Abstract
The analysis of regulatory regions in genome sequences is strongly based on the detection of potential transcription factor binding sites. The preferred models for representation of transcription factor binding specificity have been termed position-specific scoring matrices. JASPAR is an open-access database of annotated, high-quality, matrix-based transcription factor binding site profiles for multicellular eukaryotes. The profiles were derived exclusively from sets of nucleotide sequences experimentally demonstrated to bind transcription factors. The database is complemented by a web interface for browsing, searching and subset selection, an online sequence analysis utility and a suite of programming tools for genome-wide and comparative genomic analysis of regulatory regions. JASPAR is available at http://jaspar.cgb.ki.se.
INTRODUCTION
The discovery and characterization of regulatory control sequences in completed metazoan genomes have become imperatives as the compilation of successful sequencing projects accelerates. Comparative analysis of the sequences coupled with gene identification software can reliably define the vast majority of exon positions (1), but detection of the sequences regulating gene transcription remains a significant challenge (2). To meet this need, diverse researchers are dedicated to the creation of innovative new algorithms based on phylogenetic footprinting (3,4) and analysis of regulatory modules (5,6). These emerging methods for regulatory sequence analysis are united by their dependence on the quality of available models that represent the binding specificity of transcription factors. Based on years of bioinformatics research, the preferred method for modelling the binding specificity of a transcription factor (TF) has been identified as the position-specific score matrix (PSSM) (7). While higher order models can offer modest performance improvements (8), the extensive binding data required to build such models are available for a limited number of TFs. In order for researchers to move forward with the creation of new algorithms for regulatory sequence analysis, a robust and freely accessible collection of binding models is essential.
We present JASPAR, a database of matrix-based transcription factor binding profiles with associated web interface and tightly integrated programming tools, the aim of which is to warehouse non-redundant representations of high-quality matrix-based transcription factor binding site (TFBS) profiles. In this paper, we provide an outline of the derivation of the profiles in the database, followed by a description of the database web interface and programming tools.
DERIVATION OF PROFILES
All profiles in the first public release of the database are derived from published collections of experimentally defined TFBSs for multicellular eukaryotes. The database contains a curated collection of target sequences. The binding sites were determined either in SELEX (9) experiments, or by the collection of data from the experimentally determined binding regions of actual regulatory regions; this distinction is clearly marked in the profile’s annotation. Candidate profiles were included in the database after passing three stages.
Source data acquisition
Published works describing either SELEX experiments or collections of in vivo binding sequences for a candidate transcription factor were identified by review of the scientific literature.
Quality assessment
A candidate profile was approved if the published work satisfied the following criteria: (i) the report was judged to conform to high-quality standards for the experimental detection of TFBSs; (ii) for SELEX experiments, no reports were accepted where determined sites were filtered using arbitrary criteria (such as following a certain consensus), except for exclusion of sites found to be functionally inactive in cells; (iii) bona fide binding site sequences must be unambiguously locatable and obtainable from either the publication or public sequence databases; (iv) transcription factors must originate from multicellular eukaryotes; (v) there must be at least five distinct binding site sequences available; (vi) no profile redundancy was accepted, i.e. not more than one profile describing the binding preference for a particular factor (or dimer of factors) was allowed.
Alignment of known binding sites to produce profiles
Site-containing sequences from the quality-assessment stage were aligned using the motif discovery program ANN-Spec (10). ANN-Spec uses artificial neural networks and Gibbs sampling algorithms for pattern finding and is specifically intended for DNA binding site discovery. As the algorithm is probabilistic, results vary and are impacted by initial settings for motif length. Therefore, for each candidate, ANN-Spec alignments using three random seeds, all possible strand orientations, and 80 000 iterations were visually inspected to determine an acceptable range of possible widths. Subsequent alignment selection and decision on final inclusion were based on biological knowledge about the TFs and their binding sites.
THE CONTENTS OF THE JASPAR DATABASE
At present, there are 111 profiles in the database. The contents of the JASPAR database are summarized in Table 1. The present collection is restricted to profiles from multicellular eukaryotes with enough experimental data for the derivation of robust, high-quality profiles. User guidance for the expansion and improvement of the JASPAR profile collection is strongly encouraged. Due to quality concerns and probable differences in methodologies, however, it is not possible to accept user-submitted profiles without curatorial review.
Table 1. Properties of the JASPAR database.
| Profile statistics | |
| No. profiles | 111 |
| Mean sequence depth (no. sites) | 34.6 |
| Mean information content (bits) | 11.9 |
| No. vertebrate profiles | 81 |
| No. plant profiles | 15 |
| No. insect profiles | 13 |
| No. SELEX profiles | 91 |
| No. compiled profiles | 20 |
| Class coverage (no. profiles) | |
| AP2 | 1 |
| bHLH | 6 |
| bHLH-ZIP | 5 |
| bZIP | 9 |
| CAAT-BOX | 1 |
| ETS | 7 |
| FORKHEAD | 7 |
| HMG | 6 |
| HOMEO | 6 |
| HOMEO-ZIP | 2 |
| IPT/TIG | 2 |
| MADS | 5 |
| NUCLEAR RECEPTOR | 8 |
| P53 | 1 |
| PAIRED | 3 |
| PAIRED-HOMEO | 1 |
| REL | 6 |
| RUNT | 1 |
| TATA-BOX | 1 |
| T-BOX | 1 |
| TEA | 1 |
| TRP-CLUSTER | 5 |
| ZN-FINGER, C2H2 | 17 |
| ZN-FINGER, DOF | 4 |
| ZN-FINGER, GATA | 4 |
A JASPAR WEB INTERFACE AND ONLINE SERVICES
We have developed a streamlined web interface to the JASPAR database. Through the web system, users may perform the following tasks.
(i) Browse the TF binding profile collection. The purpose of the profile browser is to present the user with a quick overview of the contents of JASPAR. The profiles can be viewed in groups based on specified criteria (identifiers, structural class of the binding domain, species or names).
(ii) Search the profiles by identifiers or annotations. The search engine is designed to quickly locate subsets of the profile based on their annotation.
(iii) Compare user-entered profiles with existing profiles in the database. This function enables users to determine whether a newly discovered profile is similar to an existing profile, using a modified matrix-to-matrix Needleman–Wunsch (11) comparison algorithm (12). This service is intended for the growing number of researchers confronted by putative binding site patterns shared by genes found to be co-expressed in expression profiling studies.
(iv) Search a user-specified nucleotide sequence with selected transcription factor profiles. The JASPAR web interface provides only basic search and graphical output here—for more sophisticated sequence searches, users are directed to the ConSite web service (http://www.phylofoot.org/consite) for TFBS detection (3).
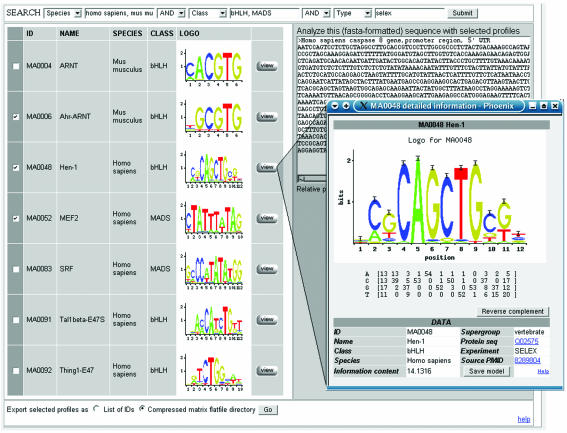
Selected profiles are presented using sequence logos (13), alongside basic annotations, each connected with a pop-up window presenting fully detailed annotation. A sample of search results with accompanying profile annotation data is given in Figure 1.
Figure 1.
Screenshot of search results from a JASPAR query.
A JASPAR APPLICATION PROGRAMMING INTERFACE
To maximize the utility of the JASPAR profiles for genome-wide sequence analysis, the database is tightly integrated with the TFBS Perl framework for TFBS analysis (14), available at http://forkhead.cgb.ki.se/TFBS. The TFBS system has been upgraded to take full advantage of the new JASPAR database. We have introduced new database modules and expanded existing ones to enable the storage and exchange of fully annotated profiles from JASPAR and other resources. In effect, TFBS is a fully functional application programming interface to the JASPAR database.
In addition to JASPAR-specific refinements, the creation of the new profiles database motivated further functional enhancements of TFBS. TFBS now can accept word-based motifs and supports the use of a leading word-based pattern discovery algorithm (15). The new and expanded modules are listed in Table 2.
Table 2. New and enhanced modules in the TFBS framework for TFBS analysis, which serves as a full-featured JASPAR application programming interface (internal and base class modules are omitted).
| New modules | |
| Name | Function |
| TFBS::Word::Consensus | A pattern expressed as fixed width IUPAC DNA string |
| TFBS::Word::RegEx | A sequence pattern expressed as Perl-style regular expression |
| TFBS::DB::Matrix::SQL | A generalized relational database format for storing annotated profiles |
| TFBS::DB::Matrix::SimpleXML | A simple XML document with DTD for storing annotated profiles |
| TFBS::Matrix::Alignment | Matrix profile aligner (12) module |
| TFBS::PatternGen::MEME | MEME (20) pattern discovery interface |
| TFBS::PatternGen::ANNSpec | ANN-Spec (10) pattern discovery interface |
| TFBS::PatternGen::ELPH | ELPH Gibbs sampler (http://www.tigr.org/software/ELPH) pattern discovery interface |
| TFBS::PatternGen::YMF | YMF (15) pattern discovery interface. Produces word-based patterns of the TFBS::Word::Consensus type |
| TFBS::SiteSetFilter::RegulatoryModule | A module for the detection of clusters of transcription factor binding sites |
| Enhanced modules | |
| Name | New features |
| TFBS::Matrix::ICM | Postscript and PDF output of sequence logos |
| TFBS::DB::Matrix::FlatFileDir | Format definition modified to hold arbitrary annotation tags |
| TFBS::Matrix::PWM | Searches can be restricted to a subsequence or subsections of alignments—simplifies and speeds up genome-wide searches |
AVAILABILITY AND DISTRIBUTION
The entire contents of the JASPAR database can be downloaded from http://jaspar.cgb.ki.se, either as a MySQL database dump that can be used to reconstitute a local copy of the database, or as a flat-file format suitable for direct use or in conjunction with the TFBS programming framework. In addition, the web interface supports retrieval of subsets of matrices for use in emerging web-based analysis tools, such as Cluster-Buster (16), MSCAN (17) or MCAST (18). The download service and web interface may be used without restriction.
CONCLUSIONS AND FURTHER DIRECTIONS
In its present form, JASPAR offers significant advantages compared with similarly scoped resources: (i) it is a non-redundant collection of reliable binding profiles; (ii) data access is unrestricted; (iii) it is powerfully coupled to programming tools: the database is ready to be deployed quickly for genome-wide studies through the JASPAR API.
JASPAR is distinct from TRANSFAC (19), the leading TF database. TRANSFAC maintains a broader mission in data collection, contains a redundant set of binding profiles of diverse quality and is marketed as a commercial resource with only a portion of its contents available in public versions. Users will find different database composition between the two systems, as the data were independently gathered.
JASPAR will continue to expand and existing profiles will be improved as new data emerge. In addition to the ongoing efforts of the authors, the diverse and growing population of JASPAR users is providing directed feedback on literature sources for additional binding profiles. We are presently finalizing a regulatory region annotation tool that will facilitate the collection of literature-based regulatory modules from metazoan genes to include in future releases. This tool will be coupled to JASPAR to dynamically build TF binding profiles. Additional efforts are underway to incorporate yeast binding profiles (both literature and footprinting derived). For users interested in combining binding profiles from multiple resources, the TFBS framework provides seamless integration on the programming level between JASPAR and other collections.
REFERENCES
- 1.Zhang M.Q. (2002) Computational prediction of eukaryotic protein-coding genes. Nature Rev. Genet., 3, 698–709. [DOI] [PubMed] [Google Scholar]
- 2.Wasserman W.W. and Krivan,W. (2003) In silico identification of metazoan transcriptional regulatory regions. Naturwissenschaften, 90, 156–166. [DOI] [PubMed] [Google Scholar]
- 3.Lenhard B., Sandelin,A., Mendoza,L., Engstrom,P., Jareborg,N. and Wasserman,W.W. (2003) Identification of conserved regulatory elements by comparative genome analysis. J. Biol., 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boffelli D., McAuliffe,J., Ovcharenko,D., Lewis,K.D., Ovcharenko,I., Pachter,L. and Rubin,E.M. (2003) Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science, 299, 1391–1394. [DOI] [PubMed] [Google Scholar]
- 5.Alkema W. and Wasserman,W.W. (2003) Understanding the language of gene regulation. Genome Biol., 4, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krivan W. and Wasserman,W.W. (2001) A predictive model for regulatory sequences directing liver-specific transcription. Genome Res., 11, 1559–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stormo G.D. (2000) DNA binding sites: representation and discovery. Bioinformatics, 16, 16–23. [DOI] [PubMed] [Google Scholar]
- 8.Barash Y., Elidan,G., Friedman,N. and Kaplan,T. (2003) Modeling dependencies in protein–DNA binding sites. In Vingron,M., Istrail,S., Pevzner,P. and Waterman,M. (eds), Proceedings of the Seventh Annual International Conference on Computational Molecular Biology. ACM Press, New York, NY, pp. 28–37. [Google Scholar]
- 9.Pollock R. and Treisman,R. (1990) A sensitive method for the determination of protein–DNA binding specificities. Nucleic Acids Res., 18, 6197–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Workman C.T. and Stormo,G.D. (2000) ANN-Spec: a method for discovering transcription factor binding sites with improved specificity. Pac. Symp. Biocomput., 467–478. [DOI] [PubMed] [Google Scholar]
- 11.Needleman S.B. and Wunsch,C.D. (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol., 48, 443–453. [DOI] [PubMed] [Google Scholar]
- 12.Sandelin A., Hoglund,A., Lenhard,B. and Wasserman,W.W. (2003) Integrated analysis of yeast regulatory sequences for biologically linked clusters of genes. Funct. Integr. Genomics, 3, 125–134. [DOI] [PubMed] [Google Scholar]
- 13.Schneider T.D. and Stephens,R.M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenhard B. and Wasserman,W.W. (2002) TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics, 18, 1135–1136. [DOI] [PubMed] [Google Scholar]
- 15.Sinha S. and Tompa,M. (2003) YMF: a program for discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res., 31, 3586–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frith M.C., Li,M.C. and Weng,Z. (2003) Cluster-Buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res., 31, 3666–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson O., Alkema,W., Wasserman,W.W. and Lagergren,J. (2003) Identification of functional clusters of transcription factor binding motifs in genome sequences: the MSCAN algorithm. Bioinformatics, 19 (Suppl. 2), I169–I176. [DOI] [PubMed] [Google Scholar]
- 18.Bailey T.L. and Noble,W.S. (2003) Searching for statistically significant regulatory modules. Bioinformatics, 19 (Suppl. 2), II16–II25. [DOI] [PubMed] [Google Scholar]
- 19.Matys V., Fricke,E., Geffers,R., Gossling,E., Haubrock,M., Hehl,R., Hornischer,K., Karas,D., Kel,A.E., Kel-Margoulis,O.V. et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res., 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundy W.N., Bailey,T.L., Elkan,C.P. and Baker,M.E. (1997) Meta-MEME: motif-based hidden Markov models of protein families. Comput. Appl. Biosci., 13, 397–406. [DOI] [PubMed] [Google Scholar]