Abstract
The Maize Genetics and Genomics Database (MaizeGDB) is a central repository for maize sequence, stock, phenotype, genotypic and karyotypic variation, and chromosomal mapping data. In addition, MaizeGDB provides contact information for over 2400 maize cooperative researchers, facilitating interactions between members of the rapidly expanding maize community. MaizeGDB represents the synthesis of all data available previously from ZmDB and from MaizeDB—databases that have been superseded by MaizeGDB. MaizeGDB provides web-based tools for ordering maize stocks from several organizations including the Maize Genetics Cooperation Stock Center and the North Central Regional Plant Introduction Station (NCRPIS). Sequence searches yield records displayed with embedded links to facilitate ordering cloned sequences from various groups including the Maize Gene Discovery Project and the Clemson University Genomics Institute. An intuitive web interface is implemented to facilitate navigation between related data, and analytical tools are embedded within data displays. Web-based curation tools for both designated experts and general researchers are currently under development. MaizeGDB can be accessed at http://www.maizegdb.org/.
INTRODUCTION
Maize (commonly referred to as corn in the United States or by its botanical name Zea mays L. ssp. mays) is an important crop. Not only does maize feed both the world’s people and its livestock, but its byproducts are also necessary for many industries where corn content is less apparent. Maize byproducts are used in the manufacture of diverse commodities including glue, soap, paint, insecticides, toothpaste, shaving cream, rubber tires, rayon, molded plastics and others [see (1) for review]. The Maize Genetics and Genomics Database (MaizeGDB; http://www.maizegdb.org) is a public database that serves the community of maize researchers by storing and curating data related to the genetics and genomics of maize. Such data include for example, locus information for genes, chromosomal variations (allelic diversity), map positions of genes, primers used for mapping analysis, probe sets used in mapping and phenotypic image data. These data types are intrinsically interrelated, and MaizeGDB’s web interface recapitulates these relationships. Using MaizeGDB, a researcher can type a term (e.g., adh1) into the search field at the top of any page and should be able to navigate intuitively from the results page to pages containing the locus, related stocks, variations, primers, the position of the locus on a variety of maps and additional information. Also available on results pages are links to contact information for maize researchers who are experts on the query topic and who can provide valuable research materials (see Usage Example below).
HISTORICAL BACKGROUND
Maize is an organism of historical importance to all biologists. A sample of seminal discoveries made by maize geneticists include: Emerson’s contributions to the concepts of epistasis and quantitative genetics (2,3); Stadler’s research showing that X-rays cause mutation (4,5); Beadle’s doctoral dissertation describing how irregular behavior of meiotic chromosomes causes heritable pollen production defects (6); Creighton and McClintock’s work showing that genetic crossing over is accompanied by physical crossovers between chromosomes (7); Rhoades’ discovery of the cytoplasmic inheritance of male sterility (8); and McClintock’s description and characterization of transposable elements, which ultimately won her the Nobel prize [reviewed in (9)].
In the late 1920s it was recognized by the community of maize geneticists that the data they were recording needed organization, publication and curation. To this end, R. A. Emerson and others began publishing the Maize Genetics Cooperation Newsletter (MNL), which is compiled and published on a yearly basis. To further the same goals, in 1991 the US Department of Agriculture-Agricultural Research Service (USDA-ARS) charged Ed Coe, then editor of the MNL, to develop a maize genome database (10). MaizeDB was one of the first biological databases to exist online, and it became an indispensable research tool utilized by maize geneticists worldwide.
In 1998, the Maize Gene Discovery Project (MGDP) was funded by the National Science Foundation, led by Virginia Walbot and including 10 research groups [reviewed in (11)]. The MGDP discovers new maize genes and develops tools for characterizing maize mutants. The microarray slides, EST clones, library plates of indexed transposon insertions and seed generated by MGDP necessitated the implementation of a resource to make these materials publicly available and to organize the data generated by the project team. This need was met by ZmDB (12). In addition to making MGDP materials available, ZmDB also encompasses all public maize ESTs, GSSs and protein sequences. ZmDB’s embedded similarity search tools and services (providing multiple sequence alignments, protein domain determination and spliced alignments) simplify sequence analysis, thus allowing researchers to spend more time making scientific discoveries at the bench. ZmDB was scheduled to shut down in September 2003 at the termination of the MGDP.
In September of 2001 the USDA-ARS began an initiative to combine MaizeDB and ZmDB, thus creating a single maize genetics and genomics database using state-of-the-art database architecture and web design protocols. As of September 1, 2003, this goal was realized. MaizeGDB supersedes MaizeDB, and makes available all data and resources that previously existed at either MaizeDB or ZmDB. Researchers working at MaizeGDB seek to serve the maize community’s database resource needs by making maize data and materials available and by collaborating with researchers to store and display their important scientific findings.
DATABASE COMPONENTS
The records contained within MaizeGDB can be grouped into four general classes of related information: genetic data, genomic and other DNA sequence files, gene product or functional characterization records, and literature reference and person or organization listings. Some of the connections within and among these four general classes are described below. (For a detailed depiction of how the data centers at MaizeGDB are interconnected see http://www.maizegdb.org/MaizeGDBSchema.pdf.)
Genetic data centers
Maps, loci, quantitative trait loci, traits, variations and seed stocks constitute the genetic data centers. Since the first maize linkage maps were compiled and published in 1935, mapping data have been crucial to maize geneticists (13). At MaizeGDB, map queries can be restricted to a particular chromosome, map source, inbred line or background. Loci along the chromosome are linked to their respective locus records, and the coordinates of and bins containing each locus are listed. For maps that are also present at Gramene (a resource for comparative analysis of grass genomes) (14) and the National Center for Biotechnology Information (NCBI) (15), links are provided for navigation to those visualization resources. Individual loci or clusters of loci that are physically linked and that act together to modulate quantitative traits are called quantitative trait loci (QTL). QTL records can be searched by experiment, identified by the person who performed the experiment and year, or by trait. Selecting an individual QTL experiment creates a page showing the experimental overview, which includes the mapping panel, progeny for genotype evaluation, progeny for trait evaluation and marker summary. The page also will list trait evaluations, QTL detected by the experiment and links to any references describing the experiment. Alternative forms of QTL, loci, chromosomes and other genetic elements are called variations. Variation searches at MaizeGDB can be restricted to type, locus, viability, progenitor stock, dominance, mutagen, mutation, expressed phenotype and stock. Selecting a particular variant from the list of records matching the search criteria creates a page including the variant’s name, allele descriptor, dominance, type (allele, QTL variant, transposition, etc.), phenotype(s) and a list of stocks known to carry the variation. To obtain seed for analysis of variations, links for ordering stocks are embedded within the variation and trait pages. Alternatively, specific seed stock searches can be performed and can be restricted by identifier, type (BA translocation, hybrid, inbred line, etc.), focus linkage group, genotypic variation, karyotypic variation, phenotype, availability and parentage. Results pages list the stock name, a descriptive name, type, focus linkage group and source. Stocks available from the Maize Genetics Cooperation Stock Center (http://www.uiuc.edu/ph/www/maize/) or NCRPIS (http://www.ars-grin.gov/ars/MidWest/Ames) can be ordered by following links that are embedded throughout MaizeGDB.
Genomic/sequence data centers
Maize sequences, SSRs, probes, BACs and overgo probes are found within MaizeGDB’s genomic and other DNA sequence data centers. Sequence searches query the database for ESTs (derived from http://www.ncbi.nlm.nih.gov/dbEST), GSSs (http://www.ncbi.nlm.nih.gov/dbGSS), HTGs (http://www.ncbi.nlm.nih.gov/HTGS), STSs (http://www.ncbi.nlm.nih.gov/dbSTS), complementary DNAs (cDNAs) and proteins using the sequence’s accession number, GI number or a part of the sequence title. Sequences also can be searched using BLAST (16) and the GeneSeqer gene discovery tool (17). Links on sequence record pages make it possible to carry out BLAST searches at MaizeGDB, PlantGDB (18) or GenBank. Simple sequence repeats (SSRs) can be identified at MaizeGDB by repeat pattern [e.g., (AAAT)3 represents AAATAAATAAAT], and the SSR browser allows researchers to examine SSR records by name, bin location along a given chromosome and base sequence. Some sequence data are also included in MaizeGDB’s probe data set, which is made up of a mix of both probe and sequence data types including amplified fragment length polymorphisms (AFLPs), restriction fragment length polymorphisms (RFLPs), non-EST cDNAs, DNA probes, genomic DNA, miniature inverted-repeat transposable elements (MITEs), random amplified polymorphic DNAs (RAPDs), yeast artificial chromosomes (YACs) and a small assortment of other probe types. Bacterial artificial chromosome (BAC) records can also be found within this data center. Contigs formed from multiple BACs can be visualized through links to WebFPC [http://www.genome.arizona.edu/fpc/maize and (19)]. Overlapping oligonucleotide pairs (overgos that were used to detect BACs) can be identified at MaizeGDB by name or using a short DNA sequence as the database query. Overgo results pages list primer pairs as a single sequence with the overlapping portions of the two sequences highlighted. Selecting a single overgo from the list creates a screen showing the two primers, their names, alignment and a list of the BACs detected by the overgo. Also listed are any ESTs that are known to contain either overgo sequence.
Gene product/functional characterization data centers
MaizeGDB stores and curates detailed descriptions of gene products, metabolic pathways, and mutant or variation phenotypes. Gene product records display gene product type (storage protein, signal receptor, transcription factor, etc.), Enzyme Commission number(s) (as assigned by IUPAC-IUBMB; http://www.chem.qmw.ac.uk/iupac/jcbn), a list of motifs and features, a list of related gene products, and a link to references that describe the gene product. Searches can be limited by environmental or chemical induction conditions; subcellular localization; metabolic pathway; metabolic constituent; and sequence, structural, and gel migration rate information. Metabolic pathway records can be searched by pathway name, metabolic process (e.g., cell division) and key enzyme. Over 7000 phenotype records can be searched by name, trait, presence of related images and body part (plant organ) exhibiting the phenotype. Selecting a phenotype search result creates a page displaying links to related traits and associated stocks.
Reference and person/organization data centers
Not all references important to maize researchers can be found using conventional journal search engines like the NCBI’s ‘Entrez Pubmed’ (http://www.ncbi.nlm.nih.gov/PubMed). MaizeGDB contains both mainstream and cryptic references including MNL references (which cannot be cited without the author’s permission but contain invaluable data, nonetheless) and references from other journals not supported by typical reference search engines. In addition, MaizeGDB maintains reference information and abstracts for works published in the annual Maize Genetics Conference Proceedings. To facilitate interactions among maize researchers, MaizeGDB also stores and curates data related to maize people and organizations. Because these records are tied to many other records in the site (such as references, probes and sequences) researchers can easily identify others with similar interests. The people or organizations search page allows for searches to be conducted by name, and the person or organization browser can be used to select information to be included in output tables.
Major data centers are interconnected
To illustrate how the major groupings of data centers are interconnected, maize mutations containing a RescueMu transposable element and corresponding seed stocks and plant phenotypes can be used as a case in point. The MGDP (11) recovered small genomic libraries of DNA derived from a grid-based field of up to 2304 RescueMu plants. From these genomic sequences they made plasmid library plates (organized in rows and columns that mirrored the organization of the field) that can be screened for RescueMu insertions using PCR. By sequencing out from the transposon, sequences of genomic DNA flanking RescueMu were recovered from many grids. In addition, seed stocks were generated by self-pollination of each RescueMu grid plant and are maintained by the Maize Genetics Cooperation Stock Center. MaizeGDB facilitates searches for specific RescueMu sequences, plant phenotypes and seeds in the following way. BLAST (15) searches can be carried out from http://www.maizegdb.org/blast.php against the maize GSSs using a DNA or protein sequence as the query. Significantly similar sequences are identified, and links to retrieve particular sequence records from MaizeGDB are given. For GSSs that were produced by MGDP, links to view plant phenotypes associated with the RescueMu insertion are provided. Phenotype browser pages link to seed stock order forms from the Maize Genetics Cooperation Stock Center. Hence search tools and data centers warehousing sequence and phenotype data as well as forms for ordering seed stock online are logically interconnected, recapitulating the biological interrelationships conserved among these related data.
USAGE EXAMPLE
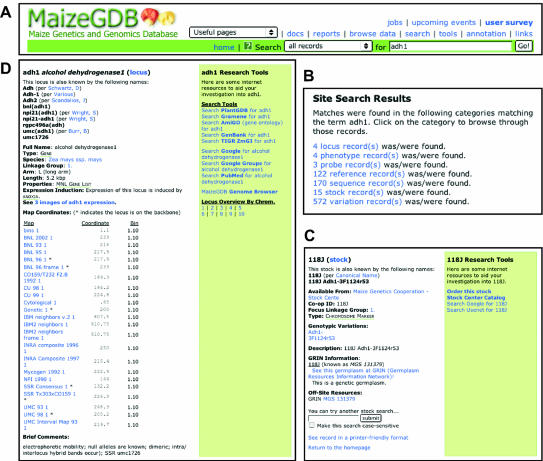
A typical researcher seeking information related to his or her gene of interest (e.g., alcohol dehydrogenase 1 or adh1) might go to MaizeGDB to gather information about the gene and to order seed for plants bearing mutations within the gene. (This conscientious researcher has already visited the first three pages shown in Table 1 and has learned how to use MaizeGDB.) The researcher proceeds to http://www.maizegdb.org to find out what information is available for adh1 by searching all records using ‘adh1’ as her query (see Fig. 1A; also note that maize researchers always italicize loci and gene names; however, searches at MaizeGDB do not require italics and will tolerate the use of all upper- or lower-case). She finds links to locus, phenotype, probe, reference, sequence, stock and variation records (Fig. 1B), and then selects the first stock. This stock happens to carry a genotypic variation called ‘Adh1-3F1124r53’ (Fig. 1C). Clicking the link to genotypic variations of ‘Adh1-3F1124r53’ would create a page (not shown) telling the researcher that the phenotypes associated with the ‘Adh1-3F1124r53’ allele are low and null activity of adh1, and that the allele is dominant. From the page shown in Figure 1C, the researcher orders the seed for ‘Adh1-3F1124r53’ from the Maize Genetics Cooperation Stock Center using the link labeled ‘Order this stock’ in the list of tools on the right of the record.
Table 1. Links for helpful and interesting sites at MaizeGDB.
| MaizeGDB tutorial | http://www.maizegdb.org/tutorial/index.php | A tutorial explaining how to use many of the features of MaizeGDB |
| Site tour | http://www.maizegdb.org/site_tour.php | A brief tour describing the highlights of MaizeGDB |
| Documentation | http://www.maizegdb.org/doc.php | A summary of documentation for the site including schema, credits, documentation of important maize projects, links and other resources |
| Credits | http://www.maizegdb.org/credit.php | A page providing credit to the software providers and other groups and individuals essential to MaizeGDB |
| Maize Genome Browser | http://www.maizegdb.org/cgi-bin/bin_viewer.cgi | A utility enabling researchers to browse through the maize chromosomes to find sequences, genes, BACs, SSRs and other genetic elements in various regions of the maize genome |
| Mapped BLAST | http://www.maizegdb.org/blast.php | A BLAST utility that also returns known map locations of sequence matches; linked throughout the site |
| GeneSeqer | http://www.maizegdb.org/geneseqer.php | A web interface to the GeneSeqer gene discovery tool that is also interlinked throughout the site for researchers to use dynamically |
| Image browser | http://www.maizegdb.org/cgi-bin/imagebrowser.cgi | A collection of image-browsing tools to help researchers locate images of maize appropriate for their educational needs |
| Educational resources | http://www.maizegdb.org/education.php | A selection of maize-related educational resources for both researchers and the general public |
| Job board | http://www.maizegdb.org/jobs.php | A page that allows members of the community to post or find jobs |
| Database statistics | http://www.maizegdb.org/cgi-bin/database_stats.cgi | A review of the counts of particular records of various types stored in MaizeGDB |
| Site statistics | http://www.maizegdb.org/cgi-bin/awstats.pl | Information on site usage |
Figure 1.
Example database search at MaizeGDB using the query ‘adh1’. The group of screenshots shows an example of how researchers can search MaizeGDB for alcohol dehydrogenase 1 (adh1). (A) All MaizeGDB pages have the same search bar at the top of the page. In this example all records are searched for the term ‘adh1’. (B) Records of various data types are retrieved. (C) Selecting one of the seed stock records creates a page showing information about the stock. In the list of tools to the right is a link for ordering it from the Maize Genetics Cooperation Stock Center. (D) Selecting the first of four locus records identified by the search creates a page (truncated here for space) showing data related to the locus adh1. Beyond ‘Brief Comments’ there are information and links to: related gene products (both internal and offsite), ESTs, probes, SSRs, primers, variations, phenotypes, nearby and related loci, sequences, map scores, recombination data, extensive comments, references and related offsite resources. A list of tools to the right links this page to search engines and other databases.
Going back to the list of original search results (Fig. 1B), the researcher decides to view the first locus record shown in the list (which represents the gene adh1). Browsing the locus page (Fig. 1D) she finds that the gene resides on the long arm of chromosome 1 and that images of adh1 mutant phenotypes exist. By going to the phenotype images (not shown), the researcher finds that expected phenotypes for adh1 mutant plants include failure to germinate in anaerobic conditions and pollen tube germination defects. This information tells the researcher what phenotypes to expect from the seed that will soon arrive in the mail from the Maize Genetics Cooperation Stock Center. Going back to the locus page (Fig. 1D) she scrolls down (not shown) to find the names of cooperators from whom RFLP probes for adh1 can be obtained. Sequences for primers that the researcher can use to amplify the adh1 gene are also displayed. Other data of interest she finds on this page include links to the genomic and EST sequences of adh1, recombination data and detailed comments about the gene product encoded by the adh1 gene. For more information on how to navigate the MaizeGDB website, please visit each of the links listed in Table 1.
FUTURE DIRECTIONS
The MaizeGDB team is dedicated to seeking out new data sources for evaluation as materials to be archived in support of research applications. Data types and efforts currently under evaluation include: (i) listing the availability of and contact information for tissue, organ and inbred line-specific cloned sequence libraries, (ii) creating a searchable maize transposon and repeat sequence database, (iii) utilizing an editorial board to provide in-depth annotation of selected publications and (iv) launching storage and display endeavors for chromosome fluorescence in situ hybridization (FISH) image data as it becomes available from the Cytogenetic Map of Maize project (ISGA-PGR; https://www.fastlane.nsf.gov/servlet/showaward?award=0321639). Researchers interested in helping to make these or other data types of interest available to the maize community through MaizeGDB are encouraged to contact the MaizeGDB team at mgdb@iastate.edu.
AVAILABILITY
MaizeGDB is accessible at the URL http://www.maizegdb.org/. Inquiries concerning the database should be directed by email to mgdb@iastate.edu.
Acknowledgments
ACKNOWLEDGEMENTS
We thank our national steering committee (Tom Slezak, Ed Buckler, Vicki Chandler, Mike Freeling, Sarah Hake, Marty Sachs, Cari Soderlund, Lincoln Stein and Virginia Walbot) and our local steering committee (Phil Becraft, Steve Howell, Robert Jernigan, Mike Lee, Alan Myers, Tom Peterson, Pat Schnable and Jonathan Wendel) for their collaboration, direction and helpful comments. We also thank Darwin Campbell (database developer), Sanford Baran (web developer) and Mike Brekke (systems support specialist) for their contributions to the success of the MaizeGDB project. This work is funded by USDA-ARS specific cooperative agreement no. 58-3625-1-192.
REFERENCES
- 1.Fussell B. (1999) The Story of Corn. 2nd edn. North Point Press, New York, NY. [Google Scholar]
- 2.Emerson R.A. and East,E.M. (1913) The inheritance of quantitative characters in maize. Bull. Agric. Exp. Sta. NE, 2, 1–120. [Google Scholar]
- 3.Emerson R.A. (1918) A fifth pair of factors, Aa, for aleurone color in maize, and its relation to the C and Rr pairs. Cornell Univ. Agric. Exp. Sta. Mem., 16, 225–289. [Google Scholar]
- 4.Stadler L.J. (1928) Genetic effects of x-rays in maize. Proc. Natl Acad. Sci. USA, 14, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stadler L.J. (1928) Mutations in barley induced by x-rays and radium. Science, 58, 186–187. [DOI] [PubMed] [Google Scholar]
- 6.Beadle G.W. (1930) Genetic and Cytological Studies of Mendelian Asynapsis in Zea mays. Doctoral Dissertation, Cornell University, Ithaca, NY. [Google Scholar]
- 7.Creighton H.B. and McClintock,B. (1931) A correlation of cytological and genetical crossing-over in Zea mays. Proc. Natl Acad. Sci. USA, 17, 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhoades M.M. (1932) Cytoplasmic inheritance of male sterility in Zea mays. Proc. Natl Acad. Sci. USA, 18, 481–484. [Google Scholar]
- 9.Federoff N. and Botstein,D. (eds) (1992) The Dynamic Genome: Barbara McClintock’s Ideas in the Century of Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 10.Polacco M. and Coe,E. (1999) MaizeDB: the maize genome database. In Letovsky,S.I. (ed.), Bioinformatics: Databases and Systems. Kluwer Academic Publishers, Norwell, MA, pp. 151–162. [Google Scholar]
- 11.Lunde C.F., Morrow,D.J., Roy,L.M. and Walbot,V. (2003) Progress in maize gene discovery: a project update. Funct. Integr. Genomics, 3, 25–32. [DOI] [PubMed] [Google Scholar]
- 12.Dong Q., Roy,L., Freeling,M., Walbot,V. and Brendel,V. (2003) ZmDB, an integrated database for maize genome research. Nucleic Acids Res., 31, 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emerson R.A., Beadle,G.W. and Fraser,A.E. (1935) A summary of linkage studies in maize. Cornell Univ. Agric. Exp. Sta. Mem., 180, 1–3. [Google Scholar]
- 14.Ware D.H., Jaiswal,P., Ni,J., Yap,I.V., Pan,X., Clark,K.Y., Teytelman,L., Schmidt,S.C., Zhao,W., Chang,K. et al. (2002) Gramene, a tool for grass genomics. Plant Physiol., 130, 1606–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. et al. (2003) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlueter S.D., Dong,Q. and Brendel,V. (2003) GeneSeqer@PlantGDB: gene structure prediction in plant genomes. Nucleic Acids Res., 31, 3597–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Q., Schlueter,S.D. and Brendel,V. (2004) PlantGDB, plant genome database and analysis tools. Nucleic Acids Res., 32, D354–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soderlund C., Humphray,S., Dunhum,A. and French,L. (2000) Contigs built with fingerprints, markers and FPC V4.7. Genome Res., 10, 1772–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]