, 23–32. [DOI] [PubMed] [Google Scholar]
- 12.Coyle C., Kent,M., Tanowitz,H.B., Wittner,M. and Weiss,L.M. (1998) TNP-470 is an effective antimicrosporidial agent. J. Infect. Dis., 177, 515–518. [DOI] [PubMed] [Google Scholar]
- 13.Denning D.W., Anderson,M.J., Turner,G., Latge,J.P. and Bennett,J.W. (2002) Sequencing the Aspergillus fumigatus genome. Lancet Infect. Dis., 2, 251–253. [DOI] [PubMed] [Google Scholar]
- 14.Clamp M., Andrews,D., Barker,D., Bevan,P., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V. et al. (2003) Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res., 31, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowell R.D., Jokerst,R.M., Day,A., Eddy,S.R. and Stein,L. (2001) The Distributed Annotation System. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.-C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attwood T.K., Bradley,P., Flower,D.R., Gaulton,A., Maudling,N., Mitchell,A.L., Moulton,G., Nordle,A., Paine,K., Taylor,P. et al. (2003) PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res., 31, 400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]