Abstract
Purpose
Mutations in tetraspanin 12 (TSPAN12) have recently been identified as a cause of autosomal dominant familial exudative vitreoretinopathy (FEVR). The purpose of this study was to detect TSPAN12 mutations in Chinese patients with FEVR and to describe the associated phenotypes.
Methods
Sanger sequencing was used to analyze the seven coding exons and their adjacent regions of TSPAN12 in 49 unrelated FEVR patients. Clinical phenotypes of the patients with TSPAN12 mutations were documented.
Results
Three novel heterozygous mutations in TSPAN12 were identified in three patients from unrelated families: c.146C>T (p.Thr49Met), c.313T>C (p.Cys105Arg), and c.601delC (p.Leu201PhefsX14). All three mutations involved highly conserved residues and were not present in 180 normal individuals. Ocular phenotypes included retinal folds, inferotemporal dragging of the optic disc and macula, increased vessels in the equatorial region, and a peripheral avascular zone. A father and his daughter had the same mutation but the father only had mild peripheral fundus changes while his daughter had obvious dragged disc and macular ectopia.
Conclusions
Our results suggest that TSPAN12 mutations are responsible for FEVR. Similar to patients with mutations in NDP, LRP5, or FZD4, the phenotypes associated with TSPAN12 mutations showed great variations between different individuals within a family and between the two eyes in individual patients.
Introduction
Familial exudative vitreoretinopathy (FEVR, OMIM 133780) is a hereditary vitreoretinal disorder characterized by developmental anomalies of the retinal vessels [1]. The primary anomaly is a premature arrest of the vascularization in the peripheral retina, which may lead to retinal neovascularization, vitreoretinal traction, exudates, fibrovascular masses, vitreous hemorrhages, retinal folds, and tractional retinal detachment. FEVR exhibits strikingly variable phenotypes among patients, even for patients from the same family or between the two eyes of an individual patient. The most severe form results in complete blindness, whereas minimally affected individuals can be totally asymptomatic, in which nonperfusion or avascular zones in the peripheral retina may only be identified by fluorescein angiography [2].
FEVR can be inherited as an autosomal dominant (OMIM 133780) [1,3-5], autosomal recessive (OMIM 601813) [6,7], or X-linked (OMIM 305390) [8,9] trait, and the autosomal dominant form is the most common [10,11]. Mutations in NDP (OMIM 300658) [8,12], FZD4 (OMIM 604579) [13], and LRP5 (OMIM 603506) [14,15] have been reported to be responsible for FEVR. The proteins encoded by these genes have all been shown to participate in the Wnt/Norrin signaling pathway [16-19]. Mutations in NDP, FZD4, and LRP5 account for approximately half of all FEVR cases, which indicates that additional causative genes remain to be identified [20-26].
Recently, heterozygous mutations in tetraspanin 12 (TSPAN12; OMIM 613138), which is a component of the Norrin-FZD4-LRP5 signaling complex [16,19], have been found to be responsible for autosomal dominant FEVR [27,28]. However, the frequency of TSPAN12 mutations and their associated phenotypes need further study.
In this study, Sanger sequencing was used to analyze the coding and adjacent intronic regions of TSPAN12 in 49 unrelated Chinese FEVR patients. Three novel heterozygous mutations were identified, and the associated phenotypes were described.
Methods
Patients
Probands with FEVR from 49 unrelated families were collected at the Pediatric and Genetic Eye Clinic, Zhongshan Ophthalmic Center, Guangzhou, China. Of the 49, 34 were sporadic and 15 had a family history of FEVR. Clinical diagnosis of FEVR was based on the presence of at least one of the following clinical findings that suggested primary retinal vascular developmental defects as previously described [21,23,24]: a peripheral retinal avascular zone with or without fibrous proliferation, peripheral neovascularization showing increased branching or a brushlike border, a peripheral fibrovascular mass, temporal dragging of the optic disc and/or macula, straightening of the temporal retinal vessels, falciform retinal folds, tractional retinal detachment with or without retinal exudates or vitreous hemorrhages, or total retinal detachment with fibrotic mass behind the lens. Retinal fluorescein angiography was performed in some suspicious cases to confirm the diagnosis of FEVR. Patients with a possible diagnosis of retinopathy of prematurity were excluded from the study. This study was approved by the Internal Review Board (IRB) of the Zhongshan Ophthalmic Center. It complied with the guidelines of the Declaration of Helsinki and the Guidance of Sample Collection of Human Genetic Diseases (863-Plan) of the Ministry of Public Health of China. Informed consent was obtained from the participating individuals or their guardians before the collection of clinical data and genomic samples.
Detection of TSPAN12 mutations
Genomic DNA was retrieved from our genomic DNA repository, which was established for the genetic study of hereditary eye disease. Seven pairs of primers (Table 1) were designed to amplify all the coding exons and the adjacent intronic sequences of TSPAN12 (reference sequences from NCBI: NC_000007.13 for gDNA, NM_012338.3 for mRNA, and NP_036470.1 for protein). Touchdown PCR was performed, with the annealing temperature decreased by 2 °C after the first 5 cycles and the second 5 cycles, and then down to the optimal annealing temperature (listed in Table 1) for the remaining 25 cycles. The sequences of the amplicons were determined with an ABI BigDye Terminator Cycle Sequencing Kit, v3.1, using an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequences from patients and TSPAN12 consensus sequences from the NCBI human genome database (NC_000007.13) were compared using the SeqManII program of the Lasergene package (DNAstar, Madison, WI). Each variation was initially confirmed by bidirectional sequencing and then evaluated in 180 normal individuals (360 chromosomes). The description of the mutations was based on the recommendations of the Human Genomic Variation Society. The effect of a missense mutation on the encoded protein was predicted by the PolyPhen-2 online tool [29].
Table 1. Primers used for PCR amplification and sequencing of TSPAN12.
| Exon | Forward primer(5′-3′) | Reverve primer(5′-3′) | Amplicon size (bp) | Annealing tempeture (°C) |
|---|---|---|---|---|
| 2 |
GCTGTGGGAAGCGTGAT |
TTAGCCATGCCCTTTGG |
281 |
58 |
| 3 |
TTCAAGATGCAGCAAATGGTAAT |
GCTATGGGCAGGAAAAACTT |
329 |
58 |
| 4 |
ATGTCTTGGGTGCATTT |
AAGCGTCCCTTCTTACA |
321 |
56 |
| 5 |
AGGGCTCACTGAGATGA |
TCTTGGGCAGTTCTTTC |
382 |
55 |
| 6 |
TTAGAAGACATTCCGAGTA |
AATTGCACCAGAGGTTA |
360 |
55 |
| 7 |
TCCTTTACTACATTTCTATC |
TAGCTTTCTTCTGCTTC |
462 |
52 |
| 8 | ACAGATTGTTTGCTTTC | AGGTGTTATTTTATGGC | 505 | 52 |
Results
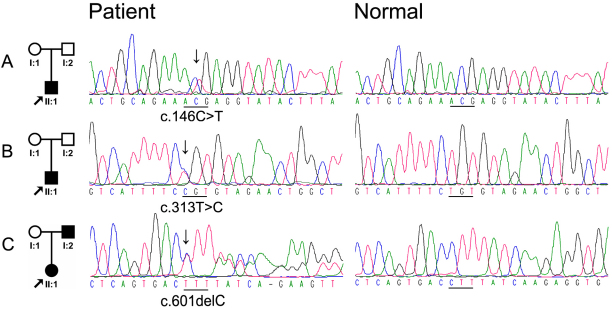
After sequencing the coding and adjacent regions of TSPAN12 in 49 unrelated FEVR patients, three novel heterozygous mutations were detected in three patients: c.146C>T, c.313T>C, and c.601delC (Figure 1). These three mutations were not present in the 180 normal individuals.
Figure 1.
Mutations identified in TSPAN12. A: The patient in the family had c.146C>T mutation. B; The patient in the family had c.313T>C mutation. C: The affected father and daughter had c.601delC mutation. The columns from left to right display the pedigree and the sequence chromatograms for these patients and the normal controls.
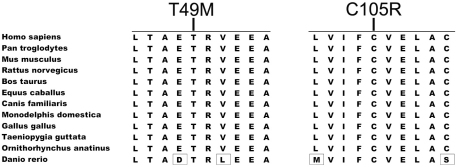
The c.146C>T mutation in exon 3 changed the encoded residue from a hydrophilic threonine to a sulfur-containing hydrophobic methionine at codon 49 (p.Thr49Met), which involves a highly evolutionarily conserved residue (Figure 2). This mutation was predicted to probably be damaging by PolyPhen-2. The c.313T>C mutation in exon 5 changed the encoded residue from a thiol-containing cysteine to a positively charged basic arginine at codon 105 (p.Cys105Arg), which is highly conserved (Figure 2). This mutation was also predicted to probably be damaging by PolyPhen-2. The c.601delC mutation in exon 7 not only changed the residue at codon 201, but also created a frame shift with 14 additional new residues before a premature termination at codon 215 (p.Leu201PhefsX14). This mutation also involved an evolutionarily conserved region.
Figure 2.
Protein alignment of 12 (TSPAN12) orthologs demonstrates conservation in the regions with mutations. The 12 orthologs are from the following 12 species: Homo sapiens (NP_036470), Pan troglodytes (XP_001142304), Mus musculus (NP_766595), Rattus norvegicus (Q569A2), Bos taurus (NP_001039977), Equus caballus (XP_001502093), Canis familiaris (XP_855095), Monodelphis domestica (XP_001364876), Gallus gallus (NP_001007850), Taeniopygia guttata (XP_002192381), Ornithorhynchus anatinus (XP_001516347), and Danio rerio (NP_957446). The regions with the two missense mutations are highly conserved. T49M stands for p.Thr49Met and C105R stands for p.Cys105Arg.
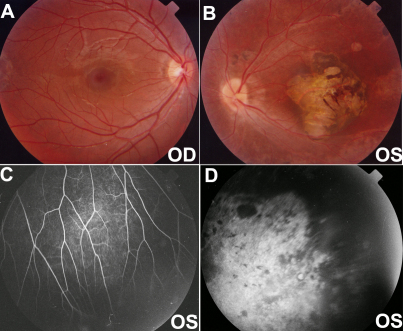
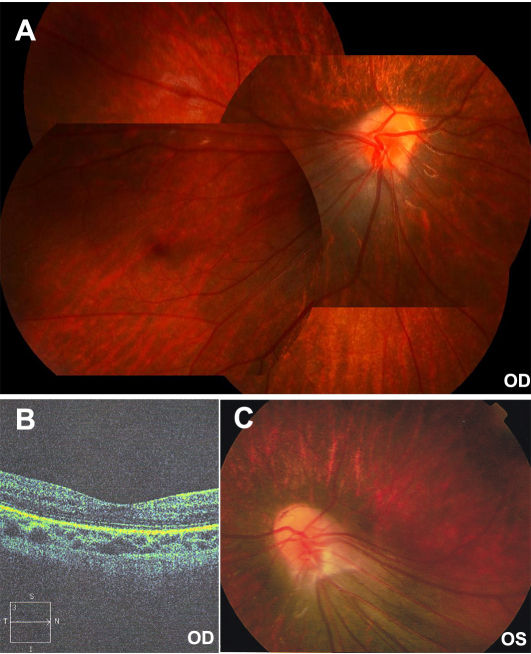
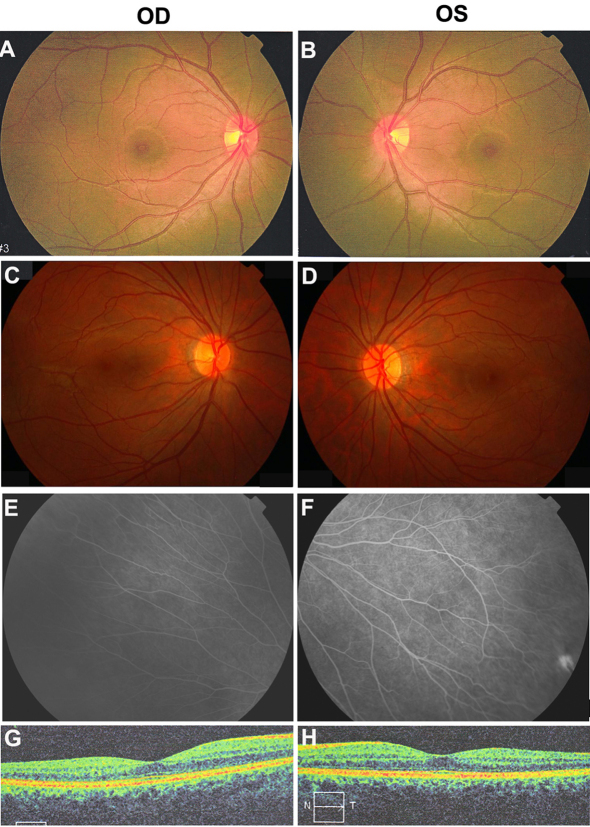
All patients with the three novel mutations showed typical signs of FEVR. The patient with the c.146C>T mutation in Family A was a six-year-old boy with strabismus. Falciform retinal folds in the left fundus were seen upon ophthalmoscopic observation. His parents were recorded to have normal visual acuity, but they were not available for additional examination. The patient with the c.313T>C mutation in Family B was a nine-year-old boy with a complaint of poor visual acuity in the left eye. He had normal visual acuity in the right eye but only recognized hand movement with the left eye. His right fundus was normal, but his left fundus showed the following changes: macular degeneration with pigment proliferation, mild temporal dragging of the optic disc, pigmentary deposits on the midperipheral retina, increased ramification of the peripheral retinal vessels, and an avascular zone on the peripheral retina (Figure 3). His parents were recorded to have normal visual acuity, but they were not available for additional examination. The heterozygous c.601delC mutation in Family C was detected in the proband and in her asymptomatic father. The proband was a three-year-old girl with a complaint of esotropia. Fundus examination showed typical inferotemporal dragging of the optic disc and macula in both eyes (Figure 4). Her mother had normal fundi (Figure 5A,B); but her asymptomatic father had typical fundus changes, although he had normal visual acuity of 1.0 in both eyes. The asymptomatic father had straightening and increased branching of his peripheral retinal vessels (Figure 5C-F) but showed normal macula on optical coherence tomography scan (Figure 5G,H).
Figure 3.
Fundus changes in the patient with the heterozygous c.313T>C mutation in Family B. A and B: These color photos demonstrate a normal right posterior fundus and traction of the retinal vessels and macular degeneration in the left fundus. C and D: Fluorescein angiography of the left eye shows straightening of the vessels with increasing branching (C) and a peripheral avascular zone (D). OD and OS represent the right and left eyes, respectively.
Figure 4.
Fundus changes in the proband with the c.601delC mutation in Family C. A: Inferotemporal dragging of the optic disc and macula is present in the right eye. B: Optical coherence tomography scan shows a flatter central macula in the right eye. C: Inferotemporal dragging of the optic disc and macula is present in the left eye. OD and OS represent the right and left eyes, respectively.
Figure 5.
Fundus photos of the asymptomatic father with the c.601delC mutation and the unaffected mother without the mutation. A and B: The mother has normal fundi. C-D: Fundus photos of the asymptomatic father shows normal posterior fundi. E and F: The father has increased vessel branching in the equatorial area and a peripheral avascular zone. G and H: Optical coherence tomography scan shows normal macula of the asymptomatic father.
Discussion
In this study, three novel TSPAN12 mutations were detected in three patients with FEVR, but not in 180 normal individuals. Segregation analysis in one family indicated a dominant role for the mutation. The two missense mutations occurred in evolutionarily conserved regions of TSPAN12 and were predicted to be pathogenic. These lines of evidence provide support for the conclusion that these mutations are the cause of FEVR in these patients.
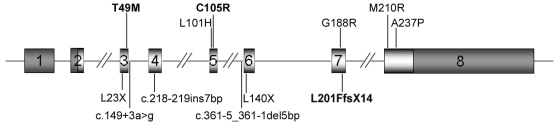
Previously, nine TSPAN12 mutations in patients with FEVR were reported. All 12 mutations, including the three reported in this study, are distributed throughout all coding exons, except for exon 2 (Figure 6). These 12 mutations can be classified into missense mutations (six), nonsense mutations (two), insertion or deletion mutations (two), and splice-site mutations (two). Of the 12 mutations, each has only been found in a single family, except for the p.Ala237Pro mutations that have been detected in four families. For the 15 families with the 12 TSPAN12 mutations, ten had a family history of FEVR, which suggests that this is an autosomal dominant trait with incomplete penetrance, whereas the other five families were isolated cases without a family history. The approximately 6.1% (3/49) mutation frequency detected in our patients is comparable to the 10% (7/70) frequency detected in one previous study [28]. The patients in that study were determined to have no mutations in the other known FEVR genes (NDP, FZD4, or LRP5) that might account for approximately half of FEVR cases [30]. In another study, two TSPAN12 mutations were present in 5 of 11 probands (45.5%), but these patients were previously excluded from having mutations in NDP, FZD4 or LRP5. Four of the five probands with the same mutation shared an at-risk haplotype of at least 5.8 Mb, suggesting a founder effect [27].
Figure 6.
Schematic representation of TSPAN12 and its familial exudative vitreoretinopathy mutations. Missense mutations are shown above the gene and others below the gene. The 12 mutations listed in this figure included the three from our study (letters in bold) and nine from two previous reports [27,28]. The dark shaded parts in exons 1, 2, and 8 represent the untranslated regions. The vertical lines in exons 2 and 8 indicate the positions of translation start and terminate, respectively.
For the patients with the previously reported TSPAN12 mutations, clinical phenotypes include the following: retinal folds, temporal dragging of the optic disc and/or macula, traction of the posterior retinal vessels, increased vascular ramifications in the equatorial area, a peripheral retinal avascular zone, retinal pigmentary disturbances that mimic retinitis pigmentosa, exudates, and retinal detachments. Retinal folds were the most common finding. FEVR in asymptomatic individuals can be detected using fluorescein angiography (Figure 5), but mild changes in the peripheral retina may be neglected, which might partly contribute to the nonpenetrance in a few families. Large phenotypic variations have been observed between different individuals within a family and between different eyes in individual patients. There is no particular genotype–phenotype correlation between particular mutations and certain clinical signs, which suggests haploinsufficiency as the underlying mechanism that induces the manifestation of the disease. Overall, the ocular manifestations caused by TSPAN12 mutations are similar to those due to mutations in FZD4, LRP5, or NDP [21,23,24].
The proteins encoded by TSPAN12, NDP, FZD4, and LRP5 are important components of the Norrin/Fz4 signaling pathway (Norrin/Fz4/Lrp5/Tspan12 signaling pathway), which controls retinal vascular development [16,19]. Recognizing additional mutations in these genes may not only provide useful information for clinical diagnosis and genetic counseling, but may also enrich our understanding of the functional domains of these proteins. Mutations in these four genes all lead to FEVR, which suggests that additional components that participate in this signaling pathway may be reasonable candidates in those FEVR patients without mutations in these four genes.
Acknowledgments
The authors thank the family members for their participation. This study was supported by the National Science Fund for Distinguished Young Scholars (30725044 to Q.Z.) National 973 Plan of China (2010CB529904 to Q.Z.).
References
- 1.Criswick VG, Schepens CL. Familial exudative vitreoretinopathy. Am J Ophthalmol. 1969;68:578–94. doi: 10.1016/0002-9394(69)91237-9. [DOI] [PubMed] [Google Scholar]
- 2.Ober RR, Bird AC, Hamilton AM, Sehmi K. Autosomal dominant exudative vitreoretinopathy. Br J Ophthalmol. 1980;64:112–20. doi: 10.1136/bjo.64.2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Nouhuys CE. Dominant exudative vitreoretinopathy. Ophthalmic Paediatr Genet. 1985;5:31–8. doi: 10.3109/13816818509007853. [DOI] [PubMed] [Google Scholar]
- 4.Feldman EL, Norris JL, Cleasby GW. Autosomal dominant exudative vitreoretinopathy. Arch Ophthalmol. 1983;101:1532–5. doi: 10.1001/archopht.1983.01040020534004. [DOI] [PubMed] [Google Scholar]
- 5.Gow J, Oliver GL. Familial exudative vitreoretinopathy. An expanded view. Arch Ophthalmol. 1971;86:150–5. doi: 10.1001/archopht.1971.01000010152007. [DOI] [PubMed] [Google Scholar]
- 6.de Crecchio G, Simonelli F, Nunziata G, Mazzeo S, Greco GM, Rinaldi E, Ventruto V, Ciccodicola A, Miano MG, Testa F, Curci A, D'Urso M, Rinaldi MM, Cavaliere ML, Castelluccio P. Autosomal recessive familial exudative vitreoretinopathy: evidence for genetic heterogeneity. Clin Genet. 1998;54:315–20. doi: 10.1034/j.1399-0004.1998.5440409.x. [DOI] [PubMed] [Google Scholar]
- 7.Shastry BS, Trese MT. Familial exudative vitreoretinopathy: further evidence for genetic heterogeneity. Am J Med Genet. 1997;69:217–8. doi: 10.1002/(sici)1096-8628(19970317)69:2<217::aid-ajmg19>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 8.Chen ZY, Battinelli EM, Fielder A, Bundey S, Sims K, Breakefield XO, Craig IW. A mutation in the Norrie disease gene (NDP) associated with X-linked familial exudative vitreoretinopathy. Nat Genet. 1993;5:180–3. doi: 10.1038/ng1093-180. [DOI] [PubMed] [Google Scholar]
- 9.Plager DA, Orgel IK, Ellis FD, Hartzer M, Trese MT, Shastry BS. X-linked recessive familial exudative vitreoretinopathy. Am J Ophthalmol. 1992;114:145–8. doi: 10.1016/s0002-9394(14)73977-7. [DOI] [PubMed] [Google Scholar]
- 10.Shastry BS, Hejtmancik JF, Hiraoka M, Ibaraki N, Okubo Y, Okubo A, Han DP, Trese MT. Linkage and candidate gene analysis of autosomal-dominant familial exudative vitreoretinopathy. Clin Genet. 2000;58:329–32. doi: 10.1034/j.1399-0004.2000.580412.x. [DOI] [PubMed] [Google Scholar]
- 11.Kondo H, Ohno K, Tahira T, Hayashi H, Oshima K, Hayashi K. Delineation of the critical interval for the familial exudative vitreoretinopathy gene by linkage and haplotype analysis. Hum Genet. 2001;108:368–75. doi: 10.1007/s004390100503. [DOI] [PubMed] [Google Scholar]
- 12.Berger W, van de Pol D, Warburg M, Gal A, Bleeker-Wagemakers L, de Silva H, Meindl A, Meitinger T, Cremers F, Ropers HH. Mutations in the candidate gene for Norrie disease. Hum Mol Genet. 1992;1:461–5. doi: 10.1093/hmg/1.7.461. [DOI] [PubMed] [Google Scholar]
- 13.Robitaille J, MacDonald ML, Kaykas A, Sheldahl LC, Zeisler J, Dube MP, Zhang LH, Singaraja RR, Guernsey DL, Zheng B, Siebert LF, Hoskin-Mott A, Trese MT, Pimstone SN, Shastry BS, Moon RT, Hayden MR, Goldberg YP, Samuels ME. Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat Genet. 2002;32:326–30. doi: 10.1038/ng957. [DOI] [PubMed] [Google Scholar]
- 14.Jiao X, Ventruto V, Trese MT, Shastry BS, Hejtmancik JF. Autosomal recessive familial exudative vitreoretinopathy is associated with mutations in LRP5. Am J Hum Genet. 2004;75:878–84. doi: 10.1086/425080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toomes C, Bottomley HM, Jackson RM, Towns KV, Scott S, Mackey DA, Craig JE, Jiang L, Yang Z, Trembath R, Woodruff G, Gregory-Evans CY, Gregory-Evans K, Parker MJ, Black GC, Downey LM, Zhang K, Inglehearn CF. Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet. 2004;74:721–30. doi: 10.1086/383202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye X, Wang Y, Nathans J. The Norrin/Frizzled4 signaling pathway in retinal vascular development and disease. Trends Mol Med. 2010;16:417–25. doi: 10.1016/j.molmed.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clevers H. Eyeing up new Wnt pathway players. Cell. 2009;139:227–9. doi: 10.1016/j.cell.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 18.Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–8. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 19.Junge HJ, Yang S, Burton JB, Paes K, Shu X, French DM, Costa M, Rice DS, Ye W. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell. 2009;139:299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 20.Kondo H, Hayashi H, Oshima K, Tahira T, Hayashi K. Frizzled 4 gene (FZD4) mutations in patients with familial exudative vitreoretinopathy with variable expressivity. Br J Ophthalmol. 2003;87:1291–5. doi: 10.1136/bjo.87.10.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toomes C, Bottomley HM, Scott S, Mackey DA, Craig JE, Appukuttan B, Stout JT, Flaxel CJ, Zhang K, Black GC, Fryer A, Downey LM, Inglehearn CF. Spectrum and frequency of FZD4 mutations in familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2004;45:2083–90. doi: 10.1167/iovs.03-1044. [DOI] [PubMed] [Google Scholar]
- 22.Qin M, Hayashi H, Oshima K, Tahira T, Hayashi K, Kondo H. Complexity of the genotype-phenotype correlation in familial exudative vitreoretinopathy with mutations in the LRP5 and/or FZD4 genes. Hum Mutat. 2005;26:104–12. doi: 10.1002/humu.20191. [DOI] [PubMed] [Google Scholar]
- 23.Nallathambi J, Shukla D, Rajendran A, Namperumalsamy P, Muthulakshmi R, Sundaresan P. Identification of novel FZD4 mutations in Indian patients with familial exudative vitreoretinopathy. Mol Vis. 2006;12:1086–92. [PubMed] [Google Scholar]
- 24.Kondo H, Qin M, Kusaka S, Tahira T, Hasebe H, Hayashi H, Uchio E, Hayashi K. Novel mutations in Norrie disease gene in Japanese patients with Norrie disease and familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2007;48:1276–82. doi: 10.1167/iovs.06-1042. [DOI] [PubMed] [Google Scholar]
- 25.Boonstra FN, van Nouhuys CE, Schuil J, de Wijs IJ, van der Donk KP, Nikopoulos K, Mukhopadhyay A, Scheffer H, Tilanus MA, Cremers FP, Hoefsloot LH. Clinical and molecular evaluation of probands and family members with familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2009;50:4379–85. doi: 10.1167/iovs.08-3320. [DOI] [PubMed] [Google Scholar]
- 26.Jia LY, Li XX, Yu WZ, Zeng WT, Liang C. Novel frizzled-4 gene mutations in chinese patients with familial exudative vitreoretinopathy. Arch Ophthalmol. 2010;128:1341–9. doi: 10.1001/archophthalmol.2010.240. [DOI] [PubMed] [Google Scholar]
- 27.Nikopoulos K, Gilissen C, Hoischen A, van Nouhuys CE, Boonstra FN, Blokland EA, Arts P, Wieskamp N, Strom TM, Ayuso C, Tilanus MA, Bouwhuis S, Mukhopadhyay A, Scheffer H, Hoefsloot LH, Veltman JA, Cremers FP, Collin RW. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:240–7. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poulter JA, Ali M, Gilmour DF, Rice A, Kondo H, Hayashi K, Mackey DA, Kearns LS, Ruddle JB, Craig JE, Pierce EA, Downey LM, Mohamed MD, Markham AF, Inglehearn CF, Toomes C. Mutations in TSPAN12 cause autosomal-dominant familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86:248–53. doi: 10.1016/j.ajhg.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikopoulos K, Venselaar H, Collin RW, Riveiro-Alvarez R, Boonstra FN, Hooymans JM, Mukhopadhyay A, Shears D, van Bers M, de Wijs IJ, van Essen AJ, Sijmons RH, Tilanus MA, van Nouhuys CE, Ayuso C, Hoefsloot LH, Cremers FP. Overview of the mutation spectrum in familial exudative vitreoretinopathy and Norrie disease with identification of 21 novel variants in FZD4, LRP5, and NDP. Hum Mutat. 2010;31:656–66. doi: 10.1002/humu.21250. [DOI] [PubMed] [Google Scholar]